Summary
Anticancer drug 5-azacytidine (aza-C) induces DNA-protein crosslinks (DPCs) between cytosine methyltransferase and DNA as the drug inhibits methylation. We found that mutants defective in the tmRNA translational quality control system are hypersensitive to aza-C. Hypersensitivity requires expression of active methyltransferase, indicating the importance of DPC formation. Furthermore, the tmRNA pathway is activated upon aza-C treatment in cells expressing methyltransferase, resulting in increased levels of SsrA tagged proteins. These results argue that the tmRNA pathway clears stalled ribosome-mRNA complexes generated after transcriptional blockage by aza-C-induced DPCs. In support, an ssrA mutant is also hypersensitive to streptolydigin, which blocks RNA polymerase elongation by a different mechanism. The tmRNA pathway is thought to act only on ribosomes containing a 3’ RNA end near the A site, and the known pathway for releasing RNA 3’ ends from a blocked polymerase involves Mfd helicase. However, an mfd knockout mutant is not hypersensitive to either aza-C-induced DPC formation or streptolydigin, indicating that Mfd is not involved. Transcription termination factor Rho is also likely not involved, because the Rho-specific inhibitor bicyclomycin failed to show synergism with either aza-C or streptolydigin. Based on these findings, we discuss models for how E. coli processes transcription/translation complexes blocked at DPCs.
Keywords: 5-azacytidine, DNA-protein crosslinks, transcription blockage, tmRNA, trans-translation, streptolydigin
Introduction
DNA-protein crosslinks (DPCs) are induced by many chemicals, including formaldehyde, and by radiation. With most of these agents, DPCs form at heterogeneous sites on the DNA with a collection of many different DNA-binding proteins. In part because of these characteristics, the study of DPCs has been difficult. Thus, compared to most forms of DNA damage, relatively little is known about the consequences and processing of DPCs in vivo.
5-azacytidine (aza-C) and 2’-deoxy-5-azacytidine (aza-dC) are used a s leukemia chemotherapeutic agents, particularly for the “pre-leukemia” condition myelodysplastic syndrome (Glover et al., 1987; Silverman et al., 2002; Kaminskas et al., 2005). These and related compounds induce DPCs that are specific both for DNA sequence and the involved protein. Aza-C is a cytidine analog with nitrogen at the C-5 position in the pyrimidine ring and is an inhibitor of DNA-cytosine methyltransferases (MTases). In a normal cytosine methylation reaction, MTase binds covalently to C-6 of cytosine at the enzyme recognition sequence in DNA. This activates transfer of the methyl group from S-adenosylmethionine to the C-5 position of cytosine, and the covalent linkage between the MTase and the DNA is reversed (Santi et al., 1983). However, when aza-C is substituted for cytosine at the enzyme recognition site, methylation is prohibited and the covalent enzyme-DNA bond becomes essentially irreversible (Santi et al., 1984; Friedman, 1985; Gabbara and Bhagwat, 1995). In this study, we have used aza-C as a reagent for a model system to study DPC consequences in vivo.
While aza-C was developed as an MTase inhibitor, its physiological effects are quite complex (Christman, 2002). In eukaryotes, inhibition of MTase leads to changes in gene expression, including reactivation of tumor suppressor genes in some cell lines (Jones, 1985; Baylin et al., 2001; Cameron et al., 1999). In both prokaryotes and eukaryotes, aza-C leads to induction of DNA damage responses due to formation of the DNA- MTase crosslinks, presumably involving some downstream damage such as DNA breaks (Barbe et al., 1986; Karpf et al., 2001). Unless aza-dC is used, aza-C residues also become incorporated into RNA molecules, which may have additional physiological consequences (Christman, 2002). We previously reported one downstream consequence of aza-C treatment: the induced DNA-MTase crosslinks block DNA replication in vivo (Kuo et al., 2007). We also presented evidence that the blocked replication forks led to DNA breaks, which could be the breaks involved in induction of the SOS DNA damage response.
Aza-C-induced DNA-MTase crosslinks have also been shown to block in vitro transcription elongation by E. coli RNA polymerase, but the in vivo consequences are unknown (Som and Friedman, 1994). Transcription elongation is also inhibited by the antibiotic streptolydigin, which binds near the polymerase active site and stabilizes a particular conformation of the enzyme (Tuske et al., 2005). Transcription complexes blocked by certain forms of DNA damage (e.g. pyrimidine dimers) can be disassembled by the Mfd helicase (Selby and Sancar, 1995; Roberts and Park, 2004), but it is not known whether Mfd can operate on RNA polymerase blocked at DPC lesions or by streptolydigin.
To further analyze the in vivo processing and consequences of aza-C-induced DPCs, we undertook a mutational screen for aza-C hypersensitive mutants. As described below, this screen uncovered mutations in genes that are connected to the co-translational quality control system for truncated and miscoding mRNAs (for review, see Karzai et al., 2000; Keiler, 2008; Moore and Sauer, 2007). The cornerstone of this system, the ssrA gene product, is a specialized RNA called tmRNA. When a ribosome reaches the end of a message without a stop codon, tmRNA binds and acts as both tRNA and mRNA. The tmRNA is associated with a protein cofactor, SmpB, which is required for all known activities of tmRNA. When the co-translational quality control system is invoked, tmRNA binds to the A site of the ribosome and the template for translation is shifted from the blocked mRNA to the mRNA portion of tmRNA. This results in addition of an 11-amino acid tag to the prematurely truncated growing peptide, with two important consequences. First, the trapped ribosome is freed by allowing a productive termination event to occur. Second, the 11-amino acid SsrA tag on the unnatural protein is a signal for its proteolysis. Several different proteases, including ClpXP, ClpAP, FtsH (HflB), Lon and Tsp, are involved in proteolysis, with ClpXP being responsible for most of the degradation (Keiler et al., 1996; Gottesman et al., 1998; Herman et al., 1998; Farrell et al., 2005; Choy et al., 2007; Lies and Maurizi, 2008).
While a major function of the tmRNA pathway is to allow completion of translation of mRNAs that lack a stop codon, the system can also be invoked when the ribosome encounters rare codons or internal RNA damage. In the latter cases, mRNA cleavage within the ribosome is thought to be required to allow tmRNA binding to the A site of the ribosome. The tmRNA system also induces degradation of the aberrant mRNAs that result in ribosome stalling, thereby preventing repeated cycles of aberrant translation and providing an mRNA quality control system (Mehta et al., 2006; Richards et al., 2006; Yamamoto et al., 2003).
At the physiological level, the tmRNA pathway is universal in eubacteria and appears to have multiple important functions. The pathway is essential for growth in some bacterial species, and even in species where it is not essential, knockout mutants (ssrA, smpB) often show increased sensitivity to stress, such as carbon starvation or oxidative stress from macrophage attack (Okan et al., 2006). The tmRNA system has also been adopted in aspects of certain gene regulatory circuits, such as in the autoregulation of LacI repressor levels (Abo et al., 2000). SsrA-deficient mutants have been shown to be hypersensitive to kanamycin and streptomycin (Abo et al., 2002), presumably because these drugs cause misreading of the genetic code including read-through of stop codons.
In this study, we show that ssrA and smpB mutants are hypersensitive to aza-C, and that aza-C treatment leads to induction of SsrA-tagged proteins in wild-type cells. We also show that an ssrA mutant is hypersensitive to streptolydigin, which blocks RNA polymerase elongation by a different mechanism. We conclude that the tmRNA pathway plays an important role in clearing stalled ribosomes that are generated after transcriptional blockage. We also found that Mfd helicase does not contribute to survival after these two forms of transcriptional blockage, and can interfere with survival when overproduced. Furthermore, inhibition of transcription termination factor Rho does not appear to increase sensitivity to either aza-C or streptolydigin. These results suggest that some other pathway is involved in clearing transcription elongation complexes that are stalled by aza-C-induced DPCs or by streptolydigin.
Results
Hypersensitivity of ssrA mutant to aza-C
To investigate the consequences and processing pathways of DPCs, we undertook a genetic screen for E. coli mutants that are hypersensitive to aza-C. The presence of a cytosine MTase expression plasmid renders E. coli more sensitive to aza-C, arguing that the DPCs are detrimental to survival (Barbe et al., 1986; Bhagwat and Roberts, 1987; Lal et al., 1988; Juttermann et al., 1994; Ferguson et al., 1997). For the mutagenesis screen, we used a strain that carries an M.EcoRII-expressing plasmid, pR215, to favor the isolation of mutants affected in some pathway relevant to DPC formation or processing. However, overexpression of MTase causes an undesirable consequence in most E. coli strains: increased levels of MTase result in methylation of sites that resemble the MTase recognition site, thereby triggering DNA damage by the McrA and McrBC restriction systems (Bandaru et al., 1996). We constructed a strain, HK21, lacking these restriction systems for the screen, and also incorporated a sulA deletion mutation (to prevent growth inhibition due to induction of the SOS system by aza-C). The presence of the M.EcoRII-expressing plasmid resulted in significantly higher aza-C sensitivity in the HK21 genetic background (Fig. S1).
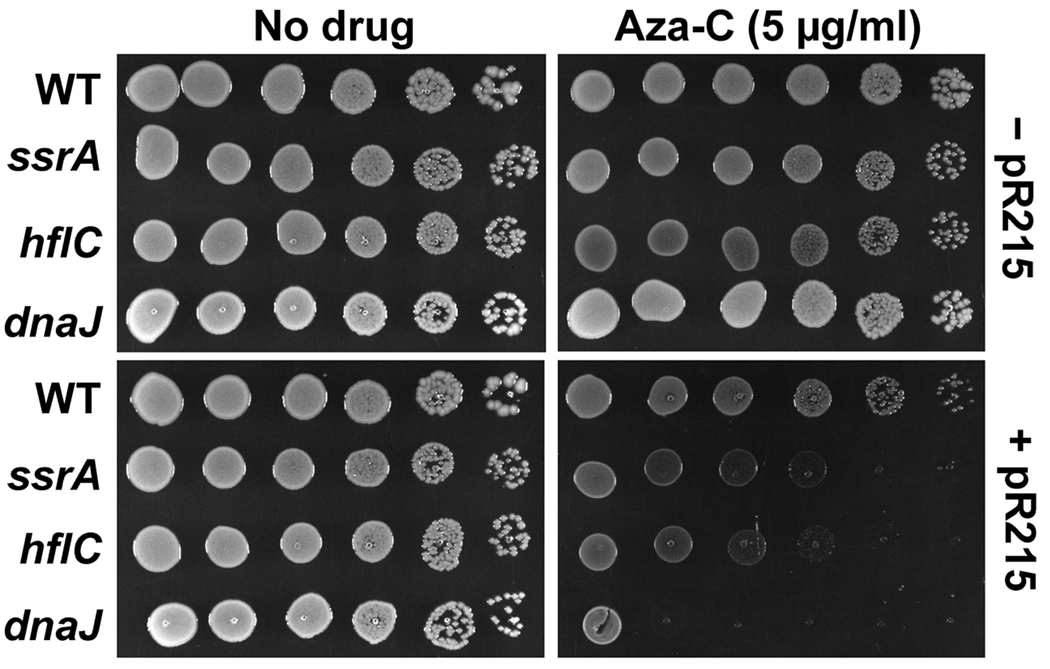
We isolated a collection of transposon insertion mutants that are hypersensitive to aza-C compared to the parental strain. Possible hypersensitive mutants were confirmed by secondary screens involving growth phenotypes on aza-C-containing plates, and the precise location of the transposon insertion in each mutant was determined by genomic DNA sequencing using a transposon-specific primer. We found an ssrA (tmRNA) mutant among the collection of aza-C hypersensitive mutants. Two other genes from the screen (hflC and dnaJ) with possible connections to the tmRNA system will be discussed below, and the complete results of the screen will be presented elsewhere. We confirmed that the transposon insertion in ssrA is causative by moving the insertion into a fresh HK21 background using phage P1-mediated generalized transduction and confirming the hypersensitivity phenotype. Hypersensitivity is evident in two different tests: growth of serial 10-fold dilutions on plates with a fixed concentration of aza-C (Fig. 1, lower panels) and minimal inhibitory concentration (MIC) measurements of growth in the presence of increasing concentrations of aza-C in liquid media (Fig. S1).
Figure 1. Aza-C hypersensitivity of mutants with M.EcoRII expression vector.
Overnight cultures of HK21 (WT) or the indicated HK21 derivatives, with (lower panels) or without (upper panels) M.EcoRII-expressing plasmid pR215, were diluted to 4 × 108 cells/ml. Ten-fold serial dilutions were generated across a microtiter plate and 5 µl of each dilution was spotted onto LB plates with no drug (left panels) or aza-C (5 µg/ml; right panels). Plates were photographed after overnight incubation at 37°C.
Aza-C hypersensitivity depends on M.EcoRII expression but is not caused by alterations in M.EcoRII expression
Because the tmRNA pathway operates on stalled ribosomes, the hypersensitivity of the ssrA mutant could conceivably be dependent on aza-C incorporation into RNA. In this model, aza-C residues in RNA might directly cause miscoding at the ribosome or lead to stalling by some indirect pathway (e.g. chemical modification of the aza-C residue into another form). This RNA-based model is strongly disfavored by the finding that the presence of the M.EcoRII-expression plasmid pR215 is required for the high level of sensitivity of the ssrA mutant (Fig. 1; Fig. S1). Therefore, aza-C hypersensitivity is dependent on expression of M.EcoRII and presumably aza-C-induced DPC formation.
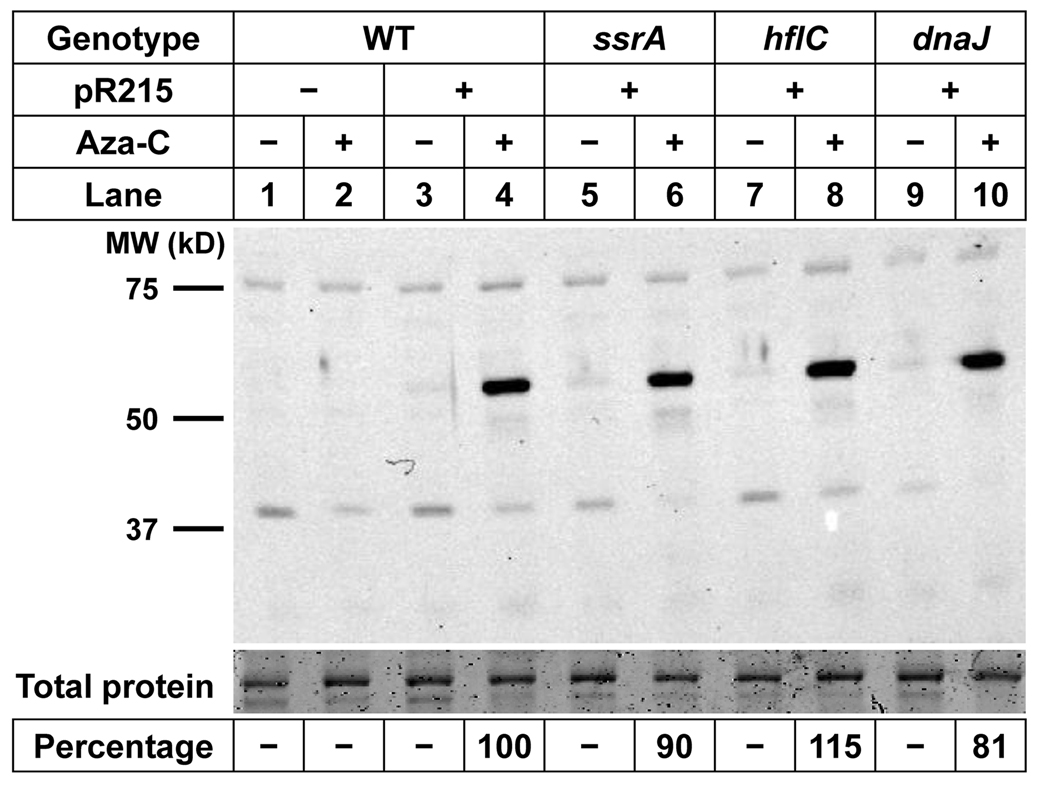
A trivial model for the hypersensitivity of the ssrA mutant to aza-C is that the mutation leads to higher levels of expression of M.EcoRII from the pR215 plasmid, thereby leading indirectly to higher levels of DPC formation. We tested this model by performing Western blots with an anti-M.EcoRII antibody, comparing protein levels in parental wild-type versus mutant cells. The polyclonal M.EcoRII antibody cross-reacts weakly with several bands present even in cells that lack any M.EcoRII-expressing plasmid (Fig. 2, lane 1 & 2). The antibody readily detected M.EcoRII protein of the expected molecular weight (~60 kDa) in cells carrying pR215 (Fig. 2, lane 4). As expected from the studies of Friedman and Som (1993), the M.EcoRII protein levels were much higher in aza-C treated cells (Fig. 2, compare lanes 3 and 4). This occurs because the M.EcoRII gene promoter is auto-regulated, and hence inhibition of the MTase by aza-C residues in DNA increases transcription of the M.EcoRII gene. The most important result of the experiment is that the induced levels of M.EcoRII protein were very similar between the parental strain and the ssrA mutant (Fig. 2, lanes 4 and 6). Therefore, hypersensitivity cannot be attributed to increased production of M.EcoRII.
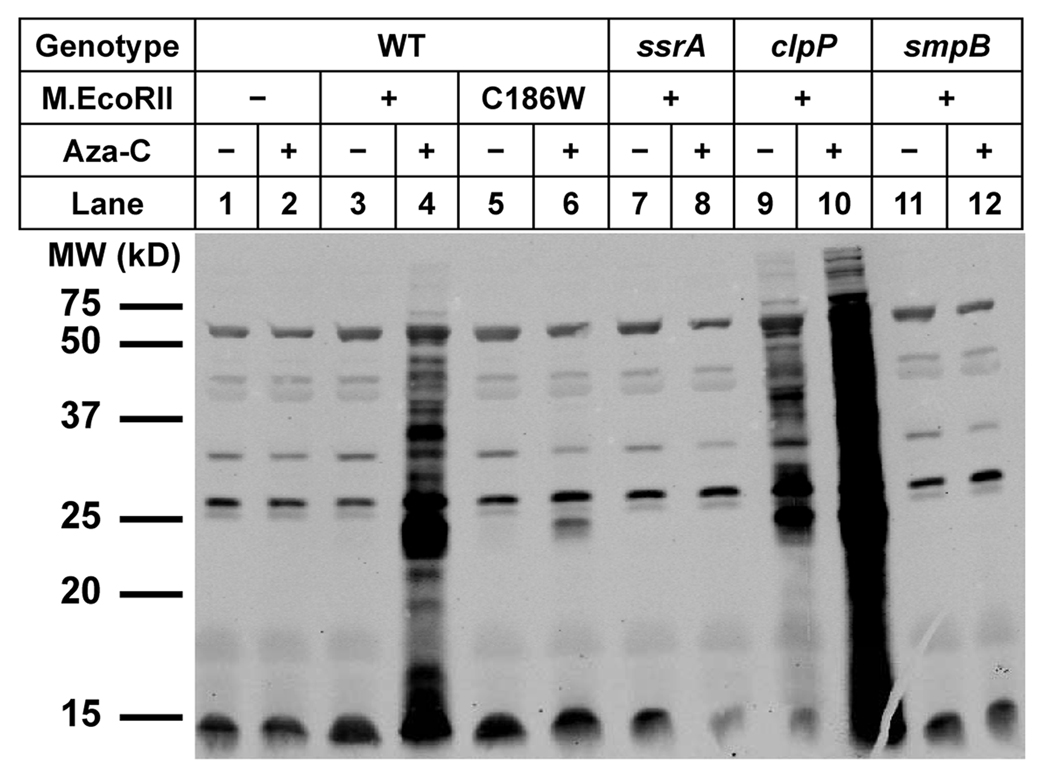
Figure 2. Expression of M.EcoRII in wild-type and mutant cells.
Extracts from HK21 (WT) or the indicated HK21 derivatives, with or without M.EcoRII plasmid pR215, were analyzed by Western blotting with polyclonal antibodies to M.EcoRII. The top portion of the gel was excised and stained with Coomassie blue as a total protein loading control. The Western antibody signals and total protein controls were quantitated using an Infrared Imaging System (see Experimental Procedures). The calculated ratio of M.EcoRII to total protein for the four lanes with significant M.EcoRII is expressed as percentage of wild-type levels at the bottom of the figure.
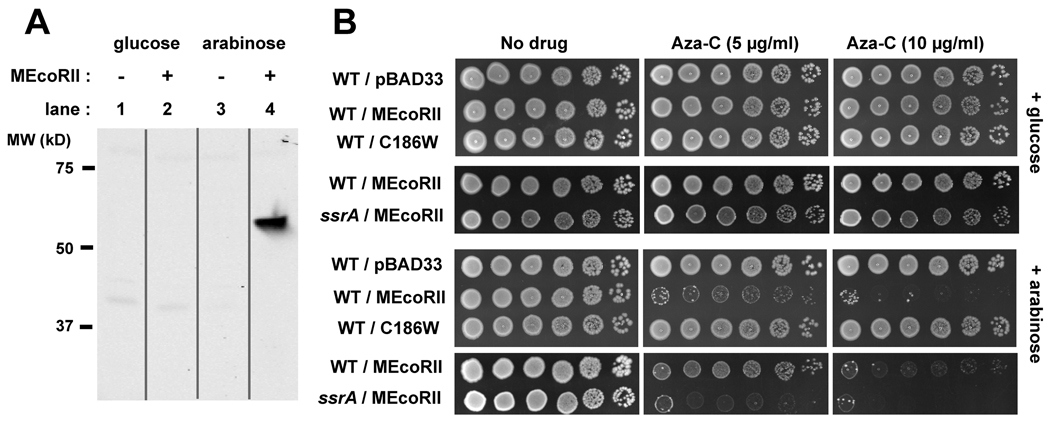
The autoregulatory nature of the M.EcoRII promoter complicates analysis of aza-C sensitivity, and we therefore sought a system to better control M.EcoRII expression. We constructed an arabinose-inducible M.EcoRII expression plasmid (pBAD-MEcoRII) from the pBAD33 vector (Guzman et al., 1995). As expected, expression of M.EcoRII protein from pBAD-MEcoRII was repressed in the presence of glucose (Fig. 3A, lane 2) and activated in the presence of arabinose (Fig. 3A, lane 4). In a corresponding manner, strain HK21 carrying pBAD-MEcoRII was sensitive to aza-C only when grown in the presence of arabinose, but cells with the empty vector pBAD33 were not affected by aza-C (Fig. 3B, lower panels; Fig. S2).
Figure 3. Arabinose-inducible M.EcoRII expression results in aza-C sensitivity.
In panel A, extracts from HK21 cells with or without M.EcoRII plasmid pBAD-MEcoRII were analyzed by Western blotting. The cells were pre-grown with glucose for 1.5 generations, pelleted and washed with fresh LB, and then resuspended in LB containing glucose (0.2%) or arabinose (0.05%). Protein was extracted after 60 minutes of incubation at 37°C. In panel B, overnight cultures of HK21 (WT) or the ssrA derivative of HK21 (ssrA), with the indicated pBAD33-derived plasmid, were diluted to approximately 4 × 108 cells/ml. Ten-fold serial dilutions were generated across a microtiter plate and 5 µl of each dilution was spotted onto LB plates with no drug or the indicated concentration of aza-C. The upper panels are plates that contained glucose (0.2%), while the lower panels are plates that contained arabinose (0.05%). Plates were photographed after overnight incubation at 37°C. The experiment comparing different M.EcoRII plasmids (panels with 3 rows) was done on a different day than the one comparing wild-type and ssrA mutant cells (panels with 2 rows); the latter plates were incubated for several hours longer to allow good growth of the ssrA mutant in the absence of aza-C. This may account for the somewhat weaker inhibition of the wild-type with M.EcoRII in row 9 compared to row 7 (however, we also detect some day-to-day variation in apparent aza-C potency in plates, perhaps due to aza-C instability).
To test whether active M.EcoRII is important for aza-C sensitivity, we introduced the C186W substitution to create plasmid pBAD-C186W; this substitution prevents both DNA binding and methylation by M.EcoRII (Wyszynski et al., 1993). HK21 cells carrying pBAD-C186W were insensitive to aza-C in the presence or absence of arabinose, arguing that active M.EcoRII is required for sensitivity (Fig. 3B).
Using pBAD-MEcoRII as a readily controlled source of M.EcoRII, we confirmed that the ssrA mutant was hypersensitive to aza-C when MTase expression was induced by arabinose but not when MTase expression was repressed by glucose (Fig. 3B; Fig. S2).
Inactivation of SmpB also causes aza-C hypersensitivity
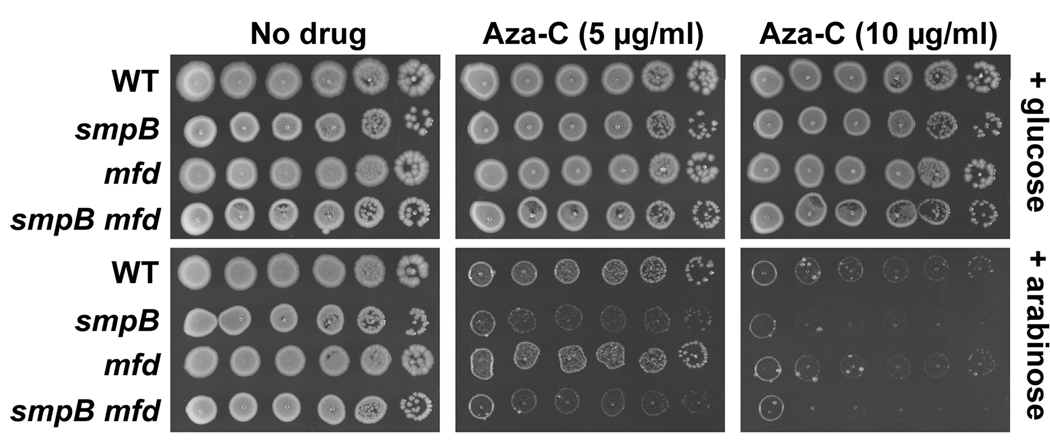
The SmpB protein associates with tmRNA and is required for all known activities of tmRNA (see Introduction). Although smpB knockout mutants were not isolated in the transposon mutagenesis screen, they should be hypersensitive to aza-C if the tmRNA system is important for survival in the presence of the drug. To test this prediction, an smpB knockout mutation from the Keio collection (Baba et al., 2006) was moved into the genetic background used above with plasmid pBAD-MEcoRII, and aza-C sensitivity of the resulting strain was assessed. As expected, the smpB mutant was hypersensitive to aza-C, and sensitivity depended on induction of M.EcoRII by arabinose (Fig. 4; Fig. S2).
Figure 4. Aza-C hypersensitivity of mutants with arabinose-inducible M.EcoRII.
HK21 (WT) or the indicated HK21 derivatives (all containing the arabinose-inducible plasmid pBAD-MEcoRII) were tested for aza-C sensitivity as described in the legend to Fig. 3B.
SsrA-tagged proteins accumulate after aza-C treatment
If the tmRNA system plays a key role in cell survival following aza-C treatment, then aza-C treatment of cells expressing M.EcoRII should lead to protein tagging with the SsrA tag. We therefore performed Western blotting of cellular proteins using antibody against the wild-type SsrA tag (provided by Dr. Tania Baker, MIT). Strikingly, aza-C led to a substantial increase in SsrA-tagged proteins in wild-type cells expressing M.EcoRII (Fig. 5, compare lane 3 to 4). The level of SsrA-tagged proteins was not increased by aza-C when the cells lacked an M.EcoRII-expressing plasmid (Fig. 5, lane 1 & 2), nor was it increased when the M.EcoRII protein contained the active site substitution that prevents DNA binding and methylation (C186W; Wyszynski et al., 1993) (Fig. 5, lane 5 and 6). The induction of SsrA-tagged proteins was a bona fide consequence of the tmRNA system, because it was abolished in both the ssrA and smpB mutants (Fig. 5, lane 7, 8, 11 and 12; the polyclonal antibody cross-reacted with a number of proteins that are not SsrA-tagged, as shown by the presence of weak bands from the ssrA and smpB knockout mutants). The induction of SsrA-tagged proteins in protease-proficient wild-type cells implies that the amount of tagging after aza-C treatment overwhelms the capacity of the downstream proteases.
Figure 5. SsrA tagging is induced by aza-C.
HK21 (WT) or the indicated HK21 derivatives were grown for 2.25 generations in the presence of glucose (0.2%) with or without aza-C, and then washed out of the aza-C-containing media into LB with arabinose (0.05%). The presence of the wild-type or C186W M.EcoRII expressing pBAD-derived plasmid is indicated just below the genotype. Extracts were made after one-hour incubation at 37°C, and equal volumes of cell extract were then analyzed by Western blotting with an SsrA tag polyclonal antibody.
ClpXP is the major protease that acts on SsrA-tagged proteins (see Introduction), and as expected, SsrA-tagged proteins were readily detected in the absence of aza-C in a clpP knockout mutant (Fig. 5, lane 9). Nonetheless, the abundance of SsrA-tagged proteins was greatly increased by aza-C treatment of the clpP mutant (Fig. 5, lane 10). The major conclusion from these studies with the anti-SsrA antibody is that SsrA-tagged proteins are induced upon aza-C treatment as a consequence of DPC formation.
Contributions of HflC and DnaJ to aza-C resistance
In the genetic screen for aza-C hypersensitivity mentioned above, we also isolated insertions in the hflC and dnaJ genes (Fig. 1; Fig. S1). The products of these two genes can play roles connected to the tmRNA pathway. The HflC gene product modulates the activity of the FtsH (HflB) protease, a highly conserved ATP-dependent protease that acts on certain SsrA-tagged polypeptides (for review, see Ito and Akiyama, 2005). Meanwhile, DnaJ is a chaperone that often acts with DnaK, and these two proteins can facilitate proteolysis of at least some substrates by FtsH (Straus et al., 1990; Yura et al., 2000); dnaJ also shows genetic interactions with ssrA (Munavar et al., 2005). While the aza-C hypersensitivity of hflC and dnaJ mutants could reflect some defect in proteolysis of SsrA-tagged proteins, these two gene products also have activities outside the tmRNA pathway.
We confirmed that the hflC and dnaJ insertions cause hypersensitivity by the P1 transduction test described above. As with the ssrA mutants, hypersensitivity of hflC and dnaJ mutants required M.EcoRII-expressing plasmid pR215 in the first genetic background (Fig. 1; Fig. S1) or arabinose treatment with the pBAD-MEcoRII plasmid (Fig. S2; Fig. S3). These results argue that M.EcoRII, and presumably aza-C-induced DPCs, are involved in the hypersensitive phenotype. Furthermore, the level of expression of M.EcoRII is very similar in the wild-type and hflC or dnaJ mutant (Fig. 2), arguing that hypersensitivity is not caused by relative overexpression of M.EcoRII.
As mentioned above, the aza-C hypersensitivities of dnaJ and hflC mutants may or may not relate to an alteration of the tmRNA pathway. If hflC and dnaJ mutants are hypersensitive to aza-C due solely to a defect in the tmRNA pathway, then the double mutants smpB hflC and smpB dnaJ should be no more sensitive than the single smpB mutant. In the presence of arabinose to induce M.EcoRII expression, each double mutant appeared more sensitive to aza-C than any of the single mutants (Fig. S3, lower panels, 5 µg/ml). In addition, the double mutants uniquely showed sensitivity to aza-C even in the presence of glucose, which should repress M.EcoRII expression (Fig. S3, upper panels). These results argue that although DnaJ and HflC might possibly contribute to alleviation of aza-C toxicity through the tmRNA pathway, their principal role must lie outside the pathway.
Proteolysis of SsrA-tagged proteins is not the critical function of the tmRNA pathway
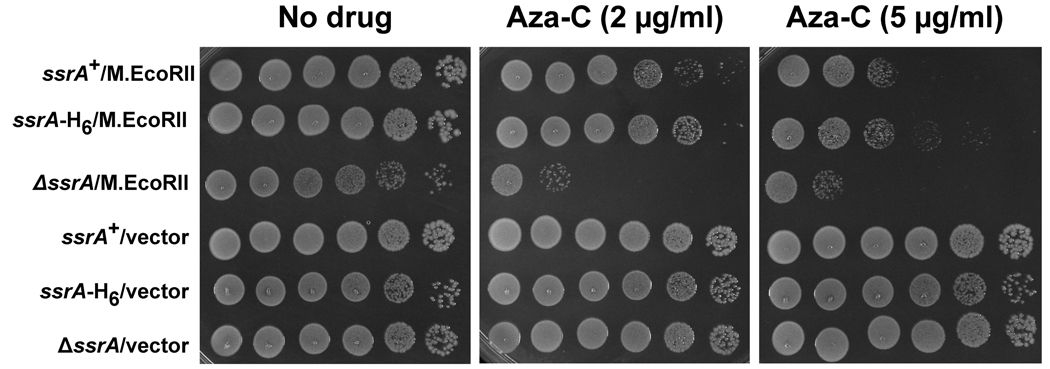
To test more directly the possible importance of proteolysis of SsrA-tagged proteins in sensitivity to aza-C-induced DPCs, we used the mutant ssrA-H6 gene, which encodes a tmRNA with a hexahistidine stretch of codons in place of the coding sequence for a functional proteolysis signal (Roche et al., 2001). This mutant tmRNA is functional for dissociating blocked ribosome complexes but causes a gross deficiency in downstream proteolysis, thus separating the two major functions of the tmRNA pathway. We introduced a p15A-derived plasmid with the ssrA-H6 mutant, wild-type ssrA, or empty vector into cells with an ssrA deletion. Due to plasmid compatibility issues, we used an M.EcoRII expression plasmid, pR234, in which the methyltransferase expression is under control of a Ptac promoter (Bandaru et al., 1996). Expression from this plasmid is leaky in the absence of IPTG, and so we also generated a set of strains in which the M.EcoRII coding sequence had been deleted from the expression plasmid to use as negative controls.
As expected, the ssrA deletion cells expressing M.EcoRII but no plasmid-borne tmRNA showed strong sensitivity to aza-C, while the same cells without M.EcoRII expression were resistant (Figure 6). Also as expected, the plasmid expressing wild-type tmRNA complemented the aza-C hypersensitivity of the ssrA knockout strain (Figure 6). The SsrA-H6 mutant tmRNA likewise complemented the aza-C hypersensitivity, showing even slightly better complementation than the wild-type tmRNA for unknown reasons (Figure 6). We conclude that the important function of the tmRNA system in protecting cells from aza-C-induced DPCs relates to rescue of stalled ribosomes rather than proteolysis of the SsrA-tagged proteins that accumulate.
Figure 6. Aza-C hypersensitivity is relieved by ssrA-H6 mutant.
Overnight cultures of strain HK22 ssrA::kan derivatives carrying two plasmids were diluted to 4 × 108 cells/ml. The identities of the plasmids are indicated on the left: ssrA+ (pKW11); ssrA-H6 (pKW24); ΔssrA (pKW1); M.EcoRII (pR234); vector (pRK1). Ten-fold serial dilutions were generated across a microtiter plate and 5 µl of each dilution was spotted onto LB plates with no aza-C (left panels), aza-C at 2 µg/ml (middle panel), or aza-C at 5 µg/ml (right panel). Note that the level of sensitivity with this IPTG-inducible M.EcoRII expression plasmid appears somewhat higher than for the other two expression plasmids in previous experiments. Plates were photographed after overnight incubation at 37°C.
Response to DPC formation parallels response to the RNA polymerase elongation inhibitor streptolydigin
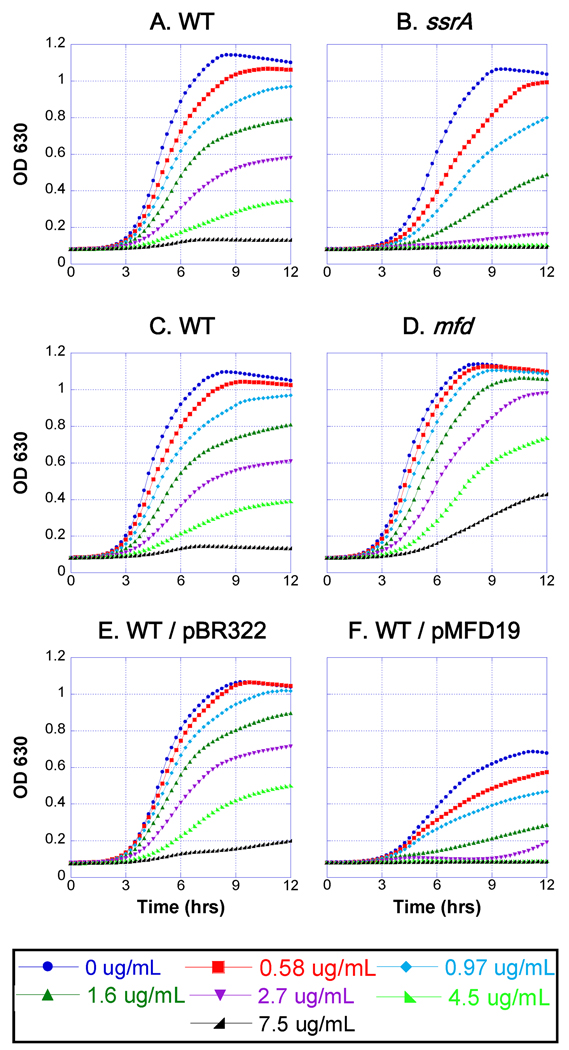
We propose below that aza-C-induced DPCs block transcription elongation, and since transcription and translation are coupled in bacteria, the translating ribosomes stall and back up along the blocked transcript. The antibiotic streptolydigin inhibits bacterial RNA polymerase at the elongation step, leading to frozen ternary complexes in vitro (Siddhiko et al., 1969; McClure, 1980; Cassani et al., 1971). We therefore asked whether the tmRNA pathway also protects against inhibition by streptolydigin. We used a tolC deletion strain (EW1B), which is deficient in a multidrug efflux system and consequently susceptible to streptolydigin (also see Tuske et al., 2005). Because of the limited availability of streptolydigin, we measured growth curves at varying streptolydigin concentrations in the wells of microtiter plates. We found that an ssrA mutation does indeed cause streptolydigin hypersensitivity, which was obvious at multiple drug concentrations (Fig. 7A and 7B). Therefore, the tmRNA system appears to be important for rescue from two different mechanisms of transcriptional blockage, supporting the generality of this model.
Figure 7. Sensitivity of ssrA and mfd strains to streptolydigin.
Cultures contained EW1b (WT; panels A, C, E and F), EW1b ssrA mutant (ssrA; panel B), or EW1b mfd mutant (mfd; panel D) with the indicated plasmid (panels E and F) or no plasmid (panels A–D). Cells were innoculated (starting titer of approximately 2 × 106 cells/ml) in wells that contained increasing concentrations of streptolydigin (see key at bottom), and grown at 37°C in a microplate reader with continuous shaking for 12 hours. Cell turbidity (OD630) was measured every 15 minutes. Plasmids pMFD19 and pBR322 confer ampicillin resistance, and these cultures also contained ampicillin at 50 µg/ml.
Mfd protein and the blocked RNA polymerase
The above results indicate that the tmRNA pathway is required to release stalled ribosomes from mRNA molecules after transcriptional blockage. However, the stalled RNA polymerase complex also presumably needs to be released from its DNA template after blockage of elongation (perhaps even before tmRNA action; see Discussion).
A well-studied mechanism for releasing RNA polymerase blocked by DNA damage (e.g. UV-induced pyrimidine dimers) involves the Mfd helicase, which also couples transcriptional blockage to excision repair (Selby and Sancar, 1993; 1994). We therefore asked whether mutational inactivation of mfd causes hypersensitivity to aza-C in cells expressing M.EcoRII. Strikingly, the mfd single mutant was no more sensitive than the wild-type control, and an mfd smpB double mutant was about as sensitive as an smpB mutant (Fig. 4).
We also found that the mfd mutant is no more sensitive to streptolydigin than the wild-type control (Fig. 7C and 7D). Indeed, the knockout mutant reproducibly showed modest levels of resistance to the drug, suggested that Mfd protein might be in competition with an alternative polymerase release mechanism. Consistent with this proposal, EW1B cells carrying an Mfd expression plasmid were significantly more sensitive to streptolydigin than control EW1B cells carrying an empty vector control (Fig. 7E and 7F). These results suggest the existence of an alternative mechanism to release RNA polymerase stalled by DPC lesions or by streptolydigin (see Discussion).
Rho does not appear to be the alternate polymerase release factor
Termination factor Rho is involved in terminating transcription at many natural terminators, and also induces termination when translation is blocked due to an upstream nonsense mutation (transcriptional polarity) (Roberts et al., 2008). Furthermore, Rho protein induces termination and release of RNA polymerase blocked in vitro by a tightly bound protein (Pavco and Steege, 1990). These results suggest Rho as a reasonable candidate for the alternative polymerase release function.
In order to test the possible involvement of Rho, we used the Rho-specific inhibitor bicyclomycin (Zwiefka et al., 1993). If Rho is involved in releasing RNA polymerase blocked by aza-C-induced DPCs or by streptolydigin, we predicted that bicyclomycin would be synergistic with aza-C and/or streptolydigin for growth inhibition. Starting with aza-C-induced DPCs, we used cells that express M.EcoRII from the pBAD-MEcoRII plasmid, and prepared microtiter plates with a double drug (aza-C and bicyclomycin) serial dilution in checkerboard fashion, thereby testing numerous combinations of drug concentrations for growth inhibition.
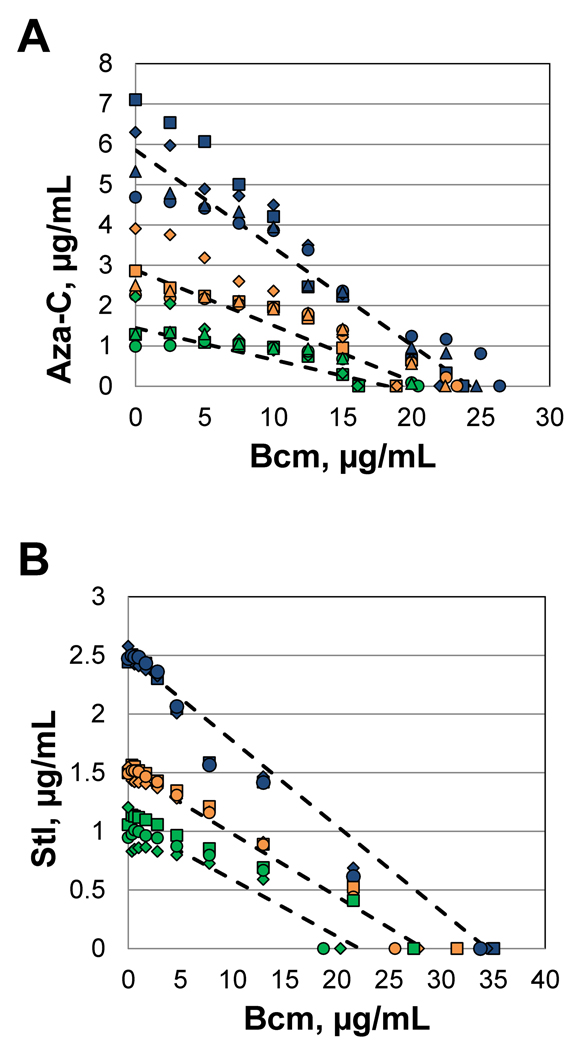
To assess whether or not the drugs act synergistically, we processed the data in two different manners. First, we looked for synergy using a graphical representation in which the two drug concentrations constitute the X and Y axes, and the amounts required to inhibit growth to 95%, 75% or 50% (isoboles) are plotted in different colors. Synergistic drug interactions are revealed by a concave shape to the isobolic curve, antagonistic interactions by a convex shape, and lack of drug interaction by a relatively straight line (Berenbaum, 1978). The data from 4 separate experiments are all plotted in Figure 8A, with the theoretical lines for no drug interaction shown as dashed lines (connecting the two experimentally determined values for each drug alone; also see Supplemental Table S1 and Fig. S4 for more detail). The multiple data points fall quite near the theoretical line for no drug interaction, with no indication of a synergistic (concave line) relationship and a slight but unconvincing hint of an antagonistic (convex line) relationship.
Figure 8. Isobolic test for synergy with Rho inhibitor bicyclomycin.
Growth curves were measured in each well of a 96-well microtiter plate, with varying concentrations of bicyclomycin (right to left) and either aza-C or streptolydigin (top to bottom). The strain for the bicyclomycin/aza-C experiment was HK22 pBAD-MEcoRII, while the strain for the bicyclomycin/streptolydigin experiment was EW1B. A detailed description of the data analysis and processing are presented in the Supplemental Material. Briefly, at each concentration of bicyclomycin (Bcm), the concentration of aza-C (panel A) or streptolydigin (Stl; panel B) necessary to inhibit growth by 95% (blue), 75% (gold) or 50% (green) was estimated. In addition, the concentration of bicyclomycin necessary for those levels of growth inhibition (in the absence of the second drug) was estimated from the bicyclomycin inhibition curve. The data from each of 4 (panel A) or 3 (panel B) repetitions (on different days) were plotted with different symbols (squares, diamonds, circles and triangles). The dashed lines connect the average determined MIC value of each drug alone.
The second method commonly used to assess synergy, supported by the American Society for Microbiology, is the so-called fractional inhibitory concentration (FIC) index (Botelho, 2000; also see Instructions to Authors for the ASM Journal Antimicrobial Agents and Chemotherapy). For each level of drug A, the FIC is calculated for the first concentration of drug B that gave the indicated level of inhibition (95%, 75% or 50%), with FIC = [(MIC of drug A tested in combination) ÷ (MIC of drug A tested alone)] + [(MIC of drug B tested in combination) ÷ (MIC of drug B tested alone)]. The American Society for Microbiology recommends that synergy is supported by FIC index values less than 0.5, while antagonism is supported by FIC index values greater than 4 (an FIC of 0.5 could reflect a situation where ¼ the concentration of each drug is required in combination to give the same growth inhibition as each drug alone at 1× concentration; the theoretical value for no interaction whatsoever is 1.0). In the same tests shown graphically in Figure 8A, we found the following FIC index values: 95% inhibition, 0.95 (+/− 0.11); 75% inhibition, 1.01 (+/− 0.16); 50% inhibition, 0.84 (+/− 0.06) (see Table S2). These FIC values clearly do not support a synergistic (or antagonistic) relationship between aza-C and bicyclomycin.
The double drug titrations with streptolydigin and bicyclomycin showed very similar results. The data points fit a straight isobolic line very well, with no indication of synergy (convex line) (Figure 8B), and the FIC index values were as follows: 95% inhibition, 0.86 (+/− 0.13); 75% inhibition, 0.94 (+/− 0.03); 50% inhibition, 0.1.00 (+/− 0.44) (see Table S3). With the caveat that multiple drug experiments need to be interpreted with caution, these results argue against an involvement of termination factor Rho in releasing RNA polymerase blocked by DPCs or by streptolydigin.
Discussion
We have shown that ssrA and smpB mutants, defective in the tmRNA pathway, are hypersensitive to aza-C in cells expressing M.EcoRII. We propose that the known crosslinking between M.EcoRII and aza-C-containing DNA leads to coupled blockage of transcription and translation, and that the tmRNA system plays important role(s) in relieving the blockage. One alternate model that might have explained the hypersensitivity involves aza-C incorporation into RNA, rendering the tmRNA system important for survival due to RNA damage. This model is refuted by the finding that the hypersensitivity of the mutants depends on M.EcoRII expression (Fig. 1, 3 and 4; Fig. S1, S2 and S3). Another alternate model is that the tmRNA system leads to degradation of M.EcoRII in wild-type cells, and therefore tmRNA-defective cells have increased levels of aza-C-induced DPCs. Western blot analysis showed similar levels of M.EcoRII induction in the presence of aza-C in the wild-type and in an ssrA mutant, providing strong evidence against this model (Fig. 2).
Our data instead support a chain-reaction model in which the coupled transcription-translation machineries of E. coli are blocked by a DPC, and the tmRNA system aids in survival by helping to clean up the resulting pile-up. We showed that aza-C causes a substantial increase in SsrA-tagged proteins in the wild-type but not in ssrA or smpB mutants (Fig. 5). SsrA-tagged proteins were not induced in cells lacking an M.EcoRII-expressing plasmid or in cells in which the EcoRII contained an active site substitution that prevents both DNA binding and methylation (C186W). Induction of SsrA-tagged proteins in protease-proficient cells implies that the amount of tagged proteins induced by aza-C overwhelms the capacity of the proteases that normally degrade SsrA-tagged proteins. SsrA-tagged proteins were also dramatically induced in protease-deficient (clpP mutant) cells. These results argue strongly that SsrA tagging is induced as a consequence of DPC formation. By using a separation-of-function mutation in ssrA, we also showed that protein degradation mediated by the SsrA tag is not important in sensitivity to aza-C-induced DPCs. This result argues that release of the blocked ribosome-mRNA complexes is the important function of the tmRNA system in sensitivity to DPCs. A similar conclusion has been reached regarding several other phenotypes of ssrA mutants (for review, see Keiler, 2008).
We also verified a second prediction of the chain-reaction model, that the tmRNA pathway should be important for surviving other transcriptional blockages. We found that the ssrA knockout mutant is hypersensitive to streptolydigin (Fig. 7), an RNA polymerase inhibitor that causes blockage of RNA polymerase elongation (Cassani et al., 1971; McClure, 1980; Siddhiko et al., 1969). A previous study (Luidalepp et al., 2005) showed that an ssrA knockout mutant is not hypersensitive to rifampicin, which blocks RNA polymerase initiation. Therefore, the role of the tmRNA system in survival with RNA polymerase inhibitors specifically involves the inhibition of elongation.
The tmRNA-SmpB complex binds to stalled ribosomes that contain a 3’ mRNA end at or very near a vacant A site (Ivanova et al., 2004; Moore and Sauer, 2007). However, when ribosomes stall at internal locations on a message, mRNA cleavage can be induced by a ribosome-associated nuclease activity to generate a new 3’ end in this location and thereby allow tmRNA binding to the A site (Hayes and Sauer, 2003; Sunohara et al., 2004a and 2004b; Ivanova et al., 2004; Li et al., 2006). Thus, one model for processing of blocked transcription/translation complexes involves ribosome-associated mRNA cleavage, which would liberate a 5’ (upstream) fragment of the mRNA bound to the stalled ribosome for tmRNA action.
Two distinct modes of A site cleavage have been analyzed. Certain toxin-antitoxin systems include a toxin that functions as a so-called RNA interferase, which can either directly or indirectly activate mRNA endoribonuclease cleavage near the A site (for reviews, see Yamaguchi and Inouye, 2009 and Dreyfus, 2009). For example, RelE protein induces mRNA cleavage at the A site, and has been directly shown to act upstream of the tmRNA system in E. coli in certain situations (Christensen and Gerdes, 2003; also see Pedersen et al., 2003). In this model, endoribonuclease cleavage liberates a 5’ fragment of the mRNA bound to the ribosome for tmRNA action, and a 3’ mRNA fragment that could still be bound to the blocked RNA polymerase. Thus, some other system would seem necessary to resolve the blocked RNA polymerase bound to the 3’ mRNA fragment end.
A second mode of ribosome-associated A site cleavage (RelE-independent) requires the 3’ to 5’ exonuclease RNase II, which digests the mRNA from the 3’ end back to the vicinity of the ribosome, thereby activating an unknown endoribonuclease for the appropriate mRNA cleavage event (Garcia-Sanchez et al., 2009). In the case of transcription/translation complexes blocked at a DPC (or by streptolydigin), the mRNA 3’ end would not be available for RNase II unless the blocked transcription complex is first disassembled by some termination mechanism. Furthermore, once this 3’ end is released from RNA polymerase, the bound ribosome(s) should be able to continue translation up to the unnatural 3’ end, which should suffice for licensing the tmRNA system. Therefore, it seems unlikely that RNase II and RelA-independent A site cleavage would be important in processing of blocked transcription/translation complexes.
It is conceivable that processing of the stalled ribosomes and the stalled RNA polymerase are somehow coupled, for example by a larger macromolecular system that includes the tmRNA complex. Intriguingly, Proshkin et al. (2010) have recently provided evidence that translating ribosomes closely follow elongating RNA polymerase, and that the processes of transcription and translation are much more tightly coupled than previously appreciated. If these two processes are directly coupled, a concerted mechanism to deal with simultaneous blockage of RNA polymerase and translating ribosomes would make great sense.
In any case, the nature of the termination event that releases the blocked RNA polymerase is of interest. Stalled RNA polymerase elongation complexes have been shown to be very stable and require some special termination mechanism for release. In vitro studies showed that E. coli RNA polymerase is blocked by covalent or tightly bound proteins, namely aza-C-induced DNA-MTase crosslinks and tightly bound (non-cleaving mutant) EcoRI endonuclease, respectively (Som and Friedman, 1994; Pavco and Steege, 1990). In the latter study, the blocked complexes were shown to be very stable and could be reactivated to resume transcription by removal of the tightly bound protein. Similarly, biochemical approaches showed that the elongation inhibitor streptolydigin freezes the transcription complex on the DNA template with the RNA product still bound. However, streptolydigin apparently causes rapid, complete and irreversible loss of transcription complexes in vivo (Von Meyenburg et al., 1978), suggesting that some system(s) exists within the cell to dislodge the frozen RNA polymerase complexes, as we are proposing here for both streptolydigin and DPCs.
A well-studied mechanism for releasing RNA polymerase complexes blocked by certain obstructions in the template DNA involves the Mfd helicase (Selby and Sancar, 1993; Chambers et al., 2003). However, an mfd knockout mutant was not hypersensitive to either agent (Fig. 4 and 7), suggesting that Mfd is not involved in releasing transcription complexes blocked by DPCs or by streptolydigin. Furthermore, overexpression of wild-type Mfd protein caused hypersensitivity to streptolydigin; one possible explanation is that Mfd competes with some other factor for access to the blocked RNA polymerase (overexpression of Mfd did not detectably affect aza-C sensitivity; data not shown). In contrast to the apparent lack of involvement of Mfd in our studies, in vitro experiments showed that Mfd can release transcription complexes blocked by a tightly (but not covalently) bound protein on the template (Selby and Sancar, 1995).
We also tested for the possible involvement of transcription termination factor Rho by asking whether the Rho inhibitor bicyclomycin acts in a synergistic fashion with either aza-C or streptolydigin. We found no evidence for any drug interaction, arguing against Rho involvement. Interestingly, Rho induces termination and release of RNA polymerase blocked in vitro by a tightly bound protein (Pavco and Steege, 1990). In that case, however, the nascent RNA was not bound by ribosomes, which are known to inhibit Rho action (see Roberts et al., 2008). In summary, neither Mfd nor Rho appears to play an important role in surviving transcriptional blockage by aza-C-induced DPCs or by streptolydigin. Further experiments are clearly needed to clarify the role of transcription termination factors in response to these transcriptional blockage events and to more directly measure the fate of the blocked RNA polymerase.
The aza-C hypersensitivity of hflC and dnaJ mutants might be related in part to the tmRNA system, but is not simply due to dysfunction of this system. We found that smpB dnaJ and smpB hflC double mutants were each more sensitive to aza-C than any of the three single mutants (smpB, dnaJ or hflC) (Fig. S3). This result implies that the tmRNA system still provides some protection from aza-C in the dnaJ and hflC mutants; otherwise the double mutants would be no more sensitive than the single dnaJ or hflC mutants. In addition, DnaJ and HflC must provide some protection from aza-C outside of their possible role in the tmRNA system; otherwise the double mutants would be no more sensitive than the single smpB mutant. One very interesting possibility is that one or both proteins are involved in processing the covalently linked M.EcoRII in the DPC as part of a repair pathway.
Our results highlight an important role of the tmRNA system in survival after transcriptional blockages, reminiscent of a demonstrated role of the tmRNA pathway in the regulation of the lac operon (Abo et al., 2000). In that case, binding of the LacI repressor to its recognition sites in the lacI gene leads to generation of truncated transcripts and SsrA tagging of the resulting truncated LacI protein fragments, and the tmRNA system appears to thereby modulate the kinetics of lac operon induction (see Abo et al., 2000). To our knowledge, the mechanism of release of the RNA from these LacI-blocked transcription complexes has not been approached, but could be the same as that used in release from DPC-blocked transcription complexes.
Recovery from transcriptional blockage could contribute to some of the diverse phenotypes of tmRNA-defective mutants. For example, tmRNA-defective mutants are hypersensitive to various stress conditions including stationary phase, heat shock and oxidative stress (see Keiler, 2008). In particular, oxidative stress presumably causes DNA lesions that block transcription, perhaps leading to an involvement of the tmRNA system. Hypersensitivity to oxidative stress correlates with an important role of the tmRNA system in bacterial pathogenesis in several species, presumably due in part due to a role of the tmRNA system in surviving oxidative attack from macrophages (Okan et al., 2006; Baumler et al., 1994; Julio et al., 2000). In addition, the tmRNA system is essential for growth in Neisseria gonorrhoeae, Shigella flexneri, Haemophilus influenza and Mycoplasma genitalium, and also for development of certain bacteriophages (see Keiler, 2008), and recovery from transcriptional blockage could be involved in one or more of these essential roles.
In summary, blockage of the concerted processes of transcription and translation leads to a requirement for the tmRNA system, presumably in conjunction with some transcription complex release mechanism. These pathways appear to play a key role in survival after transcriptional blockage by elongation inhibitors and covalently bound proteins, and could also contribute to survival after agents that cause other forms of template DNA damage and/or damage to the transcription machinery.
Experimental procedures
Materials
Aza-C was obtained from Sigma-Aldrich; nitrocellulose membrane (Protran® BA 85) was from Whatman®; Polyclonal M.EcoRII antibody was produced by Proteintech Group, Inc; Luria broth (LB) contained Bacto tryptone (10 g/l), yeast extract (5 g/l), and sodium chloride (10 g/l) and was used for all bacterial growth (with appropriate antibiotics for plasmid selection and the indicated additions); streptolydigin was the generous gift of Konstantin Severinov (Waksman Institute of Microbiology); and bicyclomycin was the generous gift of Max Gottesman and Robert Washburn (Columbia University Medical Center).
Plasmids
Plasmid pR215 is a pACYC184-derived plasmid containing the M.EcoRII gene controlled by its natural promoter, along with a tetracycline resistance gene (Bhagwat et al., 1990). Plasmid pR234 is a pKK223-3 derived plasmid with the M.EcoRII gene controlled by a Ptac promoter (Bandaru et al., 1996; Brosius and Holy, 1984). Plasmid pRK1, a vector control for pR234, was constructed by cleaving pR234 with BamH1 and religating, which removes the entire M.EcoRII coding sequence. We constructed another M.EcoRII expression plasmid, pBAD-MEcoRII, by inserting the M.EcoRII coding sequence between KpnI and SphI sites in the multiple cloning region of pBAD33, downstream from the araBAD promoter (Guzman et al., 1995). These plasmids are pACYC184-derived and contain the chloramphenicol resistance gene. Expression of M.EcoRII protein from pBAD-MEcoRII can be activated by addition of arabinose (0.05%) and is suppressed by growth in the presence of glucose (0.2%). A mutant form of M.EcoRII was created in the pBAD-MEcoRII plasmid by mutating the cysteine codon to tryptophan at position 186, using the Stratagene QuikChange Mutagenesis Kit. Plasmids carrying the ssrA+ gene (pKW11), ssrA-H6 (pKW24), or a ΔssrA control (pKW1) were obtained from Sean Moore (University of Central Florida) (Roche and Sauer, 2001).
E. coli strains
E. coli strain ER1793 [F−, fhuA2, Δ(lacZ)r1, glnV44, e14− (McrA−), trp-31, his-1, rpsL104, xyl-7, mtl-2, metB1, Δ(mcrC-mrr)114::IS10] was obtained from New England Biolabs. We introduced a sulA deletion with a kanamycin-resistance cassette from the Keio collection into this background by phage P1-mediated transduction. The kanamycin-resistance cassette was then removed using FLP recombinase, as described in Baba et al. (2006), to generate strain HK22. A dinD::lacZ fusion that contains an ampicillin-resistance gene was also moved into this background by transduction, generating strain HK21 (also contains the sulA deletion). HK21 was then transformed with M.EcoRII-expressing plasmid pR215 and this strain was used for the transposon mutagenesis screen. The same strain was also used to transform the arabinose-inducible M.EcoRII plasmid pBAD-MEcoRII for better control of M.EcoRII expression.
The transposon mutagenesis screen will be described in more detail elsewhere. Briefly, transposons were randomly inserted into the genome of HK21 pR215 using the EZ-TN™ <KAN-2> Tnp Transposome™ Kit from Epicentre (Madison) and selecting for kanamycin resistance. Repeated screens verified aza-C hypersensitive mutants, and the locations of the insertions were determined by genomic DNA sequencing using a transposon-specific primer. Transposon mutants were moved by phage P1-mediated transduction, selecting for the kanamycin-resistance gene.
EW1b [F−, lacY1 or lacZ4, tsx-64, glnV44(AS), gal-6, LAM−, hisG1(Fs), ΔtolC5, argG6, rpsL (allele 8, 104 or 17), malT1(LamR)] was obtained from the E. coli Genetic Stock Center (Yale University). EW1B derivatives were constructed by phage P1-mediated transduction.
Spot tests for aza-C sensitivity
Overnight cell cultures in LB (plus appropriate antibiotics) were diluted to roughly 4 × 108 cell/ml. Ten-fold serial dilutions were then generated across a microtiter plate and 5 µl of each dilution was spotted onto LB plates with appropriate antibiotics and the indicated aza-C concentration. The plates were incubated overnight at 37°C.
Western blot analyses
For the experiment with plasmid pR215, overnight cultures were diluted to A560 = 0.1 in LB with tetracycline (to maintain selection for the plasmid) and grown at 37°C. When the cultures reached an A560 of 0.3, aza-C (dissolved in LB) was added to one portion of the culture at a concentration of 0.05 mg/ml and LB was added to the no-drug control. After a 2-hour incubation with or without aza-C, cell amounts equivalent to 2 ml of A560 = 0.5 were collected by centrifugation and frozen in a dry ice/ethanol bath.
For the experiment with plasmid pBAD-MEcoRII, overnight cultures (grown with 0.2% glucose) were diluted to A560 = 0.1 in LB with chloramphenicol (to maintain selection for the plasmid) and glucose (0.2%; to repress M.EcoRII expression) where indicated. The cultures were grown at 37°C till they reached A560 = 0.3. Cells were collected by centrifugation, rinsed in LB, and resuspended in LB with 0.05% arabinose to activate M.EcoRII protein expression. After a 1-hour incubation, cell amounts equivalent to 2 ml of A560 = 0.5 were collected by centrifugation and frozen in a dry ice/ethanol bath. For the SsrA tagging experiment, the above procedure was followed except that aza-C (0.05 mg/ml) was added at A560 = 0.1 and the culture was grown to A560 = 0.5 before inducing M.EcoRII expression with arabinose.
For all Western blots, the frozen cell pellets were thawed and lysed by a method adapted from Sambrook et al. (1989). Cell pellets were resuspended in 200 µl of lysis buffer [50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 100 mM NaCl, 0.13 mM phenylmethylsulfonyl fluoride and 0.26 mM lysozyme] and incubated at 4°C for 30 minutes. Deoxycholic acid (3.3 mM) was added and incubation was continued at 4°C for 30 minutes. Finally, DNase I (4 units; New England Biolabs) was added and the lysates were incubated at room temperature for 30 minutes.
Lysates were loaded onto a 7.5% or 12% Ready Tris-HCl gel (Bio-Rad Laboratories) and the gels were run at constant voltage (120 V) for 2 hours in 25 mM Tris-Glycine buffer with 0.1% SDS. The portion of the gel with proteins above 75- kDa molecular weight was cut out and stained with Coomassie blue to provide a protein loading control for each sample. The remaining portion of the gel was transferred to nitrocellulose membrane (Protran® BA 85; Whatman®) for Western blot analysis. Polyclonal M.EcoRII or SsrA antibodies were used as primary antibody, and IRDye 800CW-conjugated goat anti-rabbit IgG (LI-COR®) was used as secondary antibody. Both the Coomassie blue-stained gel and the Western blot were scanned using an Odyssey Infrared Imaging System (LI-COR®), and quantitation was performed using the provided Odyssey software (version 3.0).
Inhibitor growth curves
Overnight cultures in LB were diluted to roughly 4 × 106 cell/ml in LB (containing ampicillin for plasmid-containing strains), and 75 µl was delivered to each well in a microtiter plate. For the experiments measuring streptolydigin sensitivity, 75 µl of streptolydigin at twice the indicated concentration (or drug-free control) was also added to each well, for a total volume of 150 µl/well. The plate was incubated at 37°C for 12 hours with constant shaking in a BioTek ELx808 Microplate Reader. The optical density (at 630 nm) of each well was read every 15 minutes. For the experiments measuring sensitivity to the double-drug combinations (Fig. 8), the same protocol was followed, with further details in the legend to Fig. 8 and Supplemental Material.
Supplementary Material
Acknowledgments
We are extremely grateful to Tania Baker (MIT) for providing the SsrA tag antibody, Sean Moore (University of Central Florida) for ssrA plasmids, Konstantin Severinov (Waksman Institute of Microbiology) for streptolydigin, Max Gottesman and Robert Washburn (Columbia University Medical School) for bicyclomycin, and Tao-Shih Hsieh (Duke University Medical School) for important discussions. We also greatly appreciate the helpful suggestions of anonymous reviewers. This research was supported by NIH grant GM72089.
References
- Abo T, Inada T, Ogawa A, Aiba H. SsrA-mediated tagging and proteolysis of Lacl and its role in the regulation of lac operon. EMBO J. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes to Cells. 2002;7:629–638. doi: 10.1046/j.1365-2443.2002.00549.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Sys Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru B, Gopal J, Bhagwat AS. Overproduction of DNA cytosine methyltransferases causes methylation and C->T mutations at non-canonical sites. J Biol Chem. 1996;271:7851–7859. doi: 10.1074/jbc.271.13.7851. [DOI] [PubMed] [Google Scholar]
- Barbe J, Gibert I, Guerrero R. 5-Azacytidine - Survival and induction of the SOS response in Escherichia coli K-12. Mutat Res. 1986;166:9–16. doi: 10.1016/0167-8817(86)90035-0. [DOI] [PubMed] [Google Scholar]
- Baumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infection and Immunity. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–692. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- Berenbaum MC. A method for testing for synergy with any number of agents. J Infect Diseases. 1978;137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- Bhagwat AS, Johnson B, Weule K, Roberts RJ. Primary sequence of the EcoRII endonuclease and properties of its fusions with beta-galactosidase. J Biol Chem. 1990;265:767–773. [PubMed] [Google Scholar]
- Bhagwat AS, Roberts RJ. Genetic analysis of the 5-azacytidine sensitivity of Escherichia coli K-12. J Bacteriol. 1987;169:1537–1546. doi: 10.1128/jb.169.4.1537-1546.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho MG. Fractional inhibitory concentration index of combinations of antibacterial agents against cariogenic organisms. J Dentistry. 2000;28:565–570. doi: 10.1016/s0300-5712(00)00039-7. [DOI] [PubMed] [Google Scholar]
- Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6931. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Cassani G, Burgess RR, Goodman HM, Gold L. Inhibition of RNA polymerase by streptolydigin. Nat New Biol. 1971;230:197–200. doi: 10.1038/newbio230197a0. [DOI] [PubMed] [Google Scholar]
- Chambers AL, Smith AJ, Savery NJ. A DNA translocation motif in the bacterial transcription--repair coupling factor, Mfd. Nucleic Acid Res. 2003;31:6409–6418. doi: 10.1093/nar/gkg868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK, Gerdes K. RelE toxins from Bacteria and Archaea cleave mRNAs on trnslating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Christman JK. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- Dreyfuss M. Killer and protective ribosomes. Progress in Molecular Biology and Translational Science. 2009;85:423–466. doi: 10.1016/S0079-6603(08)00811-8. [DOI] [PubMed] [Google Scholar]
- Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- Ferguson AT, Vertino PM, Spitzner JR, Baylin SB, Muller MT, Davidson NE. Role of estrogen receptor gene demethylation and DNA methyltransferase DNA adduct formation in 5-aza-2'-deoxycytidine-induced cytotoxicity in human breast cancer cells. J Biol Chem. 1997;272:32260–32266. doi: 10.1074/jbc.272.51.32260. [DOI] [PubMed] [Google Scholar]
- Friedman S. The irreversible binding of azacytosine-containing DNA fragments to bacterial DNA (cytosine-5) methyltransferases. J Biol Chem. 1985;260:5698–5705. [PubMed] [Google Scholar]
- Friedman S, Som S. Induction of EcoRII methyltransferase - Evidence for autogenous control. J Bacteriol. 1993;175:6293–6298. doi: 10.1128/jb.175.19.6293-6298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbara S, Bhagwat AS. The mechanism of inhibition of DNA (cytosine-5) methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem J. 1995;307:87–92. doi: 10.1042/bj3070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez F, Shoji S, Frederick K, Hayes CS. RNase II is important for A-site mRNA cleavage during ribosome pausing. Mol Microbiol. 2009;73:882–897. doi: 10.1111/j.1365-2958.2009.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover AB, Leyland-Jones BR, Chun HG, Davies B, Hoth DF. Azacitidine: 10 years later. Cancer Treat Rep. 1987;71:737–746. [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Akiyama Y. Cellular functions mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J Mol Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Jones PA. Altering gene expression with 5-azacytidine. Cell. 1985;40:485–486. doi: 10.1016/0092-8674(85)90192-8. [DOI] [PubMed] [Google Scholar]
- Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000;182:1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2'-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- Karpf AR, Moore BE, Ririe TO, Jones DA. Activation of the p53 DNA damage response pathway after inhibition of DNA methyltransferase by 5-aza-2'-deoxycytidine. Mol Pharmacol. 2001;59:751–757. [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kuo HK, Griffith JD, Kreuzer KN. 5-azacytidine-induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248–8254. doi: 10.1158/0008-5472.CAN-07-1038. [DOI] [PubMed] [Google Scholar]
- Lal D, Som S, Friedman S. Survival and mutagenic effects of 5-azacytidine in Escherichia coli. Mutat Res. 1988;193:229–236. doi: 10.1016/0167-8817(88)90033-8. [DOI] [PubMed] [Google Scholar]
- Li X, Hirano R, Tagami H, Aiba H. Protein tagging at rare codons is caused by tmRNA action at the 3' end of nonstop mRNA generated in response to ribosome stalling. RNA. 2006;12:248–255. doi: 10.1261/rna.2212606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lies M, Maurizi MR. Turnover of endogenous SsrA-tagged proteins mediated by ATP-dependent proteases in Escherichia coli. J Biol Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H, Hallier M, Felden B, Tenson T. tmRNA decreases the bactericidal activity of aminoglycosides and the susceptibility to inhibitors of cell wall synthesis. RNA Biol. 2005;2:70–74. doi: 10.4161/rna.2.2.2020. [DOI] [PubMed] [Google Scholar]
- McClure WR. Mechanism of streptolydigin inhibition of Escherichia coli RNA polymerase. J Biol Chem. 1980;255:1610–1616. [PubMed] [Google Scholar]
- Mehta P, Richards J, Karzai AW. tmRNA determinants required for facilitating nonstop mRNA decay. RNA. 2006;12:2187–2198. doi: 10.1261/rna.247706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Munavar H, Zhou YN, Gottesman S. Analysis of the Escherichia coli Alp phenotype: Heat shock induction in ssrA mutants. J Bacteriol. 2005;187:4739–4751. doi: 10.1128/JB.187.14.4739-4751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan NA, Bliska JB, Karzai AW. A role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathogens. 2006;2:50–62. doi: 10.1371/journal.ppat.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9969. [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudley E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- Roberts J, Park J-S. Mfd, the bacterial transcription repair coupling factor: translocation, repair and termination. Curr Opinion in Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Roberts JW, Shankar S, Filter JJ. RNA polymerase elongation factors. Annu Rev Microbiol. 2008;62:211–233. doi: 10.1146/annurev.micro.61.080706.093422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche ED, Sauer RT. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J Biol Chem. 2001;276:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. 17.38. [Google Scholar]
- Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev. 1994;58:317–329. doi: 10.1128/mr.58.3.317-329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. J Biol Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- Siddhiko C, Erbstoeszer JW, Weisblum B. Mode of action of streptolydigin. J Bacteriol. 1969;99:151–155. doi: 10.1128/jb.99.1.151-155.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- Som S, Friedman S. Inhibition of transcription in vitro by binding of DNA (cytosine-5) methylases to DNA templates containing cytosine analogs. J Biol Chem. 1994;269:25986–25991. [PubMed] [Google Scholar]
- Straus D, Walter W, Gross CA. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma-32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem. 2004a;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004b;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuske S, Sarafianos SG, Wang XY, Hudson B, Sineva E, Mukhopadhyay J, et al. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Meyenburg K, Nielsen LD, Johnsen K, Molin S, Svenningsen B, Miozzari G. Re-evaluation of mode of action of streptolydigin in Escherichia coli: Induction of transcription termination in vivo. Antimicrob Agents Chemother. 1978;13:234–243. doi: 10.1128/aac.13.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski MW, Gabbara S, Kubareva EA, Romanova EA, Oretskaya TS, Gromova ES, et al. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 1993;21:295–301. doi: 10.1093/nar/21.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Progress in Molecular Biology and Translational Science. 2009;85:467–500. doi: 10.1016/S0079-6603(08)00812-X. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Sunohara T, Jojima K, Inada T, Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T, Kanemori M, Morita MT. The heat shock response: Regulation and function in bacterial stress responses. Washington, D.C: ASM Press; 2000. pp. 3–18. [Google Scholar]
- Zwiefka A, Kohn H, Widger WR. Transcription termination factor Rho: the site of bicyclomycin inhibition in E. coli. Biochemistry. 1993;32:3564–3570. doi: 10.1021/bi00065a007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.