Abstract
Harvest index, defined as the ratio of reproductive yield to total plant biomass, and early ripening are traits with important agronomic value in processing tomatoes. The Solanum pennellii introgression-line (IL) population shows variation for harvest index and earliness. Most of the QTL mapped for these traits display negative agronomic effects; however, hi2-1 is a unique QTL displaying improved harvest index and earliness. This introgression was tested over several years and under different genetic backgrounds. Thirty-one nearly isogenic sub-lines segregating for the 18 cM TG33–TG276 interval were used for fine mapping of this multi-phenotypic QTL. Based on this analysis the phenotypic effects for plant weight, Brix, total yield and earliness were co-mapped to the same region. In a different mapping experiment these sub-lines were tested as heterozygotes in order to map the harvest index QTL which were only expressed in the heterozygous state. These QTL mapped to the same candidate region, suggesting that hi2-1 is either a single gene with pleiotropic effects or represents linked genes independently affecting these traits. Metabolite profiling of the fruit pericarp revealed that a number of metabolic QTL co-segregate with the harvest index trait including those for important transport assimilates such as sugars and amino acids. Analysis of the flowering pattern of these lines revealed induced flowering at IL2-1 plants, suggest that hi2-1 may also affect harvest index and early ripening by changing plant architecture and flowering rate.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1412-8) contains supplementary material, which is available to authorized users.
Introduction
Harvest index, defined as the ratio of reproductive yield to total plant biomass, has been taken as a measure of efficiency in partitioning assimilated photosynthates to harvestable product. This parameter was first considered in 1914 by Beaven, who described it as the ratio of grain yield to total plant weight and termed it the “migration coefficient” [reviewed in (Sinclair 1998)]. Later on the term “harvest index” was suggested and recommended as an important reference to assess progress in germplasm development towards improved yield potential. Harvest index has been commonly used as a parameter for plant breeding, particularly in cereals. Domestication of grain crops during the twentieth century caused continuous improvement in harvest index along with increasing crop yields (Hay 1995). However, it is not that harvest index of ancestral grain crops was generally low. Indeed, there is considerable evidence that some ancestral species of wheat and rice had very high harvest indices (Sinclair 1998; Doebley et al. 2006). The most dramatic improvement of harvest index in wheat and rice was due to the exploitation of a single gene mutation that causes dwarfism and the use of this gene in the breeding of high-yielding short varieties (Khush 2001; Thornsberry et al. 2001; Salamini 2003).
Earliness can be defined in many ways. Basically, it represents the time it takes the plant from sowing to harvestable product. The variation in earliness can be due to an earlier switch from vegetative to reproductive growth or due to faster ripening of the fruit (Doganlar et al. 2000; Tanksley 2004). The first stage is highly related to the harvest index because it involves changes in the growth habit of the plant. Breeding for increased harvest index and earliness in crop plants is of major importance for several reasons:
-
(i)
The harvest index represents the efficiency of using both natural and man-made resources (water, carbon dioxide, soil and artificial fertilizers) to produce harvestable product, for that reason it is important both from economical and ecological aspects.
-
(ii)
High harvest index and early maturation represent two mechanisms for plants to deal with abiotic stresses. Improved harvest index represent enhanced partitioning of the limited assimilated photosynthates, under the stress conditions, into harvestable product. From the plants perspective, early maturation is, on the other hand, an escape mechanism to ensure its propagation under conditions of stress.
-
(iii)
The development of mechanical harvesting techniques for most crops species provides an additional preference for plants displaying a relatively small canopy.
Intensive research concerning the developmental genes for the transition from vegetative to reproductive has been performed on the model plant Arabidopsis thaliana (Alonso-Blanco et al. 1998; Koornneef et al. 1998; Samach et al. 2000; El-Din El-Assal et al. 2001, 2003; Cremer and Coupland 2003; Pineiro et al. 2003; Valverde et al. 2004; Alonso-Blanco et al. 2009) and in other plant species (Murai et al. 2003; Izawa 2007; Jimenez-Gomez et al. 2007). In addition, other factors such as source–sink relations and photosynthetic efficiency have been considered to influence in this trait (Bugbee and Salisbury 1988; Nunes-Nesi et al. 2005; Hackel et al. 2006). An example of the implementation of such an approach is a work done in tobacco over-expressing a phytochrome gene was produced (Robson et al. 1996). The transgenic plants showed reduced shade avoidance causing proximity-conditional dwarfing and increased harvest index. Another example is the over-expression of the arabidopsis LFY gene in aspen (Weigel and Nilsson 1995) that caused floral development induction. In rice (Oryza sativa), which is a model crop plant, earliness is defined as “heading date”. Genetic research is being made, mainly using natural variation, to identify and characterize QTL involved in ‘heading date’ (Yamamoto et al. 2000; Monna et al. 2002; Yu et al. 2002). Some of these QTL were already cloned (Yano et al. 2000, 2001; Doi et al. 2004; Xue et al. 2008) and these improving alleles can be introduced in breeding programs for creation of early maturing rice varieties. In tomato a single gene mutation is responsible for converting indeterminate plant into determinate, which is a very dramatic change in the plant growth habit (Pnueli et al. 1998; Carmel-Goren et al. 2003). This recessive mutation (sp/sp) at the SP locus is the basis for all mechanically harvested processing varieties. However, variation in plant growth habit is still present among determinate processing tomato varieties. Recent studies have identified that S and AN, which encode a homeobox transcription factor and an F-box protein, respectively as well as single flower truss as important genes in the determination of inflorescence development and thus of heterosis (Lipmann et al. 2009; Krieger et al. 2010). Likewise the majority of the variation in fruit size has recently been associated to fw2-2 (Frary et al. 2000), and to a lesser extent to a YABBY like transcription factor (Cong et al. 2008). Despite these important gains in knowledge the objective of processing tomato breeders remains to obtain compact “bushy” plants that have concentrated uniform ripening, so that yield can be harvested at one time point, and the mechanical harvest can be as efficient as possible (Atherton and Rudich 1986).
The objective of our current work was to use natural variation in wild tomato, for the discovery of alleles that can improve harvest index and earliness of processing tomatoes. The introgression-line (IL) population of S. pennellii in a processing-tomato variety (M82) is an efficient tool for identification and mapping of QTL (Eshed and Zamir 1994). Yield associated QTL were previously identified and mapped (Eshed and Zamir 1995) but not much attention was paid to QTL affecting growth habit and the relationship between reproductive and vegetative growth. In this work we used the ILs population to look for QTL that modify the reproductive/vegetative ratio and transition timing.
Materials and methods
Plant material and field trial
Whole genome phenotypic survey for yield and vegetation-related traits was performed in different field experiments: first on summer 1993, using the 50 ILs population (Eshed and Zamir 1995). This original set of 50 ILs was extended to create the 75 ILs population (Pan et al. 2000). The extended population was phenotyped in the field on summer 2000. Both ILs and ILHs were evaluated on these trials.
Thirty-one sub-NILs were extracted from the M82 × IL2-1 F2 population. Distal markers from both extremes of the introgression were used to detect those recombinant lines following RFLP analysis (using TG33 and TG276). Detailed genotyping of those lines was performed to determine the exact region of recombination, using 24 RFLP markers. Genetic distances in cM were calculated by the ratio of recombinant progenies between adjacent markers out of the 1,600 parental gametes that were screened, and a genetic map of this region was created. Ninety percent reduction in recombination was observed compared to map distances as calculated from the S. lycopersicon × S. pennellii F2 population (Tanksley et al. 1992). This trend is in agreement with results of other comparisons of recombination frequencies between early crosses and late backcrossing in segregating populations (Rick 1969, 1972; Ji and Chetelat 2003). The effect of IL2-1p on the yield related traits were also tested in the background of the line 9,225 which is an F7 selection from an F2 population between two processing tomato inbreds. The fragment IL2-1p was introgressed using molecular markers until the F6 generation.
Three mapping trials were conducted: (1) progeny tests for the F3 families in summer 1999, (2) in summer 2000 using fixed homozygous sub-NILs and (3) in summer 2002 using sub-NILs × M82 F1 plants. Tests for the introgression effect under different genetic backgrounds were performed in summer 1999 and 2000. All open-field experiments were performed at the Western Galilee Experimental Station in Akko, Israel. Seedlings (35 days old) were transplanted in the field with 50 cm between plants and 2 m between rows (1 plant/m2). All the plants were sprinkler-irrigated immediately after transplanting with 30 m3 of water for every 1,000 m2 of field area. For the rest of the growing period, the wet treatment was drip-irrigated with 250 m3 of water per 1,000 m2 while no water was applied to the dry treatment. Inflorescence counting was performed in two different experiments: (1) on the open-field experiment in Akko in summer 1999 as described by Eshed and Zamir (1995) and (2) in a greenhouse experiment at Rehovot in winter 2001/2002. In the greenhouse experiment plants were grown in 4 liter pots. 35 days seedlings were transplanted on November 2001 and were grown until March 2002.
Results for the comparison between IL2-1 and M82 under irrigated and dry conditions were obtained from a genome-wide scan field experiment that was conducted under these two irrigation regimes in summer 2000 (Gur et al. 2010). The sub-lines were re-grown in an open-field experiment in Akko in summer 2008 with pericarp tissue being harvested for metabolite profiling exactly as described in Schauer et al. (2006).
Extraction, derivatization, and analysis of polar metabolites using GC–MS
Metabolite analysis by gas chromatography–mass spectrometry (GC–MS) was carried out essentially as described by Fernie et al. (2004) and Lisec et al. (2006). The mass spectra were cross-referenced with those in the Golm Metabolome Database (Kopka et al. 2005).
Nucleic acid analysis
The parental and recombinant lines were genotyped using RFLP analysis as described by Bernatzky and Tanksley (1986). A large genetic gap remained at the TG31–CT106 interval. This 11.5 cM gap is a result of either random lack of markers, or, more likely, due to ‘hot-spot’ of recombination in this region. In order to enrich this region with more markers we surveyed a set of RAPD markers on the two parents (IL2-1 and M82). The DNA from both parents was amplified using 300 random RAPD primers (Operon Technologies, Alameda, CA, USA) to select those that generated polymorphic bands. The RAPD procedure was as described by Doganlar et al. (2000). Six RAPD primers produced polymorphic bands and were then used to amplify DNA from the recombinant sub-lines of IL2-1, in order to map them more precisely within the region.
The BAC clones that contained the TG31 sequence were identified using hybridization on the tomato BAC library filters. The corresponding clones were ordered from the tomato BAC library at Clemson University (http://www.genome.clemson.edu/cgi-bin/orders?page=productGroup&service=bacrc&productGroup=166). The BAC ends were sequenced and used as RFLP markers to place them on the genetic map.
Phenotyping
In all open-field experiments, fruits were harvested when 80–100% of the tomatoes were red, preferably when the M82 was 80–90% so that the early ripening effect of the IL2-1p allele could be detected. Red and green fruits were weighed separately to estimate the earliness (earliness is defined as the number of days from sowing to the appearance of the first ripe fruit). Plant vegetative weight (PW) was determined by weighing only the vegetative tissue (after harvesting the fruits) without the roots. Total fruit yield (TY) per plant included both the red (RY) and the green (GY) fruits. Mean fruit weight (FW) was calculated from a random sample of 20 fruits per plant. Concentrations of total soluble solids (BX, measured in degrees Brix) were measured from a random sample of 10 fruits per plant. Harvest index (HI) was calculated as the ratio between the total yield and total biomass (TY + PW). Inflorescence labeling and counting was done at intervals of 3–7 days from beginning of flowering over a 40-day time period, on plants grown both on the open field and in a greenhouse. At each counting time, all new inflorescences that contained at least one post-anthesis flower were labeled and counted.
Mode of inheritance
The additive effect (a) was half of the difference between each IL and M82, and its significance level was determined by the comparison between the IL and M82. The dominance deviation (d) is the difference between ILH and the mid-value of its parents. Its significance level was calculated by contrasting the ILH (+1) with M82 (−0.5) and the appropriate IL (−0.5). The degree of dominance for each introgression (d/[a]) was calculated by dividing the mean dominance deviation by the mean additive effect.
Statistical analyses
Statistical analyses were performed on the JMP V.5 software package for Macintosh (SAS institute). Mean values for the parameters measured for the tested genotypes were compared to the common control using the “Fit Y by X” function and “Compare with control” with an alpha level of 0.05 (Dunnett 1955). All calculations were performed with the phenotypic values while the results are presented as the percentage difference from M82. Interactions were calculated by multi-factorial analysis of variance (ANOVA) using the “fit model” function and correlation was determined by Pearson’s analysis.
Results
Correlation between vegetative and reproductive growth and identification of QTL that modify this relation in the ILs population
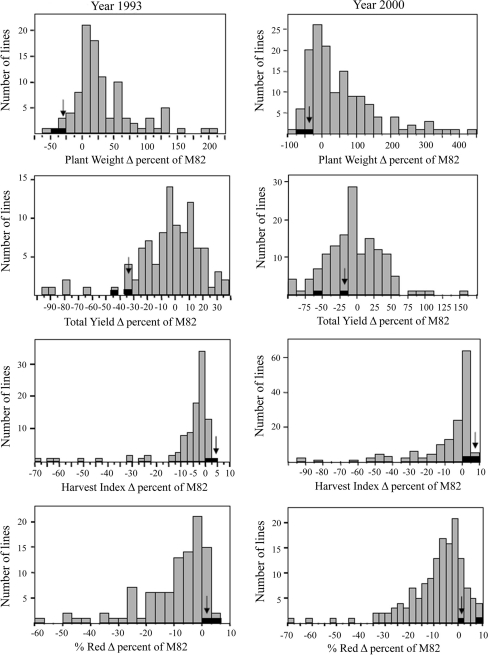
Analysis of data from whole genome phenotypic surveys on field trials, over 2 years: (1) summer 1993 (Eshed and Zamir 1995) and (2) summer 2000 (Gur et al. 2010), demonstrate that there is large variation for growth habit in the ILs population. The 66 and 65% in 1993 and 2000, respectively, of the phenotypic variation is explained by genetic variation after exclusion of the lines that contain the S. pennellii SELF-PRUNING (SP) allele from the analysis (Fridman et al. 2002). Our interest in the current study was to detect QTL that modifies the relationship between vegetative growth and yield production in such a way that the harvest index is improved. The frequency distribution for plant weight is not normal since the variation is biased towards an increase in comparison to M82 (Fig. 1). Twenty-five ILs were observed to display an increased plant weight, whilst only two displayed a reduced plant weight in comparison to the M82 control (Dunnet; P < 0.05). That said the frequency distribution of total yield is normal—with M82 being invariant from the population mean (Fig. 1). There are 12 lines in which the total yield was reduced with respect to the control and 8, which improved the total yield (Dunnet; P < 0.05). The parameters of plant weight and total yield appear to be highly correlated on the basis of measurements of a homogenous population of M82 plants (for PW: M82, 1.2 kg; 75ILs, 2.2 kg; 75ILHs, 1.7 kg; for TY: M82n=100, 8.5 kg; 75ILsn=500, 6.7 kg; 75ILHsn=430, 10 kg; R = 0.74, N = 107).Given that the harvest index is the ratio between total yield and plant weight it follows that the correlation between these traits can afford a good estimate for the variation of harvest index within a population. The low correlation coefficient within the S. pennellii IL library (for HI: M82n=100, 0.71; 75ILsn=500, 0.08; 75ILHsn=430, 0.61; R = 0.08, N = 500) indicates large variability for this trait. Intriguingly, the majority of variation apparent in this trait is expressed as a reduction of harvest index in comparison to M82 (Fig. 1). Critical assessment of these data revealed that 23 lines display a reduced harvest index in comparison to M82, while only a few lines displayed transgressive segregation and a consequent improvement of the harvest index. For earliness, the picture is quite similar to that of the harvest index in terms of the frequency distribution and numbers of increasing and decreasing lines. IL2-1 is a unique genotype, which displayed consistent transgressive segregation for reduction of total plant weight. When tested as a heterozygote (ILH2-1; IL2-1 × M82), it improved the harvest index (Fig. 1), and was, therefore, chosen for detailed analysis.
Fig. 1.
Frequency distributions of the relative performance for means of ILs and their hybrids as measured on the replicated trails on 1993 and 2000 (expressed in percent difference (Δ%) of M82). Black bars represent the IL2-1 genotypes. IL2-1 × M82 is indicated in black arrow
Phenotypic characterization of IL2-1
Yield-related traits
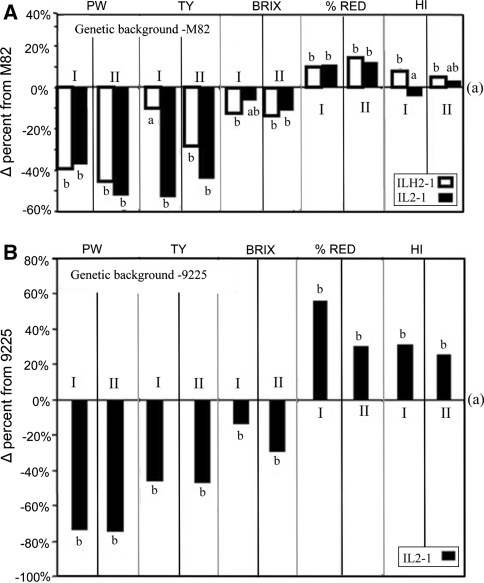
IL2-1 was tested over 3 years and in different genetic backgrounds. Two genetic backgrounds of inbred lines diverse in their phenotypic characteristics were used: (1) the core background of M82, and (2) a semi-determinate processing tomato inbred; 9,225. The effect of the introgression on plant weight (PW) was consistent, 40–60% reduction compared to the near-isogenic control at the M82 background (Fig. 2a) and 70% reduction at the 9,225 background (Fig. 2b). For total yield (TY) the effect in the homozygous lines was a reduction of 45–55% at both genetic backgrounds whilst the IL2-1 × M82 hybrid displayed a non-significant yield reduction of 10% in the 2000 trial but a substantially larger (and significant) yield reduction of 28% in 2002 (Fig. 2a). Total soluble solids content (BX) was reduced by 7–15% both in IL2-1 and in the hybrid with respect to M82 and by 10 and 30% in the 1999 and 2000 trials with respect to the 9,225 control. Both harvest index and earliness are derived parameters calculated from the measurements taken at the field. Earliness (EA) is estimated as the percentage of red yield from the total harvestable yield (green and red fruit). The harvest was performed at 80–95% red yield (depending on the experiment), of the M82 control. There was a general effect of increase in this parameter following introgression: 10–15% increase in percentage of red yield in the M82 background (Fig. 2a), and a dramatic 30–50% in the 9,225 background (Fig. 2b). Harvest index (HI) is calculated by dividing the total harvestable yield by the total plant biomass. Intriguingly, there was a consistent 5–7% increase in HI in the IL2-1 × M82 hybrid whilst the homozygote IL2-1 displayed no significant change. In the 9,225 background, there was 30% increase in HI attributable to the IL2-1 introgression.
Fig. 2.
The effects of IL2-1 and IL2-1 × M82 (ILH2-1) on different traits at the M82 (a) and 9225 (b) genetic backgrounds over 2 years. At the M82, I-Akko 2000 and II-Akko 2002. At the 9,225, I-Akko 1999 and II-Akko 2000. Values are presented in percent difference (Δ%) of nearly isogenic control. For each trait, means were compared and different letters represent means that are significantly different at P < 0.05. Number of replications for each genotype was: 1999 n = 7, 2000 n = 20, 2002 n = 15. M82 is represented the 0% (indicated as “a”). Values indicated with different letters were determined significantly different by ANOVA (P < 0.05). PW plant vegetative weight, TY total fruit yield, HI harvest index
Using two-way ANOVA, we calculated the IL2-1 × genetic background interactions for the QTL effect in the two diverse genetic backgrounds. A significant interaction (P < 0.05) was found for all traits in at least one season, since the IL2-1p (S. pennellii allele at the IL2-1 segment) effect was stronger in the 9,225 background than in M82.
Table 1 presents the mode of inheritance of the IL2-1 QTL for different traits in the M82 background. For PW, the negative effect of the QTL is dominant (d/a = 0.8 and 1.1 in 2002 and 2000, respectively). TY is additively reduced (d/a = 0.3 and −0.65 in 2002 and 2000, respectively) and HI, which combines these two traits accordingly displays an overdominant mode of inheritance (d/a = 2.2 and 5.2 in 2002 and 2000, respectively). BX, an integrative trait reflects plant source–sink relationships (Schauer et al. 2006) and is strongly correlated with HI, similarly displayed an overdominant mode of inheritance (d/a = 1.5 and 3.4 in 2002 and 2000, respectively). The earliness (percentage of red fruits) is essentially similar in the IL and the ILH and displayed a dominant mode of inheritance (d/a = 1.3 and 0.88 in 2002 and 2000, respectively).
Table 1.
Mode of inheritance of IL2-1 QTL for different traits on 2000 and 2002
| Genotype | From M82 to Akko 2002 (%) | From M82 to Akko 2000 (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PW | BX | TY | %RED | HI | PW | BX | TY | %RED | HI | |
| q | −25.90 | −5.30 | −22.00 | 6.20 | 1.50 | −18.54 | −72.00 | −26.49 | 5.44 | −1.85 |
| p (a) | 0.00 | 0.01 | 0.01 | 0.02 | 0.32 | 0.01 | 0.12 | 0.00 | 0.00 | 0.28 |
| d | −19.50 | −8.00 | −6.40 | 8.00 | 3.40 | −20.59 | −9.15 | 17.13 | 4.80 | 9.61 |
| p (d) | 0.09 | 0.01 | 0.57 | 0.04 | 0.09 | 0.09 | 0.00 | 0.08 | 0.15 | 0.00 |
| d/[a] | 0.80 | 1.50 | 0.30 | 1.30 | 2.20 | 1.10 | 3.40 | -0.60 | 0.90 | 5.20 |
PW plant vegetative weight, BX total soluble solids content, TY total fruit yield, HI harvest index
Additive effect (a), dominance deviation (d), significance of the additive effect (P(a)), significance of dominant deviation (P(d)), relative dominance deviation d/[a]
Flowering pattern
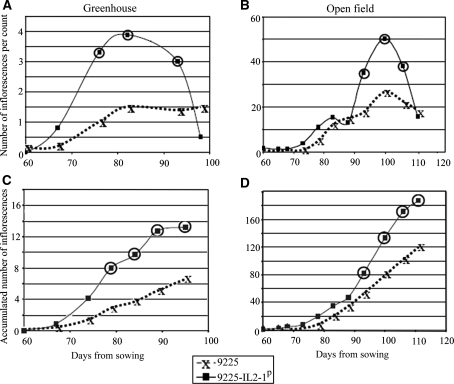
In order to perform in-depth characterization of the diverse phenotypic effects observed in IL2-1, inflorescence-counting experiments were performed in two diverse environments. The first experiment was conducted in the open field whilst the second took place in a greenhouse. The 9,225 genetic background was chosen for this analysis since the phenotypic effect of the IL2-1p allele in this background was considerably more pronounced (as observed for the yield-related traits; Fig. 2). The basic differences between plants grown in open-field conditions and those grown in 4 l pots in a greenhouse are the number of branches produced and the time to determination. Plants grown in the field have considerably more branches and their growth period is substantially longer prior to determination. Consequently, field-grown plants are bigger in size with more flowers and fruits. This difference is, moreover, evident from our measurements of the total number of inflorescences. Plants in the field had between 120 and 190 inflorescences whilst plants in the greenhouse were characterized as having between 7 and 13 (Fig. 3c, d). In spite of this large difference in plant development, we observed the same trend of flowering enhancement caused by the IL2-1p allele in both environments. The 9225-IL2-1p plants in the greenhouse displayed significantly more inflorescences between counting days 77 and 92 (Fig. 3a). In the field the 9225-IL2-1p plants displayed significantly more inflorescences between counting days 92 and 108 (Fig. 3b). Figure 3c and d describe the total number of inflorescences accumulated across the growing period. In both environments, an increasing difference in the total number of inflorescences was observed across the growth period. In the greenhouse the number of inflorescences in 9225-IL2-1p was almost double that seen in its near-isogenic control (Fig. 3c). In the field, there is more than 50% increase in the number of inflorescences 9225-IL2-1p in comparison to its near-isogenic control (Fig. 3d).
Fig. 3.
The effect of the IL2-1 introgression on flowering pattern at the 9,225 genetic background. The numbers of inflorescence per count are presented from two different experiments: greenhouse and open field. Each point represents the mean value of 10 plants. Means of the nearly isogenic lines were compared on each counting point using t test. Mean values of 9225-IL2-1p that were found significantly different from their nearly isogenic control (at P < 0.05) are circled
Using fine mapping for characterization of the multiple phenotypes displayed by IL2-1
The strategy of substitution mapping in combination with linkage analysis was performed in order to try and more precisely locate the genetic factor or factors, which are responsible for the relevant phenotypic variation associated with IL2-1. Phenotypic characterization of IL2-1 revealed that traits such as plant weight, total yield, brix and earliness could readily be mapped in a sub-NILs mapping population, homozygous for the S. pennellii introgression, while mapping the harvest index effect had to be performed in the heterozygous condition. Mapping trials were made independently for each population.
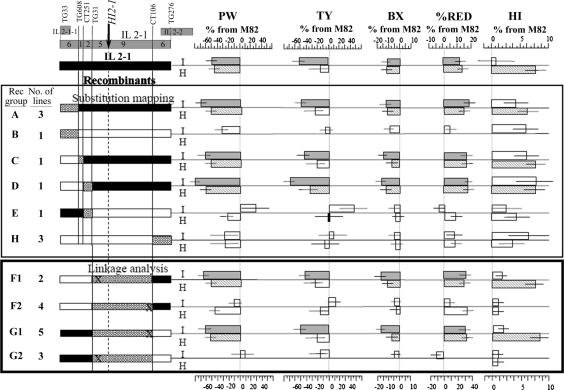
Thirty one nearly isogenic sub-lines segregating for the 18 cM TG33–TG276 interval were extracted from 800 plants screened in F2. These sub-lines were tested in field experiments as F3 families (progeny test) in summer 1999 for preliminary mapping. Twenty-four lines were further tested on a replicated trial as fixed genotypes in summer 2000. Each of the traits (PW, TY, FW, BX and %RED) was mapped independently. Twenty-one sub-lines were crossed to M82 to produce lines heterozygous for the S. pennellii introgressions. These heterozygous sub-lines were tested in a replicated trial in summer 2002 for the harvest index mapping. Recombinant sub-NILs were divided into eight genotypic groups according to their position of recombination and allelic composition. Figure 4 summarizes the mapping data collected in 2000 and 2002. For example, significant reduction of more than 50% was observed in PW in groups A, C and D whilst the non-significant effects in groups B, E and H locate the PW QTL to the CT251–TG276 interval. Fourteen lines contained recombination between TG31 and CT106 (recombination distance of 5 cM; the physical distance is currently not possible because the markers land on two independent scaffolds). They were divided into groups F (six lines) and G (eight lines) on the basis of their reciprocal allelic composition. Individual lines of each group were further subdivided according to their phenotypic values (F1, F2, G1 and G2), which allowed us to narrow down the mapping to the TG31–CT106 interval. Unfortunately, either a shortage of markers or a “hot-spot” of recombination left us with this large genetic gap. The phenotypic segregation of lines within the same genotypic group (following our genetic map resolution), forced us to further fine map the QTL on the basis of linkage analysis as opposed to substitution mapping. Five lines contained recombination between TG31 and the QTL (2 on group F and 3 on G, Fig. 4). Nine recombinants were found between CT106 and the QTL (4 on F and 5 on G, Fig. 4). These results locate the QTL to a map distance of 0.3 cM from TG31. All the other traits (TY, BX, %RED and HI), were mapped accordingly and they all displayed a complete co-segregation.
Fig. 4.
Fine mapping of QTL for plant weight (PW), total yield (TY), brix (B), percent red yield (%RED) and harvest index (HI) at the 2000 and 2002 field trials. A schematic genetic map of the IL2-1 chromosomal region is presented including neighboring ILs (IL2-1-1 and IL2-2), selected RFLP markers along the region (in top of the chromosome) and genetic distances, presented as the number of recombinant progenies between adjacent RFLP markers at the IL2-1 × M82 F2 population (N = 800). Genotype and phenotype of IL2-1 and eight genotypic groups (recombinant groups A–G) are presented. On each genotypic group, white bars represent the S. lycopersicum allele. The S. pennellii chromosomal segments introgressed into the listed lines are marked by the black bars. Stripped gray bars on the chromosomes represent regions of recombination between adjacent RFLP markers. Independent lines that shared the same recombination region and allelic composition were included in the same genetic group. The number of such independent recombinant lines included in each group is noted. Phenotypic values for all traits are expressed as percent difference from the common control; M82. All independent lines on each group were compared to M82. Based on the common trend observed for these lines, mean effect was calculated for each group. Top bar on each group (labeled as I) represent the phenotype of homozygous genotypes, where gray bars are significantly different from M82 (Dunnet; p < 0.05). Bottom bars (labeled as H) represent the phenotype of heterozygous genotypes, where stripped bars are significantly different from M82. On the linkage section (groups G and F) the ‘X’ on the stripped region represents the side of recombination between the flanking RFLP markers and the QTL
The IL2-1 phenotype is partially correlated with tolerance to drought stress
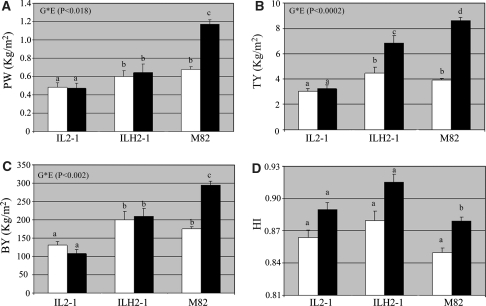
We next compared IL2-1 and ILH2-1 to M82 both under irrigated and dry conditions (Fig. 5). The purpose of this comparison was to evaluate the potential of IL2-1 for the improvement of drought tolerance, based on the rationale that harvest index and earliness are proposed mechanisms for drought tolerance (Hsieh et al. 2002; Kalifa et al. 2004 ; Bartels and Sunkar 2005) and on the observed improved performance of this genotype under optimal irrigation. In the M82, PW in the dry field was significantly reduced by 40% compared to the irrigated field. In contrast, in both IL2-1 and ILH2-1 there was no significant difference in PW between the two irrigation regimes (Fig. 5a). For TY, there was significant reduction of 55% in M82 grown under dry field conditions, whilst in IL2-1 and ILH2-1 the reduction was only of 10 and 35%, respectively (Fig. 5b). For Brix × Yield (BY), which represents the sugar output per unit area there was a significant reduction of 40% in M82, whilst both IL2-1 and ILH2-1 were invariant to the wet field controls (Fig. 5c). Moreover, ILH2-1 had a 25% higher BY value compared to M82, in the dry field, although it must be noted that this difference was not statistically significant. This differential response to the drought stress of IL2-1 and ILH2-1 compared to M82 is supported by the significant genotype × environment interactions that were found for these traits (Fig. 5). In contrast, when analyzing the harvest index (HI) minor increase in both dry and wet conditions was observed (Fig. 5d). Surprisingly in this harvest there was only a significant increase in ILH2-1 (irrigated conditions) and only a minor increase in IL2-1 itself. As such we were unable to access this parameter reliable in this harvest and further studies are thus required in order to achieve this goal.
Fig. 5.
Phenotypic values for total fruit yield (TY), plant vegetative weight (PW), Brix yield (BY) and harvest index (HI) on the dry and wet fields. Mean values ± SE for IL2-1, ILH2-1 and M82 are presented. Black bars represent values from the irrigated field. Empty bars are the dry field values. P value for the genotype × environment interaction (G × E) is presented for each trait. Values indicated with different letters were determined significantly different by ANOVA. Black bars wet conditions and white bars dry conditions
Metabolic QTL co-segregating with the IL2-1 phenotype
In order to gain further insight into the physiological basis of the phenotype we next performed GC–MS-based metabolic profiling of the lines grown in the field under the same design in Summer 2008. The yield associated traits were the same as those described above for other harvests (data not shown), whilst the metabolite profiles were in good accordance with those previously noted for the entire 2-1 introgression (compare Table 2 and Supplementary Table 1 with Schauer et al. 2006, 2008). Notable changes were that the four sub-lines from groups A, C and D with S. pennellii chromosomal segments introgressed between TG31 and CT106 interval exhibited significantly higher content of the amino acids glutamine, histidine, homoserine, lysine, tryptophan, tyramine, tyrosine, S-methyl cysteine. They additionally displayed significantly higher content of galacturonic acid, maltotriose and glycerol-3-phosphate in comparison sub-lines compare with the three sub-lines from groups B, E and H with S. lycopersicum background in hi2-1 position. Moreover, they displayed significantly lower levels of glucose, fructose and trehalose than M82, however, these changes did not completely segregate with respect to the sub-lines. These changes did however correlate with elevated levels of hexose phosphates and as such suggest that these fruits utilize their sugars more efficiently in support of growth processes.
Table 2.
Metabolite profiles in red fruits
| Rec. groups | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | D | B | E | H | |||||||||
| Il2-1-12 | SE | Il2-1-17 | SE | Il2-1-8 | SE | Il2-1-1 | SE | Il2-1-20 | SE | Il2-1-6 | SE | Il2-1-18 | SE | |
| Amino acids | ||||||||||||||
| Cysteine, S-methyl | 4.14 | 0.05 | 4.14 | 0.30 | 3.97 | 0.26 | 5.47 | 0.23 | 1.16 | 0.33 | 1.43 | 0.32 | 1.66 | 0.02 |
| Glutamine | 6.84 | 0.00 | 2.02 | 0.11 | 6.35 | 0.38 | 4.89 | 0.28 | 0.94 | 0.32 | 1.63 | 0.46 | 1.61 | 0.04 |
| Histidine | 2.27 | 0.15 | 1.65 | 0.11 | 3.55 | 0.47 | 1.97 | 0.13 | 0.96 | 0.27 | 1.60 | 0.34 | 1.63 | 0.12 |
| Homoserine | 1.77 | 0.00 | 2.83 | 0.27 | 3.21 | 0.48 | 1.93 | 0.18 | 0.90 | 0.01 | 1.01 | 0.28 | 1.38 | 0.07 |
| Lysine | 1.91 | 0.02 | 2.97 | 0.29 | 3.63 | 0.49 | 2.30 | 0.10 | 0.79 | 0.04 | 1.23 | 0.32 | 1.71 | 0.13 |
| Tryptophan | 1.74 | 0.05 | 1.73 | 0.14 | 3.02 | 0.50 | 1.63 | 0.07 | 0.93 | 0.10 | 0.97 | 0.26 | 1.30 | 0.05 |
| Tyramine | 2.91 | 0.27 | 1.98 | 0.22 | 2.71 | 0.47 | 1.67 | 0.18 | 3.55 | 0.62 | 1.45 | 0.28 | 1.53 | 0.29 |
| Tyrosine | 2.90 | 0.02 | 1.73 | 0.20 | 4.40 | 0.52 | 3.39 | 0.14 | 0.85 | 0.28 | 2.78 | 0.18 | 1.89 | 0.34 |
| Organic acids | ||||||||||||||
| Galacturonic acid | 3.40 | 0.21 | 1.68 | 0.11 | 2.98 | 0.53 | 3.00 | 0.00 | 1.96 | 0.37 | 2.14 | 0.20 | 1.10 | 0.08 |
| Sugar and sugar alcohols | ||||||||||||||
| Fructose | 0.75 | 0.17 | 0.83 | 0.12 | 0.81 | 0.10 | 0.71 | 0.06 | 0.95 | 0.22 | 0.98 | 0.16 | 0.77 | 0.06 |
| Fructose-6-phosphate | 2.03 | 0.08 | 1.84 | 0.14 | 1.70 | 0.21 | 1.69 | 0.07 | 1.09 | 0.00 | 1.00 | 0.18 | 1.40 | 0.00 |
| Glucose | 0.69 | 0.14 | 0.72 | 0.17 | 1.04 | 0.20 | 0.53 | 0.07 | 1.00 | 0.29 | 1.16 | 0.23 | 0.87 | 0.09 |
| Glucose-6-phosphate | 1.46 | 0.00 | 1.59 | 0.12 | 1.30 | 0.09 | 1.56 | 0.11 | 0.89 | 0.09 | 1.02 | 0.17 | 1.29 | 0.13 |
| Maltotriose | 1.96 | 0.18 | 4.63 | 0.19 | 7.04 | 0.10 | 5.70 | 0.26 | 0.78 | 0.48 | 1.26 | 0.18 | 3.64 | 0.22 |
| Trehalose | 0.41 | 0.09 | 0.61 | 0.16 | 0.42 | 0.23 | 0.44 | 0.09 | 0.72 | 0.21 | 0.87 | 0.10 | 0.65 | 0.05 |
| Miscellaneous | ||||||||||||||
| Glycerol-3-phosphate | 1.63 | 0.04 | 1.71 | 0.09 | 1.63 | 0.11 | 1.16 | 0.13 | 1.24 | 0.22 | 0.96 | 0.19 | 1.37 | 0.02 |
Data are normalized by the mean of M82 (M82 = 1.00). Values represent the calculated fraction of M82. Data points consist of the mean out of six replicates ± SE
Bold type values were determined by the t test to be significantly different (P < 0.05) from M82
Discussion
Controlled vegetative growth in crop plants is generally a positive trait. The discovery of a recessive mutation conferring determinate habit (sp/sp) in tomato can be regarded as a small scale ‘green revolution’ for this crop as it allowed the development of processing tomato varieties that are suited for open-field mechanical practices. Most of the tomatoes grown worldwide today and used for industrial products are processing tomatoes, which contain this single-gene mutation. Nevertheless, there is still a considerable amount of variation in plant growth–habit among all these different sp/sp cultivars. M82 is an inbred processing tomato variety with a relatively small plant size and high harvest index. Searching for alleles from the wild that will reduce plant size and increase harvest index at this genetic background seemed like an ambitious objective. Indeed, most of the wild alleles that were effective among the S. pennellii ILs population caused an increase in plant weight and reduction in harvest index. In that sense, IL2-1 is a unique genotype. The enhanced expression of this QTL under a more vegetative genetic background (i.e. 9,225) confirmed our assumption that the M82 is a stringent background for detection of QTLs that improve harvest index, and provided another example for the significance of epistasis in QTL studies (Carlborg and Haley 2004; Kroymann and Mitchell-Olds 2005; Semel et al. 2006; Wentzell et al. 2007). It also highlights the importance of analyzing QTL under the genetic background that reflects their strongest effect if a detailed genetic analysis of the QTL is the focus (Fridman et al. 2002). However, when dealing with agronomically related traits, validation of a QTL effect must be performed in the most relevant genetic background (Gur and Zamir 2004; Fridman et al. 2004; Lippman et al. 2008; Krieger et al. 2010). An indirect result of breeding for improved harvest index and earliness is the potential improvement of tolerance to abiotic stress, such as drought; either by improved partitioning of assimilates to harvestable product (Yadav et al. 2002, 2004) or through avoidance mechanisms. Our results indicate that the improved harvest index and earliness that were conferred by the IL2-1p allele were correlated with some level of drought tolerance. This tolerance was expressed in relative values as the drought induced reductions in plant weight, in total yield and in Brix × Yield of IL2-1 and ILH2-1 were less than that of M82 (Fig. 5). For all these traits, a significant genotype × environment (G × E) interaction, confirming this trend, was found. However, further breeding is needed in order to test whether this improvement will be consistent also in higher yielding genetic backgrounds. Whilst the metabolite data we obtained provide a reasonable rationale to support the improved harvest index and maybe also fruit earliness there seems to be no clear link between the metabolite data and the drought tolerance phenotype since the metabolites that differentially accumulate in the pericarp are quite different from those which we associated with drought tolerance in an earlier study (Semel et al. 2007). Since source–sink relationships have been widely demonstrated to effect metabolites (Schauer et al. 2006; Prudent et al. 2010) we cannot rule out that these play a role in the metabolites changes observed. Indeed, for other traits described in this study further genetic resolution will be required to answer this question.
A major point which must be considered when using wild germplasm for plant improvement is the forced introgression of other wild alleles that sit next to the target genes that were used for selection (Frisch and Melchinger 2001; Hospital 2001). This process is termed “linkage drag” and in many instances produces negative effects since there are negative alleles linked to the selected one. For this reason detailed characterization of a QTL that affects several diverse traits is essential in order to determine if the effect is pleiotropic or rather caused by linkage drag in which case the various QTL could be dissected. The analysis of segregating populations for the QTL mapping region is a powerful strategy to address such questions since if the multiple effects are caused by linkage then a sub-line that has only the phenotype of interest should be achievable. For this reason we here screened 1,600 gametes from an F2 population segregating for the IL2-1 genomic region. No recombination was observed between any of the measured traits at any of the recombinant lines (Fig. 4). This finding is an indication that the multi-phenotypic effect of IL2-1 is most likely a result of pleiotropic effect of a single gene, rather than of linkage between independent loci. This is of course only an assumption that can only be confirmed once the gene associated with this QTL is cloned.
Concerning the development determinants for the transition from vegetative to reproductive development some genes have been characterized (Samach et al. 2000; Cremer and Coupland 2003; El-Din El-Assal et al. 2003; Pineiro et al. 2003; Valverde et al. 2004) using the natural variation in flowering time among different Arabidopsis species (Alonso-Blanco et al. 1998; Koornneef et al. 1998; El-Din El-Assal et al. 2001; Alonso-Blanco et al. 2009). Based on these results and the conservation of this mechanism across species (Laurie 1997; Andersen et al. 2003; Yamasaki et al. 2005), these genes have also be investigated in other plant species (Murai et al. 2003; Izawa 2007; Jimenez-Gomez et al. 2007). Although the modified flowering pattern was only analyzed at the full-length introgression (IL2-1) and this phenotype was not mapped on the recombinant sub-lines, we personally strongly believe that the induced flowering is the cause of the pleiotropic effects of early maturation, reduced plant weight and Brix and increased harvest index. The induction of flowering caused by the IL2-1p allele is essentially the result of an accelerated transition from vegetative to reproductive growth. In the homozygous state the introgression results in a lack of setting of most flowers, a subsequent reduction in fruit number and a consequent reduction total yield. However, plants heterozygous for the introgression maintain the reduced plant weight, but exhibit better flower setting and display only a minor reduction in total yield (in comparison to M82). As a result lines heterozygous for this introgression display an increased harvest index.
We demonstrate the detailed characterization of a QTL which impacts the relationship between vegetative and reproductive development. The genetic dissection of this multi-phenotype QTL leads us to assume that its diverse effects most likely result from pleiotropic effects following the modulation of a single gene. We propose that the initial, causal, effect underlying these changes is the enhanced floral induction. Further support for this hypothesis is provided by the fact that when IL2-1 was tested in an indeterminate background (IL2-1p SP+) there was no Brix reduction in the fruits in comparison to the near-isogenic control (SP+), and the plants were visually indistinguishable from this control line (data not shown). This suggests that SP, which is known to be strongly epistatic over other genes which affect plant development (Fridman et al. 2002) is also epistatic over hi2-1. The fact that the Brix reduction, which is a major phenotypic effect of IL2-1 under determinate growth habit, was not observed in the indeterminate background indicates that this effect is most likely a result of the altered growth habit rather than an independent unassociated effect. However, to reiterate as stated above formal evidence in go in support (or conflict), of our theory will only be available following the cloning of the gene(s) underlying these traits.
Despite the increase in earliness and harvest index these beneficial traits come at the cost of decreased yield. However, as stated above in this current study we were not able to prove if these traits resulted by the pleiotropic effects of change in a single gene or rather the close linkage of two or more genes independently influencing these traits. Future work in which recombinants harboring smaller segments of the S. pennellii genome will be required to fully dissect this locus. Obviously if these traits effect of linkage of genes it may prove possible to segregate advantageous from deleterious phenotypes.
The QTL investigated unfortunately maps to a genomic region that is lacking in markers and rich in recombinations (both in our population and in that studied by Tanksley et al. 1992). These results when taken together with the fact that none of the new RAPD markers which we developed during this study and mapped to IL2-1 fell within this region, leads us to assume that this interval is likely to be a ‘hot-spot’ of recombination which deviates from the average bp/cM ratio of tomato. At the present moment we do not have any good estimation for the physical distance between the closest marker (TG31) and the QTL (although we confirmed that they are separated by a physical distance that is more than one BAC [~100 kb], as ends of BACs that were positive for the TG31 clone did not genetically map to different sides of the QTL; see “Materials and methods”). However, we believe that the distance between hi2-1 and TG31 is less than the expected by just multiplying the genetic distance with the average bp/cM ratio of tomato. As such we are confident that the results presented here represent a good basis for further investigation (and future cloning), of this QTL especially given that many more candidate genes can be anticipated to be uncovered given the imminent release of the tomato genome. We anticipate that the cloning of this gene will bring greater understanding of understanding both the process of resource allocation and the phenomenon of heterosis in the tomato in a similar manner to that of the recent cloning of the genes ANANTHA, COMPOUND INFLORESCENCE and SINGLE FLOWER TRUSS (Lippman et al. 2008; Krieger et al. 2010).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Metabolite profiles in red fruits. Data are normalized by the mean of M82 (M82 = 1.00). Values represent the calculated fraction of M82. Data points consist of the mean out of six replicates ± SE; bold type values were determined by the t test to be significantly different (P < 0.05) from M82. (DOC 173 kb)
Acknowledgments
We thank Gabi Gera and the Akko field experiment station group for their assistance with the field experiments. Financial support of the Deutsche Forshungsgemeinschaft to SO, ARF and DZ in the frame of the trilateral project “Drought Tolerant Sun-Dried Tomatoes: A novel Product Based on Heterotic Natural Biodiversity” (FE552/3-2) is gratefully acknowledged.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics. 1998;149(2):749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, Keurentjes JJB, Raymond M, Vreugdenhil D, Koornneef M. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CH, Jensen CS, Petersen K. Similar genetic switch systems might integrate the floral inductive pathways in dicots and monocots. Trends Plant Sci. 2003;9:105–107. doi: 10.1016/j.tplants.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Atherton JG, Rudich J. The tomato crop. London: Chapman and Hall; 1986. [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- Bernatzky R, Tanksley SD. Methods for detection of single or low-copy sequences in tomato on Southern blots. Plant Mol Biol Rep. 1986;4:37–41. doi: 10.1007/BF02672487. [DOI] [Google Scholar]
- Bugbee BG, Salisbury FB. Exploring the limits of crop productivity. I. Photosynthetic efficiency of wheat in high irradiance environments. Plant Physiol. 1988;88:869–878. doi: 10.1104/pp.88.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5(8):618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Carmel-Goren L, Liu YS, Lifschitz E, Zamir D. The SELF-PRUNING gene family in tomato. Plant Mol Biol. 2003;52(6):1215–1222. doi: 10.1023/B:PLAN.0000004333.96451.11. [DOI] [PubMed] [Google Scholar]
- Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- Cremer F, Coupland G. Distinct photoperiodic responses are conferred by the same genetic pathway in Arabidopsis and in rice. Trends Plant Sci. 2003;8:405–407. doi: 10.1016/S1360-1385(03)00192-4. [DOI] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Doganlar S, Mutschler MA, Tanksley SD. Identification and molecular mapping of loci controlling fruit ripening time in tomato. Theor Appl Genet. 2000;100:249–255. doi: 10.1007/s001220050033. [DOI] [Google Scholar]
- Doi K, Izawa T, Yamanouchi U, Kubo T, Shimatano Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-Iike gene expression independently of Hd1l. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnett CW. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096–1121. doi: 10.2307/2281208. [DOI] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29(4):435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Wagemaker C, Weller JL, Koornneef M. The role of cryptochrome 2 in flowering in Arabidopsis. Plant Physiol. 2003;133(4):1504–1516. doi: 10.1104/pp.103.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica. 1994;79:175–179. doi: 10.1007/BF00022516. [DOI] [Google Scholar]
- Eshed Y, Zamir D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics. 1995;141(3):1147–1162. doi: 10.1093/genetics/141.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Fridman E, Liu YS, Carmel-Goren L, Gur A, Shoresh M, et al. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol Genet Genomics. 2002;266(5):821–826. doi: 10.1007/s00438-001-0599-4. [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science. 2004;305:1786–1789. doi: 10.1126/science.1101666. [DOI] [PubMed] [Google Scholar]
- Frisch M, Melchinger AE. The length of the intact donor chromosome segment around a target gene in marker-assisted backcrossing. Genetics. 2001;157(3):1343–1356. doi: 10.1093/genetics/157.3.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004;2(10):E245. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur A, Semel Y, Osorio S, Friedmann M, Seekh S, Ghareeb B, Mohammad A, Pleban T, Gera G, Fernie AR, Zamir D (2010) Quantitative trait loci for yield in a tomato are predominately expressed by the shoot. Theor Appl Genet (submitted) [DOI] [PMC free article] [PubMed]
- Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kühn C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. Plant J. 2006;45:180–192. doi: 10.1111/j.1365-313X.2005.02572.x. [DOI] [PubMed] [Google Scholar]
- Hay RK. Harvest index: a review of its use in plant breeding and crop physiology. Ann Appl Biol. 1995;126:197–216. doi: 10.1111/j.1744-7348.1995.tb05015.x. [DOI] [Google Scholar]
- Hospital F. Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics. 2001;158(3):1363–1379. doi: 10.1093/genetics/158.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Charng YY, Chan MT. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002;130:618–626. doi: 10.1104/pp.006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J Exp Bot. 2007;58:3091–3097. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- Ji I, Chetelat RT. Homoeologous pairing and recombination in Solanum lycopersicoides monosomic addition and substitution lines of tomato. Theor Appl Genet. 2003;106:979–989. doi: 10.1007/s00122-002-1090-2. [DOI] [PubMed] [Google Scholar]
- Jimenez-Gomez JM, Alonso-Blanco C, Borja A, Anastasio G, Angosto T, Lozano R, Martinez-Zapata JM. Quantitative genetic analysis of flowering time in tomato. Genome. 2007;50:303–315. doi: 10.1139/G07-009. [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Perlson E, Gilad A, Konrad Z, Scolnik PA, Bar-Zvi D. Over-expression of the water and salt stress-regulated Asr1 gene confers an increased salt tolerance. Plant Cell Environ. 2004;27:1459–1468. doi: 10.1111/j.1365-3040.2004.01251.x. [DOI] [Google Scholar]
- Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Peeters AJ, Soppe W. Genetic control of flowering time in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:345–370. doi: 10.1146/annurev.arplant.49.1.345. [DOI] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet (in press) [DOI] [PubMed]
- Kroymann J, Mitchell-Olds T. Epistasis and balanced polymorphism influencing complex trait variation. Nature. 2005;435(7038):95–98. doi: 10.1038/nature03480. [DOI] [PubMed] [Google Scholar]
- Laurie DA. Comparative genetics of flowering time. Plant Mol Biol. 1997;35(1–2):167–177. doi: 10.1023/A:1005726329248. [DOI] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008;6(11):e288. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- Monna L, Lin X, Kojima S, Sasaki T, Yano M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor Appl Genet. 2002;104(5):772–778. doi: 10.1007/s00122-001-0813-0. [DOI] [PubMed] [Google Scholar]
- Murai K, Miyamae M, Kato H, Takumi S, Ogihara Y. WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 2003;44(12):1255–1265. doi: 10.1093/pcp/pcg171. [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 2005;137:611–622. doi: 10.1104/pp.104.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Liu YS, Budai-Hadrian O, Sela M, Carmel-Goren L, Zamir D, Fluhr R. Comparative genetics of nucleotide binding site—leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and arabidopsis. Genetics. 2000;155(1):309–322. doi: 10.1093/genetics/155.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro M, Gomez-Mena C, Schaffer R, Martinez-Zapater JM, Coupland G. Early bolting in short days is related to chromatin remodeling factors and regulates flowering in Arabidopsis by repressing FT. Plant Cell. 2003;15(7):1552–1562. doi: 10.1105/tpc.012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998;125(11):1979–1989. doi: 10.1242/dev.125.11.1979. [DOI] [PubMed] [Google Scholar]
- Prudent M, Bertin N, Génard M, Muños S, Rolland S, Garcia V, Petit J, Baldet P, Rothan C, Causse M (2010) Genotype-dependent response to carbon availability in growing tomato fruit. Plant Cell Environ. doi:10.1111/j.1365-3040.2010.02139.x [DOI] [PubMed]
- Rick CM. Controlled introgression of chromosomes of Solanum pennellii into Lycopersicon esculentum: segregation and recombination. Genetics. 1969;62:753–768. doi: 10.1093/genetics/62.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM. Further studies on segregation and recombination in backcross derivatives of a tomato species hybrid. Biol Zbl. 1972;90:209–220. [Google Scholar]
- Robson PR, McCormac AC, Irvine AS, Smith H. Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nat Biotechnol. 1996;14(8):995–998. doi: 10.1038/nbt0896-995. [DOI] [PubMed] [Google Scholar]
- Salamini F. Hormones and the green revolution. Science. 2003;302:71–72. doi: 10.1126/science.1090811. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- SAS Institute . JMP statistics and graphics guide: version 5. Cary: SAS Institute Inc.; 2002. [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippmann Z, Gur A, Zamir D. Overdominant quantitative trait loci for yield and fitness in tomato. Proc Natl Acad Sci USA. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semel Y, Schauer N, Roessner U, Zamir D, Fernie AR. Metabolite analysis for the comparison of irrigated and non-irrigated field grown tomato of varying genotype. Metabolomics. 2007;3:289–295. doi: 10.1007/s11306-007-0055-5. [DOI] [Google Scholar]
- Sinclair RS. Historical changes in harvest index and crop nitrogen accumulation. Crop Sci. 1998;38:638–643. doi: 10.2135/cropsci1998.0011183X003800030002x. [DOI] [Google Scholar]
- Tanksley SD. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 2004;16:S181–S189. doi: 10.1105/tpc.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, Ganal MW, Prince JP, de Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132(4):1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES. Dwarf8 polymorphisms associate with variation in flowering time. Nat Genetics. 2001;314:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of constans protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377(6549):495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BGm Ticconi C, Halkier BA, Kliebenstein DJ. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLOS Genetics. 2007;3:1687–1701. doi: 10.1371/journal.pgen.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, Yu SB, Xu CG, Li XH, Zhang QF. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yadav RS, Hash CT, Bidinger FR, Cavan GP, Howarth CJ. Quantitative trait loci associated with traits determining grain and stover yield in pearl millet under terminal drought-stress conditions. Theor Appl Genet. 2002;104(1):67–83. doi: 10.1007/s001220200008. [DOI] [PubMed] [Google Scholar]
- Yadav R, Hash C, Bidinger F, Devos K, Howarth C. Genomic regions associated with grain yield and aspects of post-flowering drought tolerance in pearl millet across stress environments and tester background. Euphytica. 2004;136(3):265–277. doi: 10.1023/B:EUPH.0000032711.34599.3a. [DOI] [Google Scholar]
- Yamamoto T, Lin H, Sasaki T, Yano M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics. 2000;154(2):885–891. doi: 10.1093/genetics/154.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki M, Tenaillon MI, Bi IV, Schroeder SG, Sanchez-Villeda H, Doebley JF, Gaut BS, McMullen MD. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, Sasaki T. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Kojima S, Takahashi Y, Lin H, Sasaki T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 2001;127(4):1425–1429. doi: 10.1104/pp.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Li XH, Zhang Q. Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor Appl Genet. 2002;104(4):619–625. doi: 10.1007/s00122-001-0772-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolite profiles in red fruits. Data are normalized by the mean of M82 (M82 = 1.00). Values represent the calculated fraction of M82. Data points consist of the mean out of six replicates ± SE; bold type values were determined by the t test to be significantly different (P < 0.05) from M82. (DOC 173 kb)