Abstract
The opportunistic pathogen Pseudomonas aeruginosa employs acyl homoserine lactones (AHL) as signaling compounds to regulate virulence gene expression via quorum sensing. The AHL N-3-oxo-dodecanoyl-L- homoserine lactone (3OC12-HSL) also induces mammalian cell responses, including apoptosis and immune modulation. In certain cell types the apoptotic effects of 3OC12-HSL are mediated via a calcium-dependent signaling pathway, while some proinflammatory effects involve intracellular transcriptional regulators. However, the mechanisms by which mammalian cells perceive and respond to 3OC12-HSL are still not completely understood. Here we used microarray analysis to investigate the transcriptional response of human lung epithelial cells after exposure to 3OC12-HSL. These data revealed that mRNA levels for several genes involved in xenobiotic sensing and drug transport were increased in cells exposed to 3OC12-HSL, which led us to examine the intracellular fate of 3OC12-HSL. Using radiolabeled autoinducer uptake assays, we discovered that intracellular 3OC12-HSL levels increased after exposure and achieved maximal levels after 20-30 minutes. Intracellular 3OC12-HSL decreased to background levels over the next 90 minutes and this process blocked by pre-treatment with an inhibitor of the ABC transporter ABCA1. Taken together, these data suggest that mammalian cells detect 3OC12-HSL and activate protective mechanisms to expel it from the cell.
Keywords: Quorum sensing, interkingdom signaling, Pseudomonas aeruginosa, AHL, autoinducer, microarray analysis, cell-to-cell signaling, xenobiotic, ABC transporter, drug pump
1. Introduction
Cell-to-cell communication is an essential process not only for complex multicellular organisms but also for unicellular organisms, including many bacterial species [1]. Functional cell-to-cell communication is a critical factor in the virulence of several pathogenic bacterial species and disruption of intercellular communication has emerged as a potential mechanism for the development of novel antimicrobial agents [1]. Quorum sensing (QS) is a bacterial intercellular communication system that coordinates gene expression within a population in a density-dependent manner. The signals that mediate QS, called autoinducers, can be a variety of types of small molecules, including a variety of structurally-related acyl homoserine lactones (AHLs) in many Gram-negative bacteria [2]. In the classic model of QS, autoinducers are constitutively synthesized by bacteria at low levels, which increase with increasing cell number. Upon reaching a threshold concentration, autoinducers accumulate within cells in the population, bind their cognate receptors and modulate transcription of target genes. Common target genes of QS in pathogenic bacteria include those encoding virulence factors and, under certain conditions, genes involved in biofilm formation [3].
Pseudomonas aeruginosa is a pathogenic bacterium whose utilization of QS systems to enhance its virulence has been well studied [4]. This Gram-negative opportunistic pathogen is a major cause of morbidity and mortality in cystic fibrosis patients, individuals with severe burn wounds, HIV/AIDS patients, and other immunocompromised humans [3-4]. P. aeruginosa possesses two complete AHL-based QS systems, the Las and Rhl systems [5]. The Las system consists of the autoinducer N-3-oxo-dodecanoyl-L-homoserine lactone (3OC12-HSL), which is synthesized by the LasI enzyme, and the LasR transcriptional regulatory protein. The target genes of the Las system in P. aeruginosa include a variety of virulence factors and disruption of the Las system severely diminishes P. aeruginosa virulence in a number of animal infection models [4, 6-10]. However, 3OC12-HSL also affects cell responses in eukaryotic cells, including cells derived from mammalian species. We have named this process interkingdom signaling, which can be defined as the transmission and productive reception of small chemical signals between species from different kingdoms [11].
Our group and others have described at least two distinct effects of 3OC12-HSL on mammalian cells, pro-inflammatory gene induction and apoptosis [12]. These effects have been observed in a variety of cell types, including macrophages, T cells, fibroblasts, epithelial and endothelial cells [12-19]. We recently reported that apoptotic induction by 3OC12-HSL involves a calcium-dependent pathway and appears to be mediated by a receptor located at or close to the cell membrane [12]. Alternatively, the pro-inflammatory effects of 3OC12-HSL are independent of calcium signaling and may be mediated by members of the nuclear hormone receptor superfamily, including the peroxisome proliferator activated receptors (PPARs) [17, 20].
Based on its described effects on mammalian cells, 3OC12-HSL appears to function as an independent virulence factor for P. aeruginosa. To extend these studies, and further characterize the mechanisms by which 3OC12-HSL affects eukaryotic cells we performed microarray analysis of mammalian cells exposed to bacterial autoinducers. We exposed human lung epithelial cells to the P. aeruginosa AHL autoinducers 3OC12-HSL and C4-HSL or 4,5-dihydroxy-2,3-pentanedione, or DPD. DPD is the precursor to the AI-2 family of autoinducers and is synthesized by the LuxS synthase [3]. DPD is highly reactive and spontaneously rearranges to produce the reactive AI-2 molecules, which have been referred to as ‘universal autoinducers’ because LuxS has been found in several different bacterial species [3].
Only 3OC12-HSL appeared to have a significant effect on the mammalian cell transciptome. Not surprisingly, many of the gene transcripts that were altered have been previously identified as targets of 3OC12-HSL. However, several new sub-sets of transcripts, including many involved in xenobiotic sensing and drug metabolism, were also affected, leading us to postulate that mammalian cells detect 3OC12-HSL and activate protective mechanisms involving membrane pumps to expel it from the cell.
Despite the extensive studies of its effects on a variety of mammalian cell types, it is still not completely understood how mammalian cells perceive and respond to 3OC12-HSL. Elucidating the mechanisms of 3OC12-HSL action on host cells requires an understanding of its fate upon coming into contact with these cells. Its ability to enter eukaryotic cells, localization upon entering, and further processing or transport are all aspects of the interkingdom signaling mediated by 3OC12-HSL. In previous bacterial studies, AHLs with relatively short side chains have been demonstrated to freely diffuse across the bacterial membrane in both directions [21]. Alternatively 3OC12-HSL, a longer side chain AHL, is subject to active efflux from P. aeruginosa [22]. Our previous work using artificial bacterial receptor constructs demonstrated that 3OC12-HSL can enter eukaryotic cells, although we did not define the mechanism of entry [23]. Recent work demonstrated that 3OC12-HSL freely enters immune cell types, including T cells and dendritic cells, quickly achieving stable intracellular levels, with a majority localized to the cytoplasm rather than the nucleus or membrane fractions [24]. Here we further assess the fate of 3OC12-HSL in mammalian cell types sensitive to its effects. By employing tritium labeled 3OC12-HSL (3H-3OC12-HSL), we were able to follow the uptake, subcellular localization, and release of 3OC12-HSL from eukaryotic cells, using both fibroblasts and epithelial cells as model systems for 3OC12-HSL interaction with mammalian cells in a P. aeruginosa infection.
2. Methods and methods
2.1 Cell Culture
NIH3T3 immortalized murine fibroblast cells and A549 human alveolar epithelial cells were sourced from ATCC (Manassas, VA). Primary MEF cells were a gift from Peter Johnson (National Cancer Institute, Fredrick, MD). NIH3T3 and MEF cells were cultured in Dulbecco's modified Eagle's Medium (DMEM, BioWhittaker, Walkersville, MD) supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT), and 100 U ml-1 penicillin and 100 μg ml-1 streptomycin (BioWhittaker, Walkersville, MD). A549 cells were cultured in Kaighn's modification of F-12 Nutrient Mixture (Mediatech, Herndon, VA) supplemented with 10% FBS and 100 U ml-1 penicillin and 100 μg ml-1 streptomycin. For microarray experiments, A549 cells were plated at 200,000 cells/well on 6-well plates (Becton Dickinson, Franklin Lakes, NJ) the day before the experiment in Kaighn's modification of F-12 Nutrient Mixture, supplemented with 10% FBS and 100 U ml-1 penicillin and 100 μg ml-1 streptomycin. The next day, the medium was removed and 50 μM autoinducer or vehicle (see below) was added with new medium, lacking FBS. For 3H-3OC12-HSL uptake analysis and ATP depletion experiments, cells were plated at 50,000 cells/well on a 24-well plate (Nunc, Denmark) the day before the experiment was performed. For ABCA1 inhibition studies, cells were pre-treated with 250 μM Glyburide (Sigma, St. Louis, MO) or the equivalent amount of vehicle (DMSO) for one hour prior to addition of autoinducer. All cells were maintained at 37°C in a humidified environment containing 5% CO2.
2.2 Autoinducer Preparation
3OC12-HSL was synthesized as previously described [25-26] and N-butyryl-homoserine lactone (C4) was purchased from Sigma (St Louis, MO). Both autoinducers were dissolved in ethyl acetate containing 0.001% glacial acetic acid for stable storage at −20°C. Custom synthesis of tritium labeled 3OC12-HSL (5, 6-3H-3OC12-HSL) with a specific activity of 25 Ci/mmol was performed by ViTrax (Placentia, CA). The 3H-3OC12-HSL was also stored in ethyl acetate at -20° C. For addition to cell culture, autoinducers were placed under a gentle stream of nitrogen gas to evaporate the ethyl acetate solvent and were dissolved into the cell culture medium immediately prior to addition to the cell culture. For vehicle controls, an equivalent volume of ethyl acetate containing 0.001% glacial acetic acid alone was placed under a gentle stream of nitrogen until evaporation, then cell culture medium was added.
2.3 Microarray and Analysis
RNA was prepared from cells exposed for 6 hours to equivalent concentrations of each chemical and two color microarray hybridizations were performed using Agilent human whole genome arrays (Agilent Technologies, Santa Clara, CA). RNA samples from four independent experiments were compared to a Universal human reference RNA sample (Stratagene) and the data were analyzed using the Agilent Feature Extraction Software. Genes whose RNA levels were altered in response to each autoinducer were computed after conversion of data to log2. The log2 ratios for each array were examined using boxplot to ensure that substantial scale differences did not exist among microarrays [27]. Differentially-expressed genes were identified by testing whether two populations (control and a treatment) exhibited statistical differences using either one-way ANOVA as a parametric test or the Wilcoxon rank sum test [28] as a non-parametric test. Normal distributions of the data were assessed according to the Kolmogorov-Simirnov goodness-of-fit hypothesis test [29]. The rank sum test was more stringent and yielded a dataset that was essentially a subset of genes identified by ANOVA. The correction for multiple testing was applied using Benjamini and Hochberg false discovery rate procedure [30]. Genes were identified to be differentially expressed using ANOVA if their corrected p-values were less than or equal to 0.05 (5% significance level). Matlab software (The MathWorks Inc., Natick, MA) was used for statistical analysis.
2.43H-3OC12-HSL Uptake Assays
For each well of a 24-well plate, 0.45 μl of tritium-labeled 3OC12-HSL was dried and then resuspended in 0.5 ml serum-free culture medium. Cells were washed twice with PBS prior to addition of the autoinducer-containing medium. The cells were incubated with the 3H-3OC12-HSL for the times indicated. At the designated time, the medium was removed, cells were washed once with PBS, and then the cells were collected by scraping in 0.5 ml PBS. 100 μl of each the medium and the cells for each time point were added to 5 ml liquid scintillation fluid and analyzed in a Beckman LS-6500 liquid scintillation counter (Beckman-Coulter, Fullerton, CA).
2.5 Reverse transcription-PCR
RNA was isolated from cells using the Versagene RNA Cell Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer's specifications. cDNA was prepared by combining 2 μg of total RNA, 400 U Superscript Reverse Transcriptase (Invitrogen, Carlsbad, CA) and 500 ng Oligo-(dT) and incubating at 37°C for 1 hour. PCR reactions were performed in a T3 Thermocycler (Biometra, Goettingen, Germany) with Taq DNA polymerase (New England Biolabs, Ipswich, MA). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). PCR products were run on 1.5% agarose gels containing 10 ng of ethidium bromide (Fisher Biosciences Lafayette, CO) and gels were visualized under UV light. The sequences of the PCR primers for the CYP1A1 and L19 genes are as follows: CYP1A1 (forward) 5’-GTTGGACCTCTTTGGAGCTG-3’, (reverse) 5’-GGTTGATCTGCCACTGGTTT; L19 (forward) 5’- GAAATCGCCAATGCCAACTC-3’, (reverse) 5’-TCTTAGACCTGCGAGCCTCA.
2.6 ATP Depletion
To optimize conditions for ATP depletion, cells were incubated in serum-free culture medium containing 5 mM sodium azide (Fisher Scientific) and 2-deoxy-D-glucose (Sigma, St. Louis, MO) in concentrations ranging from 5 mM to 80 mM for either 1, 2, or 4 hours. ATP levels were measured with the luciferase based ATP Determination Kit from Invitrogen (Carlsbad, CA). Luminescence was measured with a Modulus Single Tube Luminometer (Turner Biosystems, Sunnyvale, CA). The determined optimal condition of incubation with 80 mM 2-deoxy-D-glucose and 5 mM sodium azide for 4 hours was used for all further experiments.
2.7 Subcellular Fractionation
Differential detergent fractionation was carried out according to a standard protocol with minor modifications [31]. Buffers were prepared in ddH2O and pH was adjusted by adding either 1 M NaOH or 1 M HCl. Immediately before use, phenylmethylsulfonyl fluoride (PMSF) was added to each buffer to a final concentration of 1 mM. The buffer compositions are as follows: digitonin buffer for cytosolic protein extraction (0.015% digitonin, 10 mM PIPES, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 5 mM EDTA, pH 6.8), Triton X-100 buffer for membrane/organelle protein extraction (0.5% Triton X-100, 10 mM PIPES, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 3 mM EDTA, pH 7.4), Tween 40/deoxycholate buffer for nuclear protein extraction (1% Tween 40, 0.5% deoxycholate, 10 mM PIPES, 300 mM sucrose, 10 mM NaCl, 1 mM MgCl2, pH 7.4), and SDS buffer for cytoskeletal-matrix protein extraction (5% SDS in 10 mM sodium phosphate, pH 7.4).
For fractionation experiments with radiolabeled autoinducer, a monolayer of cultured cells (~2 × 106) in a 100 mm cell culture plate was incubated in serum-free medium containing 3H-3OC12-HSL for 30 minutes, then washed twice with PBS. 1 mL of digitonin buffer was added and the plate was rocked on ice for 15 min. The extracted solution was collected and the plate was washed with 2 mL ice-cold PBS. Subsequent extractions were carried out similarly with 1 mL Triton X-100 buffer, 500 μl Tween 40/deoxycholate buffer, and 500 μl SDS buffer with rocking on ice for 30, 20, and 20 min, respectively. Between each step, the plate was washed with 2 mL ice-cold PBS by rocking on ice for 1 min. Tritium levels in each fraction were assessed by liquid scintillation counting of 100 μl extract in 5 ml liquid scintillation fluid. The remainder of the extract was stored at -80° C.
3. Results
3.1 3OC12-HSL modulates the mammalian cell transciptome
Our group and others have characterized the induction of many host products in several different cell types in response to bacterial autoinducers. 3OC12-HSL had previously been shown to modulate the expression of a relatively small number of genes encoding inflammatory products in vitro and in vivo [12-14, 17, 32-33]. In an attempt to uncover other cellular processes modulated by AHLs, we performed microarray analyses of autoinducer-dependent gene responses in mammalian cells. We analyzed gene responses to 3OC12-HSL in human A549 lung epithelial cells by comparing mRNA levels in cells exposed to 3OC12-HSL with those in cells exposed to related or unrelated autoinducers, namely the AHL autoinducer C4-HSL and the structurally unrelated “universal autoinducer” AI-2.
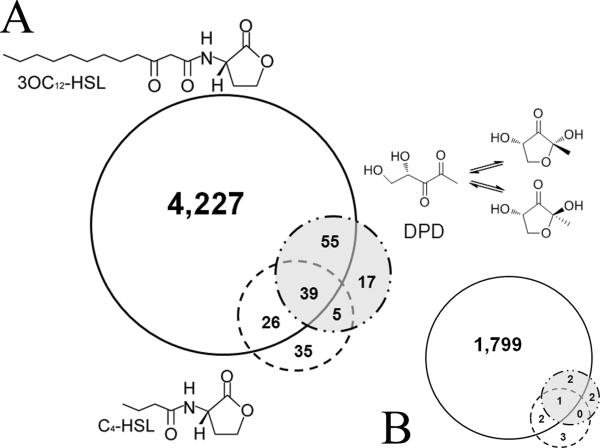
The number of genes responding to 3OC12-HSL was vastly greater than those altered in cells exposed to either C4-HSL or AI-2 and a small number of genes were common to each dataset. As shown in figure 1, the expression of 4347, 116 and 105 genes was altered by treatments with 3OC12-HSL, AI-2 and C4-HSL respectively, at a p value of 0.05 or lower. However, among those, only 1803, 5 and 6 genes, respectively for 3OC12-HSL, AI-2 and C4-HSL-treatments displayed greater than 2-fold differences (See supplemental tables 1, 2 and 3 for complete lists of these genes). These data imply that 3OC12-HSL elicits reproducible and significant changes in mRNA levels for a large number of cellular genes in A549 cells while the C4-HSL and AI-2 autoinducers had only minimal effects on mRNA levels.
Figure 1. Identification of autoinducer regulated genes in A549 cells.
Separate cultures of A549 cells were grown in six well plates and exposed to equivalent concentrations (50 μM) of 3OC12-HSL, C4-HSL, the structurally unrelated autoinducer-2 (AI-2) or to vehicle control for 6 hours. RNA was prepared from four independent cultures for each treatment. Two color microarray hybridizations were performed using Agilent human whole genome arrays in which each of these 16 RNAs was compared to a Universal human reference RNA sample (Stratagene). Differentially expressed genes were identified after data normalization (see text) and removal of genes whose expression differed between the vehicle control and the standard RNA. The gene lists were then compared in all combinations to identify genes whose expression was altered specifically in cells exposed to the three different autoinducers. A. These data are represented in the accompanying Venn diagrams showing that 3OC12-HSL treatment elicited significant changes in a greater number of genes that the other two autoinducers, and B. the respective gene populations of (A) that displayed greater than 2-fold changes.
Importantly, many of the genes altered specifically by 3OC12-HSL have previously been identified by us and others, including several involved in immune modulation (see Table 1 for examples). In addition, we detected other sub-sets of genes, including several involved in the transport and metabolic processing of xenobiotics. Among these were members of the cytochrome P450 superfamily (Cyp450), which encode protein that can metabolize drugs and other xenobiotic agents and whose transcription is often sensitive to the agents they metabolize. We chose CYP1A1 as a representative family member to examine further by RT-PCR and observed a specific dose-dependent increase in mRNA in response to 3OC12-HSL (Fig. 2). These results suggested that human cells may activate protective mechanisms involved in chemical detoxification in response to 3OC12-HSL.
Table 1.
Select human mRNAs modulated by 3OC12-HSL
| Accession Number | Gene Name | Change in 3O-C12 treated | P value* | Gene Product Description |
|---|---|---|---|---|
| Immune Response | ||||
| NM_000584 | IL8 | 47.59 | 0.0002 | Homo sapiens interleukin 8 |
| NM_000600 | IL6 | 12.81 | 0 | Homo sapiens interleukin 6 (interferon, beta 2) |
| NM_005384 | NFIL3 | 6.57 | 0.0001 | Homo sapiens nuclear factor, interleukin 3 regulated |
| NM_016584 | IL23A | 6.54 | 0.004 | Homo sapiens interleukin 23, alpha subunit p19 |
| NM_000882 | IL12A | 4.49 | 0.0223 | Homo sapiens interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| NM_000641 | IL11 | 2.80 | 0.0127 | Homo sapiens interleukin 11 |
| NM_006290 | TNFAIP3 | 12.01 | 0 | Homo sapiens tumor necrosis factor, alpha-induced protein 3 |
| NM_003807 | TNFSF14 | 4.21 | 0.0228 | Homo sapiens tumor necrosis factor (ligand) superfamily, member 14, transcript variant 1 |
| NM_020529 | NFKBIA | 12.38 | 0.0001 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| NM_004556 | NFKBIE | 7.17 | 0 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| NM_002502 | NFKB2 | 2.0 | 0 | Homo sapiens nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100), transcript variant 2 |
| NM_003955 | SOCS3 | 5.55 | 0 | Homo sapiens suppressor of cytokine signaling 3 |
| NM_003745 | SOCS1 | 2.46 | 0.0052 | Homo sapiens suppressor of cytokine signaling 1 |
| NM_002089 | CXCL2 | 13.72 | 0.0003 | Homo sapiens chemokine (C-X-C motif) ligand 2 |
| NM_002090 | CXCL3 | 9.71 | 0 | Homo sapiens chemokine (C-X-C motif) ligand 3 |
| NM_001511 | CXCL1 | 11.98 | 0.0008 | Homo sapiens chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| NM_004591 | CCL20 | 16.05 | 0.001 | Homo sapiens chemokine (C-C motif) ligand 20 |
| | ||||
| Transport/Metabolism | ||||
| AF355802 | CYP3A5 | 3.41 | 0.0151 | Homo sapiens CYP3A5 mRNA, allele CYP3A5*3, exon 5B and partial cds, alternatively spliced |
| NM_000785 | CYP27B1 | 2.30 | 0.0431 | Homo sapiens cytochrome P450, family 27, subfamily B, polypeptide 1, nuclear gene encoding mitochondrial protein |
| NM_000499 | CYP1A1 | 2.04 | 0.0277 | Homo sapiens cytochrome P450, family 1, subfamily A, polypeptide 1 |
| ENST000003 67000 | SLC30A1 | 2.01 | 0.0006 | Zinc transporter 1 (ZnT-1) (Solute carrier family 30 member 1) |
| NM_0010044 | SLC30A2 | 4.53 | 0.0123 | Homo sapiens solute carrier family 30 (zinc transporter), member 2, transcript variant 1 |
| NM_018420 | SLC22A15 | 6.04 | 0.0004 | Homo sapiens solute carrier family 22 (organic cation transporter), member 15 |
| NM_004694 | SLC16A6 | 3.29 | 0.0001 | Homo sapiens solute carrier family 16, member 6 (monocarboxylic acid transporter 7) |
| NM_194298 | SLC16A9 | 3.54 | 0.0009 | Homo sapiens solute carrier family 16, member 9 (monocarboxylic acid transporter 9) |
| BC020867 | SLC6A13 | 3.06 | 0.0148 | Homo sapiens solute carrier family 6 (neurotransmitter transporter, GABA), member 13 |
| NM_152527 | SLC16A14 | 4.78 | 0 | Homo sapiens solute carrier family 16, member 14 (monocarboxylic acid transporter 14) |
Benjamini-Hochberg corrected p-value
Figure 2.
A549 were incubated with increasing concentrations of 3OC12-HSL or C4-HSL for 6 hr and RNA was extracted. mRNA levels of the P450 CYP1A1 was assessed by RT-PCR. The mRNA for the ribosomal protein L-19 was used as a loading control.
3.2 3OC12-HSL rapidly enters multiple eukaryotic cell types
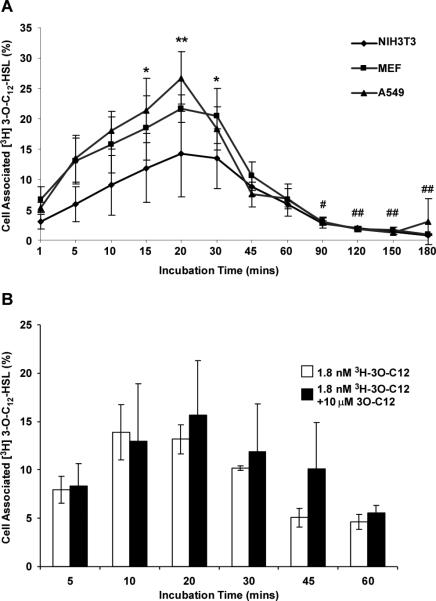
Despite the well documented effects of 3OC12-HSL on eukaryotic cells, its molecular target(s) and mechanism of action within these cells have yet to be elucidated. As an approach to this problem, we analyzed the kinetics of entry and exit of 3H-3OC12-HSL in mammalian cell types likely to be exposed to high levels of autoinducer in P. aeruginosa infections. A549 human lung epithelial cells, primary mouse embryonic fibroblasts (MEFs), and NIH3T3 mouse fibroblasts were incubated with 1.8 nM 3H-3OC12-HSL for times ranging from 1 minute to 3 hours. Cells and medium were collected at the time points indicated in Figure 3 and the level of radioactivity in each fraction was assessed by liquid scintillation counting. In each of the cell types analyzed, the time course of 3H-3OC12-HSL association with cells exhibited a similar profile, in which increasing amounts of 3H-3OC12-HSL became associated with the cells over the first 20-30 minutes of incubation. The amount of cell-associated 3H-3OC12-HSL was significantly elevated in the time period between 15 and 30 minutes after application when compared to the initial time point (1 minute after application) in all three cell types with maximal cell-associated 3H-3OC12-HSL levels reaching 25-30% percent of the total applied. Subsequently, the proportion of cell-associated 3H-3OC12-HSL levels decreased dramatically, with the values after 90 minutes being significantly lower than peak levels (Fig. 3). We interpret these data to indicate that 3H-3OC12-HSL becomes associated with target cells fairly rapidly, but that it is subsequently expelled from these cells. The rapid and efficient removal of 3H-3OC12-HSL suggests that this is likely to be an active process as passive diffusion would be unlikely to elicit a similar pattern. Although 3OC12-HSL can promote apoptosis in some mammalian cell types, this was unlikely to occur in these experiments as the concentration of applied 3H-3OC12-HSL was several orders of magnitude below the concentrations required to promote apoptosis [12].
Figure 3. Time course of tritium labeled 3OC12-HSL levels associated with mammalian fibroblast and epithelial cells.
(A) NIH 3T3, A549, and MEF cells were incubated with a defined dose of 3H-3OC12-HSL for the times indicated. (B) A549 cells were incubated with either 3H-3OC12-HSL alone or 10 μM unlabeled 3OC12-HSL spiked with 3H-3OC12-HSL for the times indicated. In both cell types, levels of radioactivity associated with the medium and the cells were analyzed by liquid scintillation counting. The data are represented as the means±s.d.; n=4.
We next assessed whether the observed pattern of 3H-3OC12-HSL cell association was due to the application of concentrations of 3OC12-HSL lower than those at which its apoptotic and immunomodulatory effects have been demonstrated. We compared the pattern of 3H-3OC12-HSL cell association in A549 cells incubated in medium supplemented with 10 mM unlabeled 3OC12-HSL spiked with 1.8 nM 3H-3OC12-HSL to cells incubated with 1.8 nM 3H-3OC12-HSL alone. The cells were processed as before and cell associated 3H-3OC12-HSL was measured by liquid scintillation counting. As shown in Figure 3b, the two treatments achieved peak levels of 3H-3OC12-HSL association with cells at similar time points. Also, the time-dependent pattern of autoinducer association with cells was very similar. These data illustrate that the uptake pattern is independent of the total amount of 3OC12-HSL present.
3.3 Subcellular localization of 3H-3OC12-HSL
The data presented above indicate that 3OC12-HSL becomes associated with cells, but do not differentiate between simple association with cellular membranes versus accessing the intracellular milieu. Therefore, we next sought to identify those subcellular compartment(s) with which 3OC12-HSL associates in mammalian cells. A549 cells were incubated with 3H-3OC12-HSL for 30 minutes, allowing the autoinducer to reach peak levels of association with the cells, and the cells were harvested and subjected to differential detergent fractionation to isolate specific subcellular compartments. The resulting cytoplasmic, membrane/organelle, nuclear, and cytoskeletal/matrix fractions were analyzed for levels of radioactivity by liquid scintillation counting. When expressed as percentages of the total cell associated radioactivity, 85.11% of 3OC12-HSL was detected in the cytoplasmic fraction, 13.71% associated with the membrane/organelle fraction, 0.84% in the nuclear fraction and 0.34% with the cytoskeletal/matrix fraction. These data show that the autoinducer does not simply associate with cell membranes but enters the cells where it may modulate a wide range of eukaryotic cell signaling pathways.
3.4 Removal of 3OC12-HSL from cells is ATP-dependent
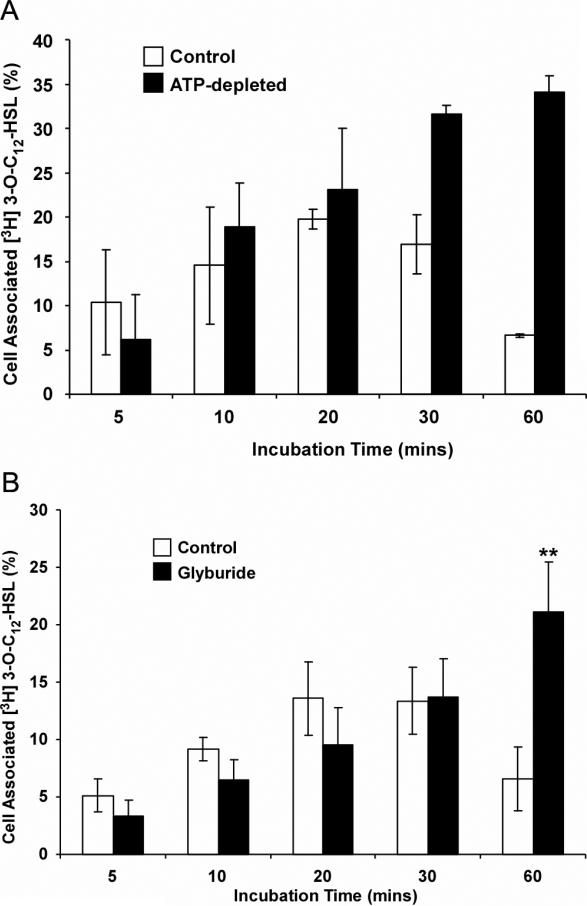
We next sought to characterize the mechanisms underlying the significant decrease in cell-associated levels of 3H-3OC12-HSL shown in Figure 3. As mentioned above, this pattern of removal from the cells does not appear to be related to cell death and thus we tested whether removal of 3H-3OC12-HSL was mediated by an active, energy-dependent mechanism. Intracellular ATP levels were depleted in A549 cells to less than 10% of control levels by incubating the cells in medium containing 5 mM sodium azide and 80 mM 2-deoxy-D-glucose for 4 hours (data not shown). Control and ATP-depleted A549 cells were then incubated with 1.8 nM 3H-3OC12-HSL and, as before, association of 3H-3OC12-HSL with cells was measured over a range of time points (Fig. 4A). As before, cell-associated 3H-3OC12-HSL levels peaked after 20 minutes of incubation and subsequently decreased (Fig. 4A). In contrast, cell-associated 3H-3OC12-HSL levels continued to increase over the full time course of the experiment in ATP-depleted A549 cells. Cell-associated 3H-3OC12-HSL levels also accumulated in ATP-depleted NIH-3T3 cells, indicating that this is not a cell-specific effect (data not shown). Taken together, these data indicate that removal of 3H-3OC12-HSL from cells is an active, ATP-dependent process in mammalian cells but that uptake does not appear to be dramatically affected by ATP depletion.
Figure 4. Removal of radioactive 3OC12-HSL from A549 cells is energy-dependent.
(A) Effect of ATP-depletion on 3H-3OC12-HSL levels associated with cells. A549 cells were ATP-depleted by incubation with 5 mM sodium azide and 80 mM 2-deoxy-D-glucose for 4 hours, while control cells received no treatment. Both groups were then exposed to 1.8nM 3H-3OC12-HSL and cells and cell associated radioactivity was assessed at the time points indicated. (B) 3H-3OC12-HSL continues to accumulate in cells treated with Glyburide. A549 cells were exposed to 250 μM Glyburide or the equivalent volume of DMSO as a vehicle control for one hour before being exposed to 1.8nM 3H-3OC12-HSL. Cell associated radioactivity was assessed at the time points indicated. The data are represented as the means±s.d.; n=3.
3.5 Removal of 3OC12-HSL from cells is inhibited by Glyburide, an inhibitor of ABCA1
The ATP-binding cassette (ABC) transporter superfamily is one of the major groups of proteins that mediate energy-dependent transport of macromolecules across the cell membrane [34]. ABC transporters are responsible for the transport of a wide variety of natural substrates such as lipids, and also participate in the expulsion of xenobiotics, including some anti-cancer drugs [35]. We therefore considered ABC transporters as excellent candidates for mediating the removal of 3OC12-HSL from mammalian cells.
Based on a recent report that the activity of the ABCA1 transporter is stimulated by P. aeruginosa in lung epithelial cells, we tested whether inhibition of ABCA1 activity affected the pattern of 3OC12-HSL cell association in A549 cells. Parallel cultures of A549 cells were either untreated or incubated in the presence of 250 μM Glyburide for one hour, prior to exposure to 3H-3OC12-HSL and cell associated radioactivity was assessed as before over a one hour time course (Fig. 4B). The accumulation of 3H-3OC12-HSL over the first 30 minutes was indistinguishable under both conditions but diverged markedly at the 60 minute time point where it had decreased in the untreated cells but continued to accumulate in the Glyburide treated cells. This result identifies a Glyburide-sensitive process in the removal of 3OC12-HSL from A549 cells.
4. Discussion
AHL autoinducers have a variety of described effects on host cells, including immune modulation and apoptosis. In order to gain additional data on the effects of autoinducers on mammalian cells, we performed microarray analysis of human lung epithelial cells treated with three different bacterial autoinducers or vehicle control. Of the 41,000+ unique human transcripts represented on the microarrays, approximately 4,500 (or 11% of the transciptome) were significantly changed in our experiments, when analyzed using a stringent statistical approach described in the methods. However, only a very small number of mRNAs exhibited significant changes in cells exposed to C4-HSL or AI-2, indicating that these autoinducers alone probably do not have major physiological effects on these types of human cells, at least at the concentration examined. This led us to conclude that the gene expression response to 3OC12-HSL is not a general response to bacterial cell products or to AHL but represents a specific response based on the unique structure of 3OC12-HSL. While the genes altered specifically by 3OC12-HSL included many previously identified by us and others, there were other affected genes that fell into numerous functional gene ontology categories. One of these groups included a number of genes involved in drug transport and metabolism. Thus we decided to further examine the fate of 3OC12-HSL in A549 cells.
We demonstrated that 3OC12-HSL enters mammalian cells, supporting our previous results using synthetic transcriptional regulatory proteins [23]. We also observed that epithelial and fibroblast cell lines accumulated 3OC12-HSL over a period of about 20-30 minutes, after which the autoinducer was expelled from the cells. Intracellular 3OC12-HSL accumulated primarily in the cytosol but was also detected in membrane-associated and nuclear fractions. There were no significant differences in the uptake of 3OC12-HSL into healthy cells versus those depleted for ATP, suggesting that energy is not required for uptake. This is consistent with studies on 3OC12-HSL uptake in dendritic and T cells, although the rate of uptake into epithelial and fibroblast cells was slower than that observed in these other cell types [24]. The preferential localization of 3OC12-HSL to the cytosol was somewhat surprising considering the hydrophobic nature of the autoinducer and suggests that it may complex with cellular proteins within the cytoplasm and other aqueous environments within the cells such as the nucleus. Thus the differences in the rate of accumulation of 3OC12-HSL in epithelial cells and fibroblasts versus dendritic cells and T cells may reflect differences in the concentration of intracellular AHL receptors, which may include NHRs such as PPARγ, in these different cell types.
Our data clearly showed that intracellular 3OC12-HSL levels peaked within about 30 minutes of application, at which time it was actively expelled from the cells in an energy-dependent manner. Mammalian cells possess mechanisms for sensing and expelling toxic, xenobiotic agents, including the activation of members of the ABC transporter superfamily. Multi-drug resistance, for example to antibiotics in bacteria [38] and to anti-neoplastic drugs in mammalian cells [35], can be caused by activation of efflux pumps that belong to this superfamily. We initially investigated two well characterized members of this family, Pgp and CFTR as potential transporters of 3OC12-HSL. Pgp has been associated with the transport of a wide variety of xenobiotic compounds [34]. However, 3OC12-HSL did not affect the activity of purified recombinant Pgp (Bryan and Urbatsch, unpublished observations). CFTR is a chloride ion channel which is altered in individuals with cystic fibrosis and its activity is regulated by phosphorylation in response to increased intracellular cyclic AMP levels [39]. Again, 3OC12-HSL efflux was not affected by treating cells with forskolin, an activator of adenylyl cyclase (Bryan, unpublished observations). However, we did determine that the sulfonylurea drug glyburide blocked 3OC12-HSL efflux, indicating that 3OC12-HSL efflux was likely mediated by a glyburide-sensitive transporter. Glyburide is an anti-diabetic drug that inhibits the activity of ABCA1 [36] as well as the scavenger receptor class B type 1 (SR-B1) protein [37]. Both ABCA1 and SR-B1 are involved in the transport of phospholipids that play key roles in the metabolism of high density lipoproteins. The combination of ATP-dependence and glyburide-sensitivity of 3OC12-HSL efflux led us to propose ABCA1 as the likely transporter in lung epithelial cells.
In summary, the microarray data presented here suggest that the human gene expression response to 3OC12-HSL is not a general response to bacterial cell products or to AHLs but represents a specific response based on the unique structure of 3OC12-HSL. Our data also demonstrate that mammalian cells may be capable of recognizing bacterial autoinducers as potentially deleterious xenobiotic compounds and subsequently activating mechanisms to expel these bacterial products. One caveat of this interpretation is that the autoinducer may be processed at an intracellular location and the processed form may be expelled. It was not possible in these experiments to assess the functional status of expelled autoinducer as the concentrations of labeled autoinducer were below those required for known functions in mammalian cells. Notwithstanding this caveat, this study identifies a series of coordinated cellular responses, consisting of transcriptome modifications and activation of efflux mechanisms, of mammalian cells to 3OC12-HSL and provides further support for this autoinducer as a multifunctional mediator of pathogen-host interactions. It will be interesting to determine whether the efflux process could represent a new therapeutic target for the amelioration of host inflammatory responses during bacterial infections by enhancing clearance of autoinducers from cells adjacent to biofilms or other accumulations of bacteria in infection scenarios.
5. Acknowledgements
This study was supported by funds from the American Lung and Diabetes Associations (KPR), the Robert A. Welch Foundation and NIH (D-1361 and R03DA026960, GL) and the Provost Scholarship of Texas Tech University (AO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parsek MR, Greenberg EP. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc Natl Acad Sci U S A. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazdunski AM, Ventre I, Sturgis JN. Regulatory circuits and communication in Gram-negative bacteria. Nat Rev Microbiol. 2004;2:581–592. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu Rev Cell Dev Biol. 2005:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Rumbaugh KP, Griswold JA, Hamood AN. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2000;2:1721–1731. doi: 10.1016/s1286-4579(00)01327-7. [DOI] [PubMed] [Google Scholar]
- 5.Smith RS, Iglewski BH. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6:56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Song Z, Givskov M, Doring G, Worlitzsch D, Mathee K, Rygaard J, Hoiby N. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology. 2001;147:1105–1113. doi: 10.1099/00221287-147-5-1105. [DOI] [PubMed] [Google Scholar]
- 7.Pearson JP, Feldman M, Iglewski BH, Prince A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun. 2000;68:4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesprit P, Faurisson F, Join-Lambert O, Roudot-Thoraval F, Foglino M, Vissuzaine C, Carbon C. Role of the Quorum-sensing System in Experimental Pneumonia due to Pseudomonas aeruginosa in Rats. Am J Respir Crit Care Med. 2003;167:1478–1482. doi: 10.1164/rccm.200207-736BC. [DOI] [PubMed] [Google Scholar]
- 9.Imamura Y, Yanagihara K, Tomono K, Ohno H, Higashiyama Y, Miyazaki Y, Hirakata Y, Mizuta Y, Kadota J, Tsukamoto K, Kohno S. Role of Pseudomonas aeruginosa quorum-sensing systems in a mouse model of chronic respiratory infection. J Med Microbiol. 2005;54:515–518. doi: 10.1099/jmm.0.46004-0. [DOI] [PubMed] [Google Scholar]
- 10.Mittal R, Sharma S, Chhibber S, Harjai K. Contribution of quorum-sensing systems to virulence of Pseudomonas aeruginosa in an experimental pyelonephritis model. J Microbiol Immunol Infect. 2006;39:302–309. [PubMed] [Google Scholar]
- 11.Shiner EK, Rumbaugh KP, Williams SC. Interkingdom signaling: Deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev. 2005;29:935–947. doi: 10.1016/j.femsre.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Shiner EK, Terentyev D, Bryan A, Sennoune S, Martinez-Zaguilan R, Li G, Gyorke S, Williams SC, Rumbaugh KP. Pseudomonas aeruginosa autoinducer modulates host cell responses through calcium signalling. Cell Microbiol. 2006;8:1601–1610. doi: 10.1111/j.1462-5822.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 13.Smith RS, Fedyk ER, Springer TA, Mukaida N, Iglewski BH, Phipps RP. IL-8 production in human lung fibroblasts and epithelial cells activated by the Pseudomonas autoinducer N-3-oxododecanoyl homoserine lactone is transcriptionally regulated by NF-kappa B and activator protein-2. J Immunol. 2001;167:366–374. doi: 10.4049/jimmunol.167.1.366. [DOI] [PubMed] [Google Scholar]
- 14.Smith RS, Kelly R, Iglewski BH, Phipps RP. The Pseudomonas autoinducer N-(3-oxododecanoyl) homoserine lactone induces cyclooxygenase-2 and prostaglandin E2 production in human lung fibroblasts: implications for inflammation. J Immunol. 2002;169:2636–2642. doi: 10.4049/jimmunol.169.5.2636. [DOI] [PubMed] [Google Scholar]
- 15.Tateda K, Ishii Y, Horikawa M, Matsumoto T, Miyairi S, Pechere JC, Standiford TJ, Ishiguro M, Yamaguchi K. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect Immun. 2003;71:5785–5793. doi: 10.1128/IAI.71.10.5785-5793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, Hooi D, Chhabra SR, Pritchard D, Shaw PE. Bacterial N-acylhomoserine lactone-induced apoptosis in breast carcinoma cells correlated with down-modulation of STAT3. Oncogene. 2004;23:4894–4902. doi: 10.1038/sj.onc.1207612. [DOI] [PubMed] [Google Scholar]
- 17.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol. 2008;190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobi CA, Schiffner F, Henkel M, Waibel M, Stork B, Daubrawa M, Eberl L, Gregor M, Wesselborg S. Effects of bacterial N-acyl homoserine lactones on human Jurkat T lymphocytes-OdDHL induces apoptosis via the mitochondrial pathway. Int J Med Microbiol. 2009;299:509–519. doi: 10.1016/j.ijmm.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Wang L, Ye L, Mao Y, Xie X, Xia C, Chen J, Lu Z, Song J. Influence of Pseudomonas aeruginosa quorum sensing signal molecule N-(3-oxododecanoyl) homoserine lactone on mast cells. Med Microbiol Immunol. 2009;198:113–121. doi: 10.1007/s00430-009-0111-z. [DOI] [PubMed] [Google Scholar]
- 20.Cooley MA, Whittall C, Rolph MS. Pseudomonas signal molecule 3-oxo-C12-homoserine lactone interferes with binding of rosiglitazone to human PPARgamma. Microbes Infect. 2010;12:231–237. doi: 10.1016/j.micinf.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SC, Patterson EK, Carty NL, Griswold JA, Hamood AN, Rumbaugh KP. Pseudomonas aeruginosa Autoinducer Enters and Functions in Mammalian Cells. J Bacteriol. 2004;186:2281–2287. doi: 10.1128/JB.186.8.2281-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritchie AJ, Whittall C, Lazenby JJ, Chhabra SR, Pritchard DI, Cooley MA. The immunomodulatory Pseudomonas aeruginosa signalling molecule N-(3-oxododecanoyl)-L-homoserine lactone enters mammalian cells in an unregulated fashion. Immunol Cell Biol. 2007:596–602. doi: 10.1038/sj.icb.7100090. [DOI] [PubMed] [Google Scholar]
- 25.Chhabra SR, Harty C, Hooi DS, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-L-homoserine lactone as immune modulators. J Med Chem. 2003;46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- 26.Shiner EK, Reddy S, Timmons C, Li G, Williams SC, Rumbaugh KP. Construction of a bacterial autoinducer detection system in mammalian cells. Biol Proced Online. 2004;6:268–276. doi: 10.1251/bpo98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- 28.Wilcoxon F. Individual comparison by ranking methods. Biometrics Bulletin. 1945;1:80–83. [Google Scholar]
- 29.Massey FJ. The Kolmogorov-Smirnov test for goodness of fit. Journal of the American Statistical Association. 1951;46:68–78. [Google Scholar]
- 30.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 31.Ramsby ML, Makowski GS, Khairallah EA. Differential detergent fractionation of isolated hepatocytes: biochemical, immunochemical and two-dimensional gel electrophoresis characterization of cytoskeletal and noncytoskeletal compartments. Electrophoresis. 1994;15:265–277. doi: 10.1002/elps.1150150146. [DOI] [PubMed] [Google Scholar]
- 32.Smith RS, Harris SG, Phipps R, Iglewski B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J Bacteriol. 2002;184:1132–1139. doi: 10.1128/jb.184.4.1132-1139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rumbaugh KP, Hamood AN, Griswold JA. Cytokine induction by the P. aeruginosa quorum sensing system during thermal injury. J Surg Res. 2004;116:137–144. doi: 10.1016/j.jss.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 35.Deeley RG, Westlake C, Cole SP. Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev. 2006;86:849–899. doi: 10.1152/physrev.00035.2005. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Kimura Y, Kioka N, Matsuo M, Ueda K. Purification and ATPase activity of human ABCA1. J Biol Chem. 2006;281:10760–10768. doi: 10.1074/jbc.M513783200. [DOI] [PubMed] [Google Scholar]
- 37.Nieland TJ, Chroni A, Fitzgerald ML, Maliga Z, Zannis VI, Kirchhausen T, Krieger M. Cross-inhibition of SR-BI- and ABCA1-mediated cholesterol transport by the small molecules BLT-4 and glyburide. J Lipid Res. 2004;45:1256–1265. doi: 10.1194/jlr.M300358-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Putman M, Van Veen HW, Degener JE, Konings WN. Antibiotic resistance: era of the multidrug pump. Mol Microbiol. 2000;36:772–773. doi: 10.1046/j.1365-2958.2000.01871.x. [DOI] [PubMed] [Google Scholar]
- 39.Gadsby DC, Vergani P, Csanady L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]