Abstract
Cognitive neuroscientists increasingly recognize that continued progress in understanding human brain function will require not only the acquisition of new data, but also the synthesis and integration of data across studies and laboratories. Here we review ongoing efforts to develop a more cumulative science of human brain function. We discuss the rationale for an increased focus on formal synthesis of the cognitive neuroscience literature, provide an overview of recently developed tools and platforms designed to facilitate the sharing and integration of neuroimaging data, and conclude with a discussion of several emerging developments that hold even greater promise in advancing the study of human brain function.
Science by synthesis
Science is, by nature, a cumulative endeavor. Scientific advances generally build directly on previous studies and issue findings that only make sense in light of existing knowledge. In cognitive neuroscience, as in many other scientific disciplines, a gold standard for scientific progress and accumulation of knowledge has historically been the ‘critical experiment’: a single empirical test that decisively disqualifies one or more hypotheses from further consideration [1]. Valuable as they can be, however, critical experiments are not the only way to make scientific progress. In fields such as genetics, cognitive science, and, we argue, functional neuroimaging, important scientific advances also result from the synthesis and modeling of existing data, in addition to the collection of new data. The overall behavior of a system as complex as the human brain cannot readily be inferred from isolated analyses of a few variables. No single experiment can control for all, or even most, extraneous variables; and even if it were possible to isolate and control a single variable—i.e., a single brain region or psychological factor—the ‘critical experiment’ would only allow for very limited inferences about human behavior. In recognition of these basic principles, a trend has emerged across disciplines towards the synthesis of data and modeling of the overall behavior of highly multivariate systems. These approaches build on accumulated evidence from hundreds or thousands of individual experiments, and provide a ‘bird's eye view’ that complements the traditional experimental approach.
In this article, we review recent efforts to accelerate progress in cognitive neuroscience through greater formal synthesis of the rapidly growing primary literature. We begin by elaborating on the motivation for such an approach by discussing several constraints that limit the ability of individual brain imaging studies to draw strong inferences about structure-function relationships. Next, we briefly review key historical developments and discuss several currently available tools and techniques for aggregating, organizing, and analyzing existing neuroimaging data. Finally, we turn a speculative eye towards the future and discuss potential developments that might accelerate the development of a cumulative cognitive neuroscience.
The rationale
Why is an increased focus on formal synthesis of cognitive neuroscience literature needed? Much of the difficulty in drawing strong and selective inferences about brain structure and function reflects fundamental statistical and methodological constraints that are difficult if not impossible for most individual studies to overcome. We focus here on a number of limitations that can be ameliorated by synthesizing results across many experiments and laboratories.
Most neuroimaging studies are underpowered
Most neuroimaging studies produce maps of brain regions that are activated by some process of interest. When researchers draw inferences about brain-behavior relationships from such maps, they often tacitly assume that these maps provide a relatively comprehensive and accurate picture of the true effects. Unfortunately, this assumption is likely to fail in most cases. The small sample sizes (typically 15 – 20 subjects) and stringent statistical thresholds (p < .001 or lower) commonly used in fMRI studies may provide little power to detect anything but extremely large effects in many circumstances [2-4]. As a result, many if not most fMRI analyses will detect only a fraction of the true effects, producing a deceptive illusion of “selective” activation. Moreover, because researchers typically report only those results that attain statistical significance, the effect sizes reported in the literature tend to be substantially inflated [2, 5]. Because pragmatic considerations suggest that sample sizes in most neuroimaging studies are likely to remain relatively small, synthesis of results via meta-analysis offers the best way to increase statistical power and, ultimately (via image-based meta-analyses discussed below), better effect size estimates. Moreover, unlike large, highly powered studies of a single paradigm, meta-analysis can capture effects that are consistent across laboratories and task variants (Box 1).
Box 1. Meta-analysis of neuroimaging: Goals and techniques.
Meta-analysis techniques provide a way of aggregating data across studies and testing the replicability of effects across laboratories, sites, and study variants; they are increasingly used to evaluate and synthesize the research literature. For example, recent meta-analyses of drug treatments for depression have explicitly examined publication bias [55], and have called into question the benefits of antidepressants for mild and moderate depression [56, 57].
Neuroimaging meta-analyses are typically based on published three-dimensional coordinates for activation locations, with dozens of published meta-analyses of various basic and clinical topics [39, 40]. Though a variety of approaches have been used [40], the dominant approaches use kernel-based reconstruction of meta-analytic statistical maps (see Figure 1a and [6, 25] for details). Such meta-analyses can serve many functions, a few of which we describe below.
Consensus a priori regions of interest (ROIs). Areas such as the “anterior cingulate” span thousands of brain voxels. Results from prior studies of even narrowly-defined tasks can span broad regions, limiting their usefulness for ROI development. Meta-analysis can provide a consensus specification of precise voxels of interest, reducing multiple comparisons and increasing power.
Evaluating psychological specificity. Quantitative discrimination among psychological states requires comparison of results across many task domains, patient groups, etc. Meta-analysis is uniquely suited for such comparisons. For example, though the amygdala is sometimes considered synonymous with fear, meta-analytic evidence suggests that it is at least as activated for disgust ([53]; Figure 1b).
Testing existing hypotheses. As in other domains, neuroimaging meta-analyses can be used to assess the accumulated evidence for existing hypotheses. Exemplar meta-analysis results include findings that amygdala responses in emotion tasks are left lateralized and are stronger for negative emotion, confirming prior hypotheses [58], coupled with lack of support for right-hemisphere dominance in emotion and limited support for differential frontal lateralization by valence.
Establishing correspondence across domains. Meta-analysis can help establish commonalities and dissociations across task types or patient groups. For example, a recent meta-analysis established common amygdala and insula hyper-reactivity across three kinds of anxiety-related disorders, but found hypoactivity in ventromedial prefrontal cortex only in post-traumatic stress disorder [52].
Developing new hypotheses. Negative emotion is sometimes considered synonymous with the amygdala, but other areas that appear critical in animal models, such as the periaqeductal gray and hypothalamus, are rarely mentioned in human studies. A recent meta-analysis found evidence for replicable activity in human studies of emotion in both regions ([51]; Figure 1c). Such findings can lead researchers to consider the importance of PAG activity in new studies.
False positive results are prevalent
Because of the large number of comparisons tested in a typical neuroimaging study, even relatively conservative uncorrected statistical thresholds (e.g., p < .001) are usually insufficient to adequately control the false positive rate across the entire volume (e.g., the brain). In recent meta-analyses, Wager and colleagues estimated the incidence of false positives among published stereotaxic coordinates (‘foci’) at approximately 15%—considerably above the conventionally expected 5% level, even with a modal threshold of p < .001 [6]. Although individual studies generally cannot distinguish true activations from false positives, meta-analyses can at least separate consistent findings from idiosyncratic ones.
Direct replication is uncommon
A hallmark of the scientific method is its emphasis on replicating findings across different studies. Unfortunately, the high cost of fMRI data collection often precludes direct replication of previous fMRI studies; more typically, researchers focus on “conceptual” replications that retain features of a previous study while introducing a new manipulation or context. In addition, there is no consensus on exactly what constitutes a replication. In voxel-wise mapping studies, which currently dominate the field, it is unclear how close an activation peak must be to a previously reported one, or how much overlap in activated voxels two maps must share, to count as a replication. These difficulties arise in part because inferences are typically made on whether voxels are activated and not directly on where activated regions are located. Meta-analyses have the potential to ameliorate these problems by defining consensus areas that can be tested in direct replications.
Selective association is difficult to establish
A primary goal of cognitive neuroscience research is to selectively associate particular functions with specific regions or networks. However, the standard cognitive neuroscience strategy of determining which brain regions are associated with a particular manipulation of cognitive function is not well suited for the identification of selective structure-function associations, because overlapping brain regions may be reliably activated by multiple psychological functions [7]. To identify truly selective mappings, one must establish both that a specific task consistently activates a given region, and that the same region is not consistently activated by many other tasks [8]—a challenging proposition for any individual study. For example, common assumptions about the mapping between neural and mental activity can be empirically tested using meta-analysis, and often prove to be incorrect — such as the notion that increased amygdala activity is a specific marker of negative emotions such as fear (see Box 1 and Figure 1).
Figure 1. Meta-analysis of neuroimaging data: methods and application.
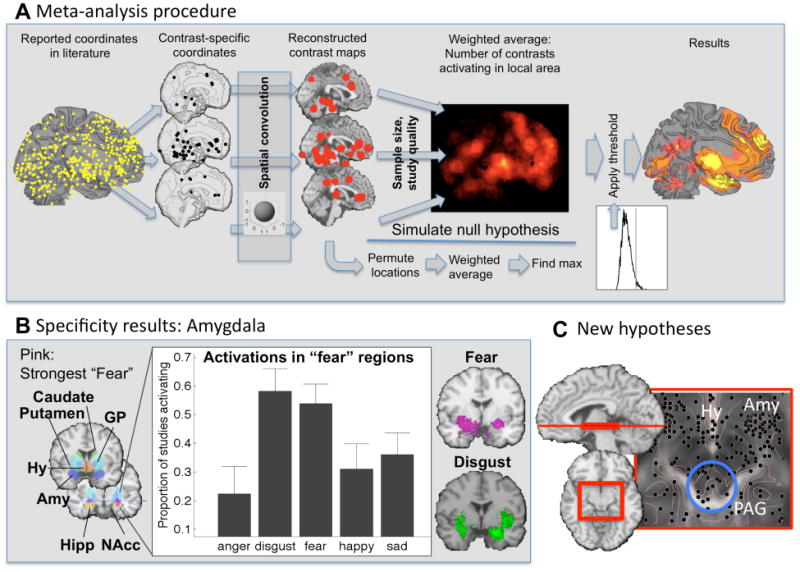
A) Coordinate-based meta-analysis. Whereas early kernel-based approaches were based on simply aggregating coordinates across a set of studies [37, 50], recent approaches explicitly test replicability across studies and allow for weighting by sample size and study quality [6, 25]. The diagram shows the procedure for one of the newer techniques, Multilevel Kernel Density Analysis [51, 52]. Reported peaks are separated by contrast map (often synonymous with study) and convolved with a spatial smoothing kernel. A weighted average map is constructed, considering sample size and other measures of study quality. The map is thresholded by randomizing the locations of the within-study activation regions many times (e.g., 10,000) and calculating the null-hypothesis distribution of the maximum across the image. This threshold provides family-wise error rate control, so that any region in the resulting thresholded map can be interpreted as more consistently activated across studies than would be expected by chance. Similar methods are available for comparing two or more task conditions (see [40]). B) Results in the sublenticular extended amygdala (Amy) from a meta-analysis comparing emotional tasks across emotion types (adapted from Table 1 in [53]). Amygdala responses are not specific for fear. C) Results in the periaqueductal gray (PAG), hypothalamus (Hy), and amygdala across studies (adapted from [51]). Replicable activation in the PAG points towards new hypothesis about PAG's previously under-appreciated role in human emotion.
Formal structure is needed
There is a growing tendency within cognitive neuroscience to move beyond simple brain-behavior mappings that focus on where in the brain activation is occurring, and toward more integrative models that seek to characterize the large-scale functional-anatomic organization of the brain. This shift in focus is reflected, for example, in efforts to identify core networks underlying human cognitive function—e.g., the ‘default’ network [9] and the ‘task-positive’ network [10]—so named for their high levels of activity at rest and responsivity to a range of tasks that require external attention, respectively. The ‘core network’ approach complements the more conventional focus on isolated brain regions. The introduction of multivariate approaches such as independent component analysis (ICA) [11] and related techniques to the fMRI arsenal has provided researchers with powerful tools for modeling networks. A major barrier to progress, however, is the relative absence of an overarching framework for describing neural and mental function. There is currently little consensus about how to classify or group different brain regions, networks, experimental tasks, or cognitive functions, let alone how to develop mappings between different levels of description. Development of suitable descriptive frameworks (or “ontologies”) of cognitive and brain function cannot be accomplished within individual studies that focus narrowly on specific experimental contrasts, and will instead require the formal synthesis of large portions of the published literature.
The methods
The importance of conducting formal syntheses of the cognitive neuroscience literature is increasingly appreciated by many researchers in the field. In this section we summarize relevant history and discuss several recently developed tools and platforms designed to facilitate the sharing and integration of neuroimaging data. Our review is by no means exhaustive; it mainly emphasizes what we view as some of the more promising recent developments.
Data aggregation, atlasing, and sharing
One way to increase the power and generalizability of neuroimaging studies is to aggregate data across multiple sites and studies [12]. This entails bringing data from different studies into a common spatial framework (an atlas) and a common data format, and also making the data readily available. Over the past two decades, there have been important advances on all three fronts. Many of these advances were fueled by the Human Brain Project [13], BIRN (Biomedical Informatics Research Network; http://www.birncommunity.org/), and other targeted funding mechanisms in the U.S. and other countries.
The Talairach atlas and its associated stereotaxic space, which allows for the reporting of stereotaxic coordinates (foci) describing the centers of brain activations or deactivations associated with various tasks, were introduced to neuroimaging in the 1980s [14, 15] as a way to compensate for individual variability in brain size, shape, and patterns of cortical folding. Efforts to improve alignment and better compensate for variability have yielded a plethora of MR-based atlases (both single-subject and probabilistic) and stereotaxic spaces, thereby posing a fresh set of challenges for comparing results across studies [16-18]. A critical factor supporting the shift toward greater integration of the cognitive neuroscience literature has been the development of large-scale online databases [17, 19, 20] that provide support for rapid data mining, visualization, and analysis of stereotaxic coordinates from many studies. Two of the most prominent such databases are the BrainMap database (http://www.brainmap.org) [21, 22] and SumsDB (http://sumsdb.wustl.edu/sums) [23, 24], each of which contains study metadata and activation coordinates for a sizable proportion of the neuroimaging literature. Both databases contain extensive functionality for searching, retrieving and analyzing neuroimaging data, though they also have somewhat different emphases (e.g., BrainMap interoperates closely with ALE (Activation Likelihood Estimate) meta-analysis software [25], whereas SumsDB has greater support for online and offline visualization [23, 26]). The emergence of such databases has greatly lowered the barrier to formal integration of the research literature, giving rise to a proliferation of studies focusing on synthesis of previous findings rather than generation of primary data.
Although stereotaxic coordinates are easy to report and communicate, they constitute a compact but impoverished distillation that belies the spatial complexity and richness of neuroimaging data. An early database infrastructure that could handle the full complexity of imaging data was developed in the 1990s by the fMRI Data Center (fMRIDC), which was devoted to storing and sharing large repositories of primary as well as processed neuroimaging data [27, 28]. The fMRIDC faced significant challenges, including infrastructure limitations, the use of seemingly incommensurable experimental paradigms and data formats, and a reluctance on the part of many researchers to freely share their data [19, 28]. Although it no longer accepts new contributions, the fMRIDC has inspired other recent developments designed to facilitate multi-site collaboration and data sharing of full image information. These include the XNAT (eXtensible Neuroimaging Archive Toolkit) software platform for the storage and dissemination of neuroimaging data [29], as well as web-based resources such as the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC) for neuroimaging tools [30] and the Neuroscience Information Framework (NIF, http://www.neuinfo.org/; [31]), which provide easy access to a rapidly growing set of databases, neuroimaging tools, and other online resources.
Collaborative efforts to aggregate and share data have also produced a number of very large, publicly accessible datasets. The recent release of the 1,000 Functional Connectomes Project—containing data from over 1,400 participants scanned at over 35 sites—provides researchers with a valuable dataset for studying brain function during the resting state [32], and can serve as a model for similar efforts using different experimental paradigms. Additional resources include the OASIS dataset (http://www.oasis-brains.org/; [33]) and the ADNI project (Alzheimer's Disease Neuroimaging Initiative; http://www.adni-info.org/; [34]), and the recently announced Human Connectome Project will soon similarly provide immense amounts of information on brain connectivity in a large population of healthy adults (http://www.humanconnectome.org).
Meta-analysis
Researchers have conducted informal qualitative and quantitative meta-analyses of functional neuroimaging data for nearly two decades [35-38]; until recently, however, published meta-analyses were relatively rare. The recent development of standardized and user-friendly meta-analysis software has led to the rapid adoption of stereotaxic coordinate-based meta-analysis as a primary tool for formal integration of neuroimaging results (Box 1; Figure 1; for review, see [6, 39, 40]). Meta-analyses combining the results of dozens or even hundreds of studies, often reflecting thousands of discrete activation peaks, have been successfully applied in many areas of cognitive neuroscience, ranging from focal analyses of specific cognitive tasks to large-scale analyses of the emotion literature to characterizations of brain disorders (for a summary, see [40]). Such meta-analyses have generally complemented, and in some cases supplanted, the conclusions drawn in individual neuroimaging studies (Box 1).
Ontology development
In order to identify relations between brain structure and mental function, it is necessary to describe both in a systematic way. In other fields such as molecular biology and genomics, cumulative progress has relied heavily upon knowledge frameworks known as “ontologies” that describe the conceptual structure of the domain and provide a basis for the annotation of data within databases [41-44]. Cognitive neuroscience currently lacks such consensus frameworks, and several recent projects have been launched with the aim of developing formal ontologies for cognitive neuroscience. One example is the Cognitive Atlas project (http://cognitiveatlas.org; Figure 2), which leverages collaborative social knowledge building to develop a broad knowledge base that characterizes the state of current thought regarding the relations between mental processes and tasks. At the level of task paradigms, the Cognitive Paradigm Ontology (CogPO) project (http://cogpo.org) is developing a framework to describe the many variable features of cognitive tasks. With regard to neural structure, there is a well-established, albeit incomplete, ontology that is now part of the Foundational Model of Anatomy [45]. All of these efforts are being united under the NeuroLex neuroscience lexicon (http://neurolex.org; [46]), which aims to provide a comprehensive lexicon of terms used in neuroscience.
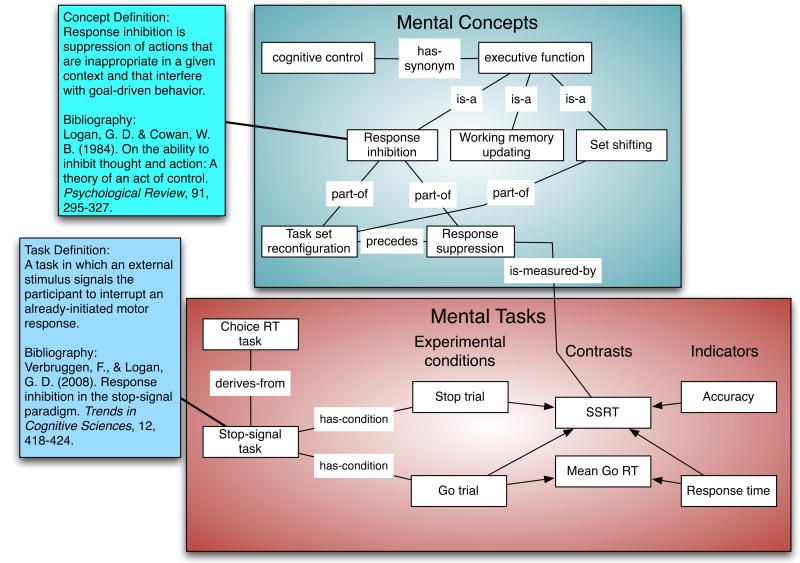
Figure 2. A schematic depicting the structure of the Cognitive Atlas.
The Cognitive Atlas (http://www.cognitiveatlas.org) aims to formally represent mental concepts and their relationships to the tasks that are meant to measure them. In this example, a subset of concepts in the domain of executive function is depicted, along with a task (the stop-signal task) that is thought to measure one of these components. Mental concepts (i.e., any process, representation, or concept related to mental function) can be related to one another in a number of ways, including basic ontological relations (such as “is-a” and “part-of”) as well as temporal precedence relations (“precedes”) and semantic relations such synonymy. Tasks are defined in terms of their particular experimental conditions, the contrasts between conditions that generally define experimental effects, and measured variables (called “indicators”). Specific contrasts (e.g., subtractions between conditions) are related to specific mental concepts by the measurement relation (“is-measured-by”), which formalizes the relations between mental constructs and task manipulations that are often left implicit in cognitive neuroscience research. Tasks can also be related to one another in a family-tree relation (derives-from), which represents the “task phylogeny” [54] describing the historical evolution of psychological tasks.
The future
The tools and techniques described in the previous section represent, in many respects, only the first steps towards a truly cumulative cognitive neuroscience. Going forward, new tools and technologies will undoubtedly continue to reshape the way cognitive neuroscientists conduct research. Here we highlight several emerging developments that could confer important benefits for the field.
Greater automation and standardization of data reporting and processing
A major limitation of extant coordinate databases and meta-analysis packages is the need for considerable human input at multiple stages of processing. This makes it difficult for databases to keep up with the current literature, let alone incorporate a large backlog of older studies [47]. Overcoming this limitation will require a shift toward greater automation of data gathering and integration. Short-term efforts toward this goal include ongoing development of software supporting the automated extraction of activation coordinates from published articles (Box 2), as well as the introduction of “best-practice” standards for reporting coordinates in published articles [48]. In the longer term, advances in natural language processing and text-mining could afford automated and machine-readable semantic tagging of studies, facilitating more precise literature searches and eventually perhaps even fully automated large-scale meta-analyses of the literature.
Box 2. The evolution of neuroimaging databases.
Databases such as BrainMap and SumsDB currently serve a vital role in facilitating synthesis of the neuroimaging literature (see main text), but they also have several important limitations. To extend the functionality of neuroimaging databases and ensure their continued evolution, an informal working group named the NeuroImaging Data Access Group (NIDAG; http://nidag.org) has recently been formed. The broad aim of NIDAG is to promote rapid, open, and efficient access to the world's neuroimaging data. Interested neuroimaging researchers are encouraged to join the group and contribute to its ongoing projects. Concretely, NIDAG aims to extend existing databases in several ways, including:
Automated extraction and coding of activation foci. The need for extensive manual entry of information has kept databases such as SumsDB and BrainMap from catching up with the explosive growth of the primary neuroimaging literature [47]. To overcome this limitation, pilot software has been developed capable of automatically extracting foci information and metadata from published journal articles with a relatively high degree of accuracy (http://nidag.org/tools/). To store and serve the extracted data, a “Neuroimaging Coordinate Warehouse” extension to SumsDB is currently being developed.
Enhanced metadata. Metadata is information associated with neuroimaging results (e.g., reported activation coordinates) that can be used to filter or organize results. As comprehensive manual annotation of the entire literature is practically impossible, an approach is being piloted that can “tag” every published article with a set of words or phrases that occur at an unusually high frequency within the article text. The resulting tags can then be used to rapidly identify relevant coordinates in a content-based manner (cf [59]). For example, one can request only coordinates from studies that prominently feature the terms “language,” “phonology,” and “lexicon”. Pilot efforts suggest that this simple approach is fast, flexible, and effective (http://nidag.org/tools).
Image storage. In the long term, we believe the success of neuroimaging databases will be tied to their ability to store, manage, and serve whole-brain image volumes rather than discrete foci (see main text). Achieving this goal will require extensions to existing databases, as well as creation of client-side tools (e.g., plug-ins for widely-used fMRI packages such as SPM and FSL) that can interface with and upload images to these databases.
Images, not foci
At present, virtually all formal syntheses of the neuroimaging literature operate on discrete foci rather than continuous whole-brain images. From an analytical standpoint, image-based analyses are unquestionably superior to foci-based analyses, as the former preserve the full range of effect sizes in the data, providing a substantial power boost while minimizing selection bias [6, 49]. There are, however, two major barriers to widespread adoption of image-based meta-analysis. On the technical side, databases are needed that can store rich image data, not just isolated foci, and provide flexible search capabilities for the complex metadata needed to describe experimental results (Box 2). On the sociological side, researchers must be convinced that the benefits of freely sharing their images outweigh the costs. These challenges are significant but not insuperable; in the long run, we believe that a shift toward image-based data sharing will offer major benefits to cognitive neuroscience research.
Open cognitive neuroscience
More generally, we see cognitive neuroscience increasingly following an open source model, in which both the data and tools used to generate scientific results are made readily available. This shift is, in many respects, the logical endpoint of current trends discussed in the preceding sections. As bandwidth and storage costs continue to fall, centralized repositories of both raw and processed data will become more practical to maintain and access. Increasing automation of data extraction and visualization, coupled with the development of new metadata standards and image-based data repositories, will make it ever easier for researchers to locate, obtain, and integrate data from multiple sites, fulfilling the promise of greater data sharing that funding agencies such as the NIH have long strived for. In the longer term, we view the development of open platforms for storing, managing, and retrieving data as a prerequisite for the development of next-generation tools that will reshape the way cognitive neuroscientists conduct research (Box 3).
Box 3. What will cognitive neuroscience look like 10 years from now?
The bulk of our discussion focuses on current and short-term developments in cognitive neuroscience. What about the longer term? How might cognitive neuroscientists accumulate scientific results and compare them with new findings 10 years from now? Here is a ‘wish list’ of some possible future developments:
Fully automated quantitative mapping between cognitive and neural states. Researchers would upload observed activation maps to a database as input and receive as output probabilistic estimates of the psychological states participants are in—essentially, pattern classification on a large scale. Conversely, one could define a novel psychological state using structured queries (based on well-developed psychological ontologies) and obtain a map of the predicted neural correlates of that state.
Intelligent preprocessing and analysis pipelines that evaluate the quality of newly acquired neuroimaging data in relation to large databases of existing data and flag problems overlooked by standard quality control tools (e.g., identifying subjects whose multivariate activation patterns are inconsistent with known distributions for the task in question).
Integration or interoperation of neuroimaging databases with other types of data—for instance, construction of large, freely accessible functional genomics repositories that combine behavioral measures, structural and functional brain imaging, and genomic data, enabling researchers to construct integrative models that span genes, brain, and behavior.
Deployment of massive data repositories capable of storing and serving raw data from tens of thousands of neuroimaging studies. Such warehouses could be coupled to high-capacity computing clusters, enabling researchers to conduct large-scale analyses currently beyond the reach of individual labs, and supported by web-based front-ends that allow real-time visualization of results.
Peer-to-peer data collaboration using distributed authoring and version control systems adopted from open source software development (such as git [http://git-scm.com]). Using these systems, data and code could be shared for each step of an analysis, along with raw data.
Integration of ontologies and data sharing methods with fMRI analysis software. Each experimental condition in an analysis would be linked to a formal description of the task in a cognitive ontology such as CogPo (Cognitive Paradigm Ontology; http://cogpo.org), with seamless integration of the ontology into the software. The cognitive paradigm would then be linked to the proposed underlying mental processes in a psychological ontology such as the Cognitive Atlas (http://cognitiveatlas.org). Because the necessary metadata regarding the task would be captured in the analysis, this would enable one-click data sharing from within the analysis software package.
Concluding remarks
The explosive growth of human brain mapping over the past two decades has raised important challenges for the field. As the primary literature expands, the need for powerful tools capable of synthesizing and distilling the findings of many different studies grows commensurately. The present article highlighted the benefits of a synthesis-oriented research strategy and reviewed several ongoing efforts to facilitate greater integration of the published literature. Going forward, such integration will undoubtedly accelerate progress in elucidating the neural mechanisms that support the full range of human thought, feeling, and action in health and disease. There is every reason to push forward energetically on efforts to develop a cumulative science of human brain function.
Acknowledgments
This work was supported by NIH award F32NR012081 to T.Y., NIH awards NIMH R01MH076136 and NIDA R01DA027794 to T.D.W, NIH award R01MH082795 to R.A.P., and NIH award R01MH60974 to D.V.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Platt J. Strong inference. Science. 1964;146:347–353. doi: 10.1126/science.146.3642.347. [DOI] [PubMed] [Google Scholar]
- 2.Yarkoni T. Big correlations in little studies. Perspect Psychol Stud. 2009;4:294–298. doi: 10.1111/j.1745-6924.2009.01127.x. [DOI] [PubMed] [Google Scholar]
- 3.Braver TS, et al. Vive les differences! Individual variation in neural mechanisms of executive control. Curr Opin Neurobiol. 2010;20:242–250. doi: 10.1016/j.conb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wager TD, et al. Elements of functional neuroimaging. In: Cacioppo JT, et al., editors. Handbook of Psychophysiology. 3rd. 2007. pp. 19–55. [Google Scholar]
- 5.Ioannidis J. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 6.Wager TD, et al. Meta-analysis of functional neuroimaging data: current and future directions. Soc Cogn Affect Neurosci. 2007;2:150–158. doi: 10.1093/scan/nsm015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poldrack RA. Mapping mental function to brain structure: How can cognitive neuroimaging succeed? Persp Psychol Sci. doi: 10.1177/1745691610388777. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poldrack RA. Can cognitive processes be inferred from neuroimaging data. Trends Cogn Sci. 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Raichle ME, et al. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox MD, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calhoun V, et al. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage. 2009;45:S163–S172. doi: 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costafreda S. Pooling FMRI data: meta-analysis, mega-analysis and multi-center studies. Frontiers in neuroinformatics. 2009;3:33. doi: 10.3389/neuro.11.033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huerta MF, et al. The human brain project: an international resource. Trends Neurosci. 1993;16:436–438. doi: 10.1016/0166-2236(93)90069-x. [DOI] [PubMed] [Google Scholar]
- 14.Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging. Thieme; 1988. [Google Scholar]
- 15.Fox PT, et al. A stereotactic method of anatomical localization for positron emission tomography. J Comput Assist Tomogr. 1985;9:141–153. doi: 10.1097/00004728-198501000-00025. [DOI] [PubMed] [Google Scholar]
- 16.Devlin JT, Poldrack RA. In praise of tedious anatomy. Neuroimage. 2007;37:1033–1041. doi: 10.1016/j.neuroimage.2006.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Essen D, Dierker D. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56:209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Van Essen DC, Dierker D. On navigating the human cerebral cortex: Response to ‘in praise of tedious anatomy’. Neuroimage. 2007;37:1050–1054. doi: 10.1016/j.neuroimage.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Horn J, Toga A. Is it time to re-prioritize neuroimaging databases and digital repositories? Neuroimage. 2009;47:1720–1734. doi: 10.1016/j.neuroimage.2009.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Essen D. Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr Opin Neurobiol. 2002;12:574–579. doi: 10.1016/s0959-4388(02)00361-6. [DOI] [PubMed] [Google Scholar]
- 21.Laird AR, et al. The Social Evolution of a Human Brain Mapping Database. Neuroinformatics. 2005;3:65–78. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- 22.Fox PT, et al. BrainMap: a database of human functional brain mapping. In: Thatcher RW, et al., editors. Advances in Functional Neuroimaging: Technical Foundations. Academic; 1994. pp. 95–106. [Google Scholar]
- 23.Van Essen DC. Lost in localization--But found with foci?! Neuroimage. 2009;48:14–17. doi: 10.1016/j.neuroimage.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson J, et al. ‘The surface management system’ (SuMS) database: a surface-based database to aid cortical surface reconstruction, visualization and analysis. Philos Trans R Soc Lond B Biol Sci. 2001;356:1277–1292. doi: 10.1098/rstb.2001.0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eickhoff S, et al. Coordinate-based activation likelihood estimation meta-analysis of neuroimaging data: A random-effects approach based on empirical estimates of spatial uncertainty. Hum Brain Mapp. 2009;30:2907–2926. doi: 10.1002/hbm.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Essen D, et al. An integrated software suite for surface-based analyses of cerebral cortex. J Am Med Inform Assoc. 2001;8:443–459. doi: 10.1136/jamia.2001.0080443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Horn JD, et al. The Functional Magnetic Resonance Imaging Data Center (fMRIDC): the challenges and rewards of largeñscale databasing of neuroimaging studies. Philos Trans R Soc Lond, Ser B: Biol Sci. 2001;356:1323. doi: 10.1098/rstb.2001.0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Horn J, et al. Sharing neuroimaging studies of human cognition. Nat Neurosci. 2004;7:473–481. doi: 10.1038/nn1231. [DOI] [PubMed] [Google Scholar]
- 29.Marcus D, et al. The extensible neuroimaging archive toolkit. Neuroinformatics. 2007;5:11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, et al. Neuroimaging informatics tools and resources clearinghouse (NITRC) resource announcement. Neuroinformatics. 2009;7:55–56. doi: 10.1007/s12021-008-9036-8. [DOI] [PubMed] [Google Scholar]
- 31.Gardner D, et al. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6:149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biswal B, et al. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marcus DS, et al. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 34.Mueller SG, et al. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koski L, Paus T. Functional connectivity of the anterior cingulate cortex within the human frontal lobe: a brain-mapping meta-analysis. Exp Brain Res. 2000;133:55–65. doi: 10.1007/s002210000400. [DOI] [PubMed] [Google Scholar]
- 36.Fox PT, et al. Beyond the single study: function/location metanalysis in cognitive neuroimaging. Curr Opin Neurobiol. 1998;8:178–187. doi: 10.1016/s0959-4388(98)80138-4. [DOI] [PubMed] [Google Scholar]
- 37.Smith EE, Jonides J. Storage and executive processes in the frontal lobes. Science. 1999;283:1657–1661. doi: 10.1126/science.283.5408.1657. [DOI] [PubMed] [Google Scholar]
- 38.Phan KL, et al. Functional neuroanatomy of emotion: a meta-analysis of emotion activation studies in PET and fMRI. Neuroimage. 2002;16:331–348. doi: 10.1006/nimg.2002.1087. [DOI] [PubMed] [Google Scholar]
- 39.Kober H, Wager TD. Meta-analysis of neuroimaging data. Wiley Interdisc Rev Cogn Sci. 2010;1:293–300. doi: 10.1002/wcs.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wager TD, et al. Evaluating the consistency and specificity of neuroimaging data using meta-analysis. Neuroimage. 2009;45:210–221. doi: 10.1016/j.neuroimage.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith B, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen LJ, Bork P. Ontologies in Quantitative Biology: A Basis for Comparison, Integration, and Discovery. PLoS Biol. 2010;8:e1000374. doi: 10.1371/journal.pbio.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bard J, Rhee S. Ontologies in biology: design, applications and future challenges. Nat Rev Genet. 2004;5:213–222. doi: 10.1038/nrg1295. [DOI] [PubMed] [Google Scholar]
- 45.Rosse C, Mejino J. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inf. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 46.Bug WJ, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6:175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derrfuss J, Mar R. Lost in localization: the need for a universal coordinate database. Neuroimage. 2009;48:1–7. doi: 10.1016/j.neuroimage.2009.01.053. [DOI] [PubMed] [Google Scholar]
- 48.Poldrack R, et al. Guidelines for reporting an fMRI study. Neuroimage. 2008;40:409–414. doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salimi-Khorshidi G, et al. Meta-analysis of neuroimaging data: A comparison of image-based and coordinate-based pooling of studies. Neuroimage. 2009;45:810–823. doi: 10.1016/j.neuroimage.2008.12.039. [DOI] [PubMed] [Google Scholar]
- 50.Turkeltaub PE, et al. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16:765–780. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- 51.Kober H, et al. Functional grouping and cortical-subcortical interactions in emotion: A meta-analysis of neuroimaging studies. Neuroimage. 2008;42:998–1031. doi: 10.1016/j.neuroimage.2008.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Etkin A, Wager T. Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry. 2007;164:1476–1488. doi: 10.1176/appi.ajp.2007.07030504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wager T, et al. The neuroimaging of emotion. In: Lewis M, et al., editors. The handbook of emotion. 2008. pp. 249–271. [Google Scholar]
- 54.Bilder R, et al. Cognitive ontologies for neuropsychiatric phenomics research. Cognitive neuropsychiatry. 2009;14:419–450. doi: 10.1080/13546800902787180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melander H, et al. Evidence b (i) ased medicine--selective reporting from studies sponsored by pharmaceutical industry: review of studies in new drug applications. Br Med J. 2003;326:1171–1175. doi: 10.1136/bmj.326.7400.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fournier J, et al. Antidepressant drug effects and depression severity: a patient-level meta-analysis. JAMA. 2010;303:47–53. doi: 10.1001/jama.2009.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirsch I, et al. Initial severity and antidepressant benefits: a meta-analysis of data submitted to the Food and Drug Administration. PLoS Med. 2008;5:e45. doi: 10.1371/journal.pmed.0050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wager TD, et al. Valence, gender, and lateralization of functional brain anatomy in emotion: a meta-analysis of findings from neuroimaging. Neuroimage. 2003;19:513–531. doi: 10.1016/s1053-8119(03)00078-8. [DOI] [PubMed] [Google Scholar]
- 59.Poldrack R, et al. Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol Sci. 2009;20:1364–1372. doi: 10.1111/j.1467-9280.2009.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]