Abstract
Retinal pigment epithelial (RPE) cells are continually exposed to oxidative stress that contributes to protein misfolding, aggregation and functional abnormalities during aging. The protein aggregates formed at the cell periphery are delivered along the microtubulus network by dynein-dependent retrograde trafficking to a juxtanuclear location. We demonstrate that Hsp90 inhibition by geldanamycin can effectively suppress proteasome inhibitor, MG-132-induced protein aggregation in a way that is independent of HDAC inhibition or the tubulin acetylation levels in ARPE-19 cells. However, the tubulin acetylation and polymerization state affects the localization of the proteasome-inhibitor-induced aggregation. These findings open new perspectives for understanding the pathogenesis of protein aggregation in retinal cells and can be useful for the development of therapeutic treatments to prevent retinal cell deterioration.
1. Introduction
Retinal pigment epithelial (RPE) cells are exposed to chronic oxidative stress. They must constantly absorb light energy and phagocyte lipid rich photoreceptor outer segment shed from neural retina due to normal physiological visual cycle. Oxidative stress refers to progressive cellular damage that contributes to protein misfolding and functional abnormalities in the RPE cells during cellular senescence [1]. The accumulation of this damage in the postmitotic RPE cells seems to be one of the key events in the development of age-related macular degeneration (AMD), the leading cause of blindness in the elderly in the developed countries. The RPE cells ensure the survival of neural cells, rod, and cones. In senescent RPE cells, this ability is reduced causing secondary adverse effects on the neural retina, ultimately leading to loss of vision. Both intra- and extracellular aggregation processes are crucial in cell degeneration and AMD [2].
Efficient removal of misfolded proteins from cytoplasm is critical for cellular survival and adaptation. However, potentially toxic misfolded protein aggregates accumulate during the aging process [3, 4]. Control of protein turnover is particularly important in postmitotic cells, since the accumulation of malfunctioning proteins may be highly detrimental to the cells [5]. Once heat shock protein-mediated protein folding fails, the misfolded proteins are usually tagged with a ubiquitin (Ub) moiety that directs the complex to the ubiquitin/proteasomal protein degradation pathway (UPP) [6]. It is believed that the aggregation of oxidized and ubiquinated proteins is due to a decline in the proteasomal activity with age, and that this also occurs in RPE cells [7–10]. Protein aggregates formed in the cell periphery are delivered along microtubulus network by dynein-dependent retrograde trafficking to a juxtanuclear location where they form aggresomes [11–13]. The development of these aggresomes is part of a cellular defence mechanism against misfolded proteins [14], and it can be inhibited by drugs that depolymerize microtubules [15, 16].
Tubulin undergoes various posttranslational modifications including polyglutamylation, polyglycylation, carboxyterminal cleavage, and acetylation [17, 18]. Acetylation is unique among the known tubulin modifications, in that it occurs on the lysine 40 of α-tubulin which can be found on stable microtubules in most cell types [19]. The deacetylation of the lysine residue in tubulin is catalyzed by enzymes called histone deacetylases (HDACs). In contrast, histone acetyltransferases (HATs) transfer acetyl groups to lysine residue to increase acetylation of tubulin [20]. This results in a balance between acetylation and deacetylation states of tubulin, and any shift in this balance results in changes in the regulation of tubulin function [21]. The HDAC6 is the major cytoplasmic tubulin deacetylase [21–23]. Moreover, it efficiently binds mono- and polyubiquitin molecules [24–26]. One interesting role of tubulin and tubulin modifying deacetylases is their influence on aggregation of misfolded proteins [12]. HDAC6 contributes to the degradation of aggregated proteins because it is able to bind to both poly-ubiquitinated and dynein motor proteins as an adaptor protein to help transport misfolded proteins along microtubules into aggresomes which are finally degraded by autophagy [13, 22, 27–29]. HDAC6 is able to sense ubiquitinated cellular aggregates and consequently induces the expression of major cellular chaperones by triggering the dissociation of a repressive HDAC6/HSF1 (heat-shock factor 1)/HSP90 (heat-shock protein 90) complex and subsequent HSF1 activation. HDAC6 therefore appears as a master regulator of the cell protective response to cytotoxic protein aggregate formation [13].
We have recently shown that proteasome inhibition evoked perinuclear protein accumulation which then leads to the autophagy-mediated removal of the deposits [29]. In the present study, we demonstrate that Hsp90 inhibition with geldanamycin can effectively decrease the proteasome inhibitor-induced aggregation. Increased tubulin acetylation was observed in response to Hsp90, proteasome, and HDAC inhibition, but the acetylation level was not correlated with the amount of the aggregates in the ARPE-19 cells. However, the tubulin polymerization state did influence the localization of the aggregates.
2. Materials and Methods
2.1. Cell Culture and Treatments
Human retinal pigment epithelial cells (ARPE-19, from ATCC, [30]) were used in this study. The cells were grown in Dulbecco's MEM/Nut MIX F-12 (1 : 1) medium (Life Technologies, Paisley, UK) containing 10% fetal bovine serum (Hyclone, Logan, UT, USA), 2 mM L-glutamine (Cambrex, Charles City, IA, USA), 100 units/ml penicillin (Cambrex), and 100 μg/ml streptomycin (Cambrex). Before any exposure, the cells were grown to confluency in a standard incubator (10% CO2, +37°C). Proteasome inhibition was accomplished with 5 μM MG-132 (Calbiochem, San Diego, CA, USA). Geldanamycin (GA, Calbiochem), at 0.25 μM concentration, was used to inhibit the function of Hsp90. Microtubules were acetylated with 1 μM trichostatin A (TSA; Sigma-Aldrich, Steinheim, Germany). Taxol (TAX, Paclitaxel; Calbiochem), at 1 μM concentration and nocodazole (NOC; Calbiochem), at 5 μM concentration, were used to disrupt to function of microtubules. The cells were exposed to the GA, TSA, TAX, or NOC for 24 h, simultaneously with MG-132 for 24 h or allowed to recover after 24 h MG-132 insult in medium with or without chemicals (GA, TSA, TAX or NOC).
2.2. Phase Contrast Microscopy
Phase contrast microscopy photographs were taken from live cells with a Nikon Eclipse TE300 (Nikon, Tokyo, Japan) microscope.
2.3. Electron Microscopy
For transmission electron microscopy (TEM), the cell culture samples were prefixed in 2.5% glutaraldehyde (in 0.1 M phosphate buffer, pH 7.4) for 2 hrs at room temperature, followed by washing (15 min in 0.1 M phosphate buffer). The samples were postfixed in 1% osmium tetraoxide (in 0.1 M phosphate buffer) for 1 hr at room temperature and washed as before prior to standard ethanol dehydration. Subsequently, the samples were infiltrated and embedded in LX-112 resin (Ladd Research Industries, Burlington, VT, USA). Polymerization was carried out at 37°C for 24 hrs and at 60°C for 48 hrs. The sections were examined in a JEOL-1200EX transmission electron microscope (Jeol, Tokyo, Japan) at 80 kV.
2.4. Western Blotting
Exposed cells were lysed in M-Per lysis buffer (Thermo Scientific, Waltham, MA, USA). Protein concentrations were analysed with the Bradford (Coomassie Brilliant Blue dye) method [31]. Whole cell extracts (20 μg of protein) were run in 10% or 15% SDS-PAGE gels and then wet blotted to nitrocellulose membranes (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The membranes were blocked for 1 hour in 3% fat-free dry milk in 0.3% Tween 20/PBS at room temperature (RT). Thereafter, the membranes were incubated for 1 hour at RT with rabbit polyclonal ubiquitin antibody (DakoCytomation, Glostrup, Denmark, cat. no. Z 0458) or rat monoclonal Hsp90 antibody (Assay Designs, Ann Arbor, MI, USA, cat. no. SPA-835) or mouse monoclonal Hsp70 antibody (Assay Designs, cat. no. SPA-810) or rat monoclonal Hsc70 antibody (Assay Designs, cat. no. SPA-815) or rabbit polyclonal LC3 antibody (Abgent, San Diego, CA, USA, cat. no. AP1802a) except for monoclonal acetylated tubulin antibody (Sigma-Aldrich, cat. no. T6793) where the incubation was for 30 minutes at RT. Primary antibodies were diluted (1 : 500, 1 : 5 000, 1 : 5 000, 1 : 5 000, 1 : 250 or 1 : 8 000 resp.) in 0.5% bovine serum albumin in 0.3% Tween 20/PBS except for acetylated tubulin which was diluted in 1% fat-free dry milk in 0.05% Tween 20/PBS. After 3 × 5 minutes washes with 0.3% Tween 20/PBS (for acetylated tubulin 0.05% Tween 20/PBS) the membranes were incubated for one hour at RT with horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG antibodies or anti-rat IgG antibodies (GE Healthcare). The secondary antibodies were diluted (for ubiquitin 1 : 20 000, for Hsp90 1 : 10 000, for Hsp70 1 : 40 000, for Hsc70 1 : 15 000 and for LC3 1 : 5000, resp.) in 3% fat-free dry milk in 0.3% Tween 20/PBS except for acetylated tubulin (1 : 6 000) which was diluted in 1% fat-free dry milk in 0.05% Tween 20/PBS. Before detection, all of the membranes were washed as before. Protein antibody complexes were detected with an enhanced chemiluminescence method (Millipore, Billerica, MA, USA). The western blots were quantified with Quantity One software 4.5.0. (Bio-Rad, Hercules. CA, USA).
3. Results and Discussion
3.1. Hsps and Protein Aggregation
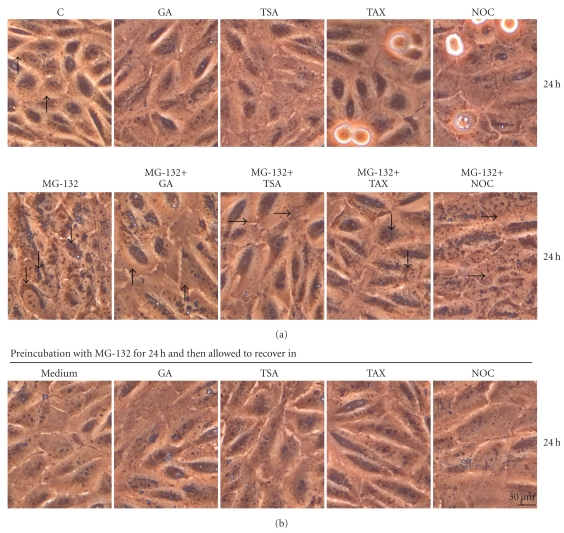
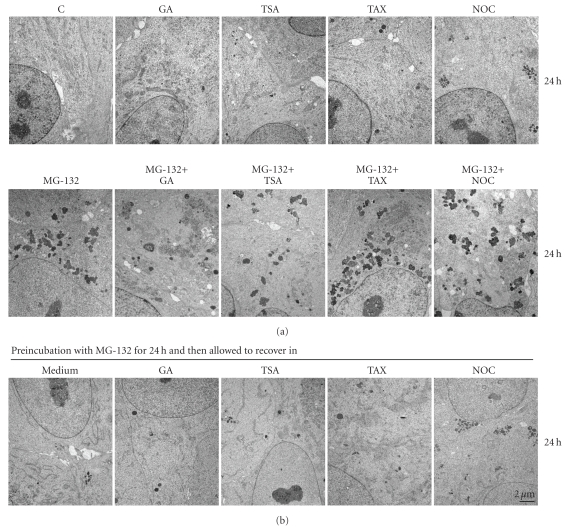
The ARPE-19 cells were either nonstressed, or exposed to drugs, that is, 5 μM MG-132 or 0.25 μM geldanamycin, 1 μM trichostatin A or 1 μM taxol or 5 μM nocodazole for 24 hours. In addition, the cells were treated simultaneously with MG-132 in conjunction with geldanamycin, trichosatin A, taxol or nocodazole up to 24 hours. Only a single MG-132 insult or combination treatment with taxol induced a typical perinuclear aggregation in ARPE cells (Figures 1(a) and 2(a)). Interestingly, Hsp90 inhibition with geldanamycin effectively suppressed MG-132 induced protein aggregation. HDAC inhibition with trichostatin A or tubulin depolymerization with nocodazole evoked dispersed the mid-peripherical aggregation process during proteasome inhibition. With all of the insults, cytoplasm underwent a similar effective aggregation clearance when exposed to MG-132 for 24 hours and then allowed to recover for 24 hours in normal cell culture medium (Figures 1(b) and 2(b)).
Figure 1.
(a) Phase contrast micrographs of control (C) cells, and cells exposed to 5 μM MG-132 (MG) or 0.25 μM geldanamycin (GA) or 1 μM trichostatin A (TSA) or 1 μM taxol (TAX) or 5 μM nocodazole (NOC) for 24 hours. In addition, the cells were treated simultaneously with GA or TSA or TAX or NOC and MG-132 for up to 24 hours, (b) and then allowed to recover under the indicated conditions for 24 hours. Protein aggregates are seen as dark granular deposits in perinuclear space (downward arrows), the examples of clear cytoplasm are marked with upward arrows. Arrows from left to right indicate disperse aggregation.
Figure 2.
(a) Transmission electron micrographs of control (C) cells, and cells exposed to 5 μM MG-132 (MG) or 0.25 μM geldanamycin (GA) or 1 μM trichostatin A (TSA) or 1 μM taxol (TAX) or 5 μM nocodazole (NOC) for 24 hours. In addition, the cells were treated simultaneously with GA or TSA or TAX or NOC and MG-132 for up to 24 hours, (b) and then allowed to recover under the indicated conditions for 24 hours.
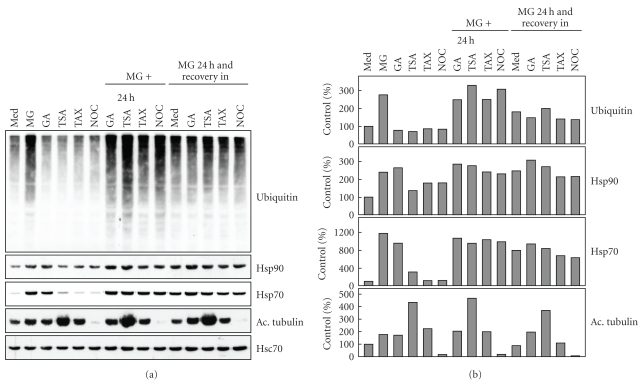
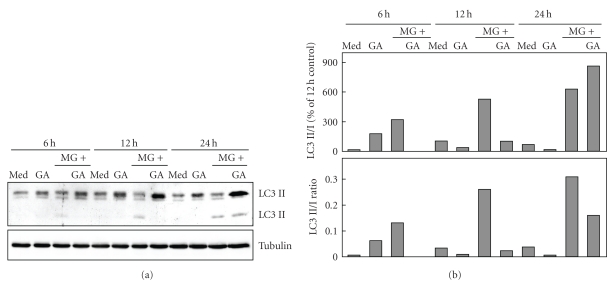
A robust elevation of Hsp70 protein expression was seen in response to MG-132 or geldanamycin exposures (Figure 3). Hsp90 levels were not markedly affected by the treatments. Abrogation of proteasome-mediated protein degradation caused a clear increase in the amount of Hsp70 which has a cytoprotective capacity in the MG-132 -treated ARPE-19 cells [29]. The classical transcriptional heat-shock gene induction is attributable to the activation of HSF1 transcription factor [32]. In the activation process Hsp90 and Hsp70 dissociate from HSF1 transcription factor [33, 34]. Geldanamycin has been shown to bind Hsp90, to inhibit its function and to elicit the Hsp90 client protein degradation in proteasomes [35, 36]. In line with previous studies [37–39], the geldanamycin was found to trigger a strong expression of Hsp70, while the Hsp90 response remained weaker. The response is likely mediated through HSF1 transcriptional activation [33, 34]. The increase in the amount of inducible Hsp70 might be one of the regulators suppressing the proteasome inhibitor-induced aggregation process [29], when Hsp90 is simultaneously inhibited. It has also been documented that a Hsp90 inhibitor may prevent the aggregation of protein by regulating client protein posttranslational modifications [40]. Since Hsp90 inhibition has been reported to trigger autophagy clearance [41, 42], we wished to analyze the autophagy induction marker LC3 I/II levels in ARPE cells treated with geldanamycin or MG-132 solely or both together for 6, 12, and 24 hours. Our findings clearly show that proteasome inhibition mildly induced autophagy, when related to LC3 II levels, but this was not involved in geldanamycin treatments (Figure 4). This indicates that autophagy clearance is not implicated in the suppression of protein aggregation during Hsp90 and proteasome inhibition in the ARPE-19 cells.
Figure 3.
(a) Total proteins (20 μg) of whole cell extracts were examined by western blot using antibodies against ubiquitin, Hsp90, Hsp70, acetylated tubulin, and Hsc70 of control (C) cells, and cells exposed to 5 μM MG-132 (MG) or 0.25 μM geldanamycin (GA) or 1 μM trichostatin A (TSA) or 1 μM taxol (TAX) or 5 μM nocodazole (NOC) for 24 hours. In addition, the cells were treated simultaneously with GA or TSA or TAX or NOC and MG-132 for up to 24 hours and then allowed to recover under the indicated conditions for 24 hours. Hsc70 was used to check the equal loading of proteins. (b) Quantifications of western blots.
Figure 4.
(a) Total proteins (20 μg) of whole cell extracts were examined by western blot using antibodies against LC3 of control (med) cells, and cells exposed 0.25 μM geldanamycin (GA) or 5 μM MG-132 (MG) or their combination for 6, 12, and 24 hours. Tubulin was used to check the equal loading of proteins. (b) Quantifications of western blots and LC3 II/I ratio.
3.2. Ubiquitination
Cellular ubiquitin-protein (Ub) conjugate levels were analyzed by western blotting from cells treated as described above. The level of Ub conjugation was not changed by geldanamycin, trichostatin A, taxol, or nocodazole treatment, but a very intensive induction of ubiquitination was seen in response to MG-132 exposure whether geldanamycin, trichostatin A, taxol, or nocodazole were present or not (Figure 3). Inhibition of Hsp90 therefore seems to have only a minor impact on the level of ubiquitination in the ARPE-19 cells, although Hsp90 has been shown to regulate both proteasomal and autophagy clearance through its client proteins [41–43]. The ubiquitin levels were slightly reduced when proteasome inhibition was removed and the cells recovered either with geldanamycin, trichostatin A, taxol, or nocodazole compared to incubations of MG-132 together with these chemicals. This is apparently explained by the autophagic clearance of proteasome-inhibitor-induced aggregates, as we have recently documented [29].
3.3. Acetylation
Protein aggregates formed at the cell periphery are delivered along the microtubulus network by dynein-dependent retrograde trafficking to a juxtanuclear location where they form aggresomes [11, 12]. The reversible acetylation of α-tubulin has been linked to the regulation of microtubule stability and function [21]. Decreased tubulin acetylation has been shown in some reports to reduce microtubule stability [21, 22], but in other experiments no association was observed [44, 45]. HDAC6 deacetylates tubulin, and Hsp90, and forms complexes with many other proteins, including ubiquitinated proteins. HDAC6 also interacts with a component in the dynein-dynactin microtubule motor complex and regulates protein aggregation trafficking [13, 45, 46]. Trichostatin A is a classical inhibitor of HDAC deacetylase activity and thus it also blocks the HDAC6 [47]. The phase contrast and transmission electron microscopic analysis revealed dispersed cytoplasmic aggregation in response to simultaneous treatment of trischostatin A or nocodazole and proteasome inhibition (Figures 1 and 2). Trichostatin A evoked a clear increase in tubulin acetylation levels, while in the situation of tubulin depolymerizator induced by nocodazole, tubulin acetylation remained at control level (Figure 3). In contrast, a tubulin stabilizer, taxol induced tubulin acetylation that was not, however, as extensive as that evoked by trichostatin A. Hsp90 and proteasome inhibition evoked a similar slight increase in the tubulin acetylation levels. All these findings indicate that total tubulin acetylation is not related to perinuclear aggregation. The HDAC inhibition by trichostatin A probably affects the dynein motor and regulates aggregation localization, but does not prevent the formation of aggregates. Moreover, the polymerization state of tubulin regulates the localization of proteasome inhibitor-induced aggregates, but this process cannot be estimated by measuring total tubulin acetylation levels. Hsp90 inhibition seems to prevent aggregation rather than regulating trafficking through the tubulin network.
4. Conclusions
Hsp90 inhibition is effectively involved in the regulation of protein aggregation that is independent of HDAC inhibition or tubulin acetylation levels in the RPE cells. These findings open new perspectives for understanding the pathogenesis of protein aggregation in retinal cells, and they may be useful in the development of therapeutic treatments to prevent retinal cell deterioration, that is, during aging.
Acknowledgments
This work was supported by the Academy of Finland, the Emil Aaltonen Foundation, the Finnish Cultural Foundation and its North Savo Fund, the Finnish Eye Foundation, the Finnish Eye and Tissue Bank Foundation, the Finnish Funding Agency for Technology and the Päivikki and Sakari Sohlberg Foundation. The authors wish to thank Dr. Ewen MacDonald for checking the language.
References
- 1.Kaarniranta K, Salminen A, Eskelinen E-L, Kopitz J. Heat shock proteins as gatekeepers of proteolytic pathways-Implications for age-related macular degeneration (AMD) Ageing Research Reviews. 2009;8(2):128–139. doi: 10.1016/j.arr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Kaarniranta K. Autophagy—hot topic in AMD. Acta Ophthalmologica. 2010;88(4):387–388. doi: 10.1111/j.1755-3768.2009.01840.x. [DOI] [PubMed] [Google Scholar]
- 3.Söti C, Csermely P. Protein stress and stress proteins: implications in aging and disease. Journal of Biosciences. 2007;32(3):511–515. doi: 10.1007/s12038-007-0050-z. [DOI] [PubMed] [Google Scholar]
- 4.Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging—a mini-review. Gerontology. 2009;55(5):550–558. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terman A, Kurz T, Navratil M, Arriaga EA, Brunk UT. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxidants and Redox Signaling. 2010;12(4):503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickart CM. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 7.Carrard G, Bulteau A-L, Petropoulos I, Friguet B. Impairment of proteasome structure and function in aging. International Journal of Biochemistry and Cell Biology. 2002;34(11):1461–1474. doi: 10.1016/s1357-2725(02)00085-7. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes AF, Zhou J, Zhang X, et al. Oxidative inactivation of the proteasome in retinal pigment epithelial cells: a potential link between oxidative stress and up-regulation of interleukin-8. Journal of Biological Chemistry. 2008;283(30):20745–20753. doi: 10.1074/jbc.M800268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Wang Y-S, Shen X-F, et al. Alterations of activity and intracellular distribution of the 20S proteasome in ageing retinal pigment epithelial cells. Experimental Gerontology. 2008;43(12):1114–1122. doi: 10.1016/j.exger.2008.08.052. [DOI] [PubMed] [Google Scholar]
- 10.Jung T, Catalgol B, Grune T. The proteasomal system. Molecular Aspects of Medicine. 2009;30(4):191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends in Cell Biology. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao T-P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 13.Boyault C, Zhang Y, Fritah S, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes and Development. 2007;21(17):2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor JP, Tanaka F, Robitschek J, et al. Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Human Molecular Genetics. 2003;12(7):749–757. doi: 10.1093/hmg/ddg074. [DOI] [PubMed] [Google Scholar]
- 15.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. Journal of Cell Biology. 1998;143(7):1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Mata R, Bebök Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. Journal of Cell Biology. 1999;146(6):1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webster DR, Borisy GG. Microtubules are acetylated in domains that turn over slowly. Journal of Cell Science. 1989;92(1):57–65. doi: 10.1242/jcs.92.1.57. [DOI] [PubMed] [Google Scholar]
- 18.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 19.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Current Opinion in Cell Biology. 2008;20(1):71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schemies J, Sippl W, Jung M. Histone deacetylase inhibitors that target tubulin. Cancer Letters. 2009;280(2):222–232. doi: 10.1016/j.canlet.2009.01.040. [DOI] [PubMed] [Google Scholar]
- 21.Matsuyama A, Shimazu T, Sumida Y, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO Journal. 2002;21(24):6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO Journal. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seigneurin-Berny D, Verdel A, Curtet S, et al. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Molecular and Cellular Biology. 2001;21(23):8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hook SS, Orian A, Cowley SM, Eisenman RN. Histone deacetylase 6 binds polyubiquitin through its zinc finger (PAZ domain) and copurifies with deubiquitinating enzymes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(21):13425–13430. doi: 10.1073/pnas.172511699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyault C, Gilquin B, Zhang Y, et al. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO Journal. 2006;25(14):3357–3366. doi: 10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. Journal of Biological Chemistry. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 28.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447(7146):859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 29.Ryhänen T, Hyttinen JMT, Kopitz J, et al. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. Journal of Cellular and Molecular Medicine. 2009;13(9b):3616–3631. doi: 10.1111/j.1582-4934.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Experimental Eye Research. 1996;62(2):155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259(5100):1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 33.Abravaya K, Myers MP, Murphy SP, Morimoto RI. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes and Development. 1992;6(7):1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 34.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 35.Sreedhar AS, Soti C, Csermely P. Inhibition of Hsp90: a new strategy for inhibiting protein kinases. Biochimica et Biophysica Acta. 2004;1697(1-2):233–242. doi: 10.1016/j.bbapap.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 36.Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell. 1997;89(2):239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 37.Sittler A, Lurz R, Lueder G, et al. Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington’s disease. Human Molecular Genetics. 2001;10(12):1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 38.Mimnaugh EG, Xu W, Vos M, et al. Simultaneous inhibition of hsp 90 and the proteasome promotes protein ubiquitination, causes endoplasmic reticulum-derived cytosolic vacuolization, and enhances antitumor activity. Molecular Cancer Therapeutics. 2004;3(5):551–566. [PubMed] [Google Scholar]
- 39.Kaarniranta K, Ryhänen T, Sironen RK, et al. Geldanamycin activates Hsp70 response and attenuates okadaic acid-induced cytotoxicity in human retinal pigment epithelial cells. Molecular Brain Research. 2005;137(1-2):126–131. doi: 10.1016/j.molbrainres.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Luo W, Dou F, Rodina A, et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(22):9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn PF, Mesires NT, Vine M, Dice JF. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1(3):141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 42.Riedel M, Goldbaum O, Schwarz L, Schmitt S, Richter-Landsberg C. 17-AAG induces cytoplasmic α-synuclein aggregate clearance by induction of autophagy. PLoS One. 2010;5, article e8753 doi: 10.1371/journal.pone.0008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress and Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palazzo A, Ackerman B, Gundersen GG. Tubulin acetylation and cell motility. Nature. 2003;421(6920):p. 230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- 46.Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends in Cell Biology. 2008;18(6):291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Jose B, Okamura S, Kato T, Nishino N, Sumida Y, Yoshida M. Toward an HDAC6 inhibitor: synthesis and conformational analysis of cyclic hexapeptide hydroxamic acid designed from α-tubulin sequence. Bioorganic and Medicinal Chemistry. 2004;12(6):1351–1356. doi: 10.1016/j.bmc.2004.01.014. [DOI] [PubMed] [Google Scholar]