Abstract
While copy number variation (CNV) is an active area of research, de novo mutation rates within human populations are not well characterized. By focusing on large (>100 kbp) events, we estimate the rate of de novo CNV formation in humans by analyzing 4394 transmissions from human pedigrees with and without neurocognitive disease. We show that a significant limitation in directly measuring genome-wide CNV mutation is accessing DNA derived from primary tissues as opposed to cell lines. We conservatively estimated the genome-wide CNV mutation rate using single nucleotide polymorphism (SNP) microarrays to analyze whole-blood derived DNA from asthmatic trios, a collection in which we observed no elevation in the prevalence of large CNVs. At a resolution of ∼30 kb, nine de novo CNVs were observed from 772 transmissions, corresponding to a mutation rate of μ = 1.2 × 10−2 CNVs per genome per transmission (μ = 6.5 × 10−3 for CNVs >500 kb). Combined with previous estimates of CNV prevalence and assuming a model of mutation-selection balance, we estimate significant purifying selection for large (>500 kb) events at the genome-wide level to be s = 0.16. Supporting this, we identify de novo CNVs in 717 multiplex autism pedigrees from the AGRE collection and observe a fourfold enrichment (P = 1.4 × 10−3) for de novo CNVs in cases of multiplex autism versus unaffected siblings, suggesting that many de novo CNV mutations contribute a subtle, but significant risk for autism. We observe no parental bias in the origin or transmission of CNVs among any of the cohorts studied.
Copy number variants (CNVs) are known to affect a wide range of human phenotypes (Lupski et al. 1991; Edwards et al. 2005; Aitman et al. 2006; Fellermann et al. 2006; The International Schizophrenia Consortium 2008; Slavotinek 2008; Weiss et al. 2008). Despite its importance, the de novo CNV mutation rate has remained elusive due to limited sample size and source material. Genome-wide estimates of the CNV mutation rate have previously been based on extrapolation from mutation rates estimated from individual loci (van Ommen 2005; Lupski 2007; Turner et al. 2008) or on indirect estimates using population genetic-based approaches (Conrad et al. 2009). However, the former is affected by locus-specific variability, while the latter assumes all CNVs are neutral mutations. Direct estimates of the genome-wide CNV mutation rate from family studies have been performed in at least two studies (Sebat et al. 2007; Xu et al. 2008) but have each been based on few observations (two or less) in relatively small sample sizes (N < 200), with one of these studies having a resolution of ∼100 kb (Sebat et al. 2007). A third study screened a large collection of trios and parent–child pairs for de novo CNVs to identify loci to test for an association to schizophrenia; however, because parent–child pairs were not ascertained for duplications (Stefansson et al. 2008), it is not possible to use this information to estimate CNV mutation rates without introducing substantial downward bias. Here, we seek to expand on previous work and estimate the genome-wide CNV formation rate through the direct identification of de novo events in a large number of trios from three different population sources (HapMap, familial autism, and asthmatic trios). We consider potential artifacts that arise from cell line versus primary tissue, and use the latter to estimate the de novo CNV mutation rate. By comparing these estimates with the population prevalence, we estimate the extent of selection operating on large CNVs (>500 kbp). Moreover, we show that different mutation processes contribute disproportionately to CNVs related to the size of the de novo event.
Results
In this study, we focused on the identification and characterization of larger de novo CNVs (median, ∼150 kbp), leveraging single nucleotide polymorphism (SNP) microarray data from three sample collections (Table 1). We consider each of these analyses independently.
Table 1.
Overview of data sets
aThe relatively large number of excluded trios is due to higher stringency for CNV discovery (see Methods).
HapMap analysis
The HapMap Phase I trios represent one of the most well-characterized data sets for human genetic variation and include about 60 trios of Northern European and Yoruban Nigerian descent (The International HapMap Consortium 2005). After quality control, 54 complete trios were analyzed on the Illumina 1MDuo platform (Table 1). There was no significant difference in the number of CNVs per individual between trio children and either parent (Supplemental Table 1). Within the complete trios, we manually screened 1366 CNVs identified in children for de novo CNVs (Supplemental Table 2). After several additional filters, we identified seven candidate de novo CNVs ranging in size from 25 to 260 kb (Supplemental Tables 2, 3). Among the inherited events that could be assigned to a single parent, there was no significant difference between maternal and paternal inheritance (374 vs. 333, P = 0.1324 binomial test) (Supplemental Table 2).
We attempted to validate four candidate de novo CNVs identified in the CEU trios by performing array comparative genomic hybridization (CGH) on DNA samples from the extended CEPH pedigrees (Supplemental Figs. 1, 2). A truly de novo CNV would be unobserved in the first generation (CEU trio parents), validated in the second generation (CEU trio children), and, assuming no selective effects, transmitted to approximately half of the individuals in the third generation. Observing transmission of a CNV would serve to distinguish between a true CNV within the germline versus a potential cell line artifact. While all four CNVs were validated by array-CGH in the second generation, transmission of these CNVs was never observed in any of the 23 grandchildren tested (Supplemental Figs. 2–4). Segregation analysis of flanking microsatellites and SNPs (CEPH genotype database, http://www.cephb.fr/en/cephdb/) in the pedigrees confirmed that in all families, both haplotypes near the predicted CNV were transmitted to at least one individual in the third generation without transmission of the associated CNV (Supplemental Figs. 5, 6). We conclude these putative de novo CNVs likely represent cell line artifacts that arose during passaging of cell cultures or represent potential somatic mosaicisms that were cloned during the establishment of the cell culture. Thus, we observed no true de novo CNVs in this data set (Table 2).
Table 2.
Summary of de novo CNVs identified
aParentheses indicate number of putative de novo CNVs if both CNVs in three instances of potential germline mosaicism are weighted equal to other CNVs.
Asthma analysis
Due to the inherent problem with utilizing cell line–derived material, we sought out a large familial DNA data set derived from primary tissue. We selected a sample collection of 492 trios (386 after CNV-specific quality control [QC]; Methods) collected for studies of asthma where both parents and children had provided blood samples as the source of DNA (Hancock et al. 2009). Cases of nonpaternity in this data set had previously been removed, and testing SNPs for Mendelian concordance additionally excluded large-scale uniparental disomy (see Methods). Although children in this sample showed mild to moderate asthma based on clinical symptoms or response to treatment of a pediatric allergist, we found no significant difference in the number of CNVs called per individual with respect to parent versus child (P = 0.65, Wilcoxon rank-sum test) (Supplemental Table 4).
Solely for examining the frequency of large CNVs, we included both children with (N = 386) or without (N = 25) complete parental data (N = 411, total). Approximately 9% (35/411) of children in these trios carried a CNV ≥ 500 kb and 3% (13/411) carried a CNV ≥ 1 Mb. Neither frequency is significantly different from previous frequency estimates in the general population (see Methods; P = 0.60 and 0.12, respectively, two-tailed Fisher's exact test). Among inherited events, there were no significant differences in the paternally versus maternally inherited CNVs (490 vs. 522, P = 0.3298, binomial test) (Supplemental Table 5). These analyses imply that asthmatics are unlikely to be enriched in large CNVs and that this cohort allows estimates of de novo rates of CNVs applicable to the general population.
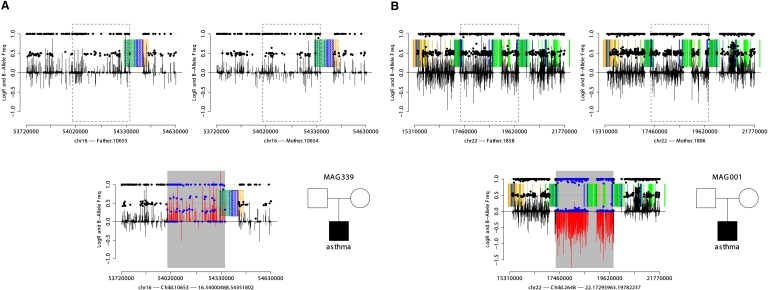
Among the children, we identified 11 out of 2025 CNVs for which a corresponding CNV was not observed in either parent (Fig. 1; Table 3; Supplemental Table 5). Validation was carried out using conventional array-CGH with custom NimbleGen arrays (Table 3; Supplemental Fig. 7; Supplemental Table 6). DNA was available to attempt the validation of nine candidate de novo CNVs. For all available parental DNA, no CNVs were detected at these loci. In the children, eight of nine loci validated as copy number changes. The smallest event, a 33-kb deletion, failed to validate and was excluded from further analysis. An additional CNV that validated was subsequently removed from further analysis based on its overlap with a known site of copy number polymorphism (CNP), leaving seven de novo CNVs validated by array-CGH. Based on the high rate of validation in samples for which DNA was available and the lack of overlap with known CNPs, the two putative CNVs for which DNA was not available likely represent true de novo CNVs and were included in along with the seven validated de novo CNVs in further analyses. In summary, manual screening for de novo CNVs recovered 11 candidate events, two of which we later excluded, yielding nine de novo CNVs from 386 trios. Finally, we note that two individuals (10421, 2648) sharing the same deletion at 22q11 (chr22:17.2–19.7M) were confirmed using SNP genotypes to represent distinct samples as opposed to mislabeled replicates.
Figure 1.
An example of de novo duplication (A) and deletion (B) detected by SNP array data. SNP array data from the father, mother, and child are displayed as shown in the pedigree in the lower-right portion of each panel. Each plot shows LogR Ratio (vertical bars), B-allele frequency (solid points), intrachromosomal segmental duplications in direct orientation (green blocks), inverted orientation (blue blocks), and interchromosomal segmental duplications (orange blocks). CNVs are highlighted by gray rectangles, contrasting the LogR ratio (red) and B-allele frequency (blue) with flanking regions (black). The de novo duplication (A) is characterized by an increase in the LogR ratio and altered clustering of heterozygote B-allele frequencies not seen in either parent. The de novo deletion (B) displays a decreased LogR ratio and loss of heterozygosity not observed in either parent.
Table 3.
de novo CNVs identified in asthma trios
ND, Not determined.
We estimate the genome-wide frequency of de novo CNVs to be 1.2 × 10−2 (nine of 772) per haploid genome per generation (μ = 6.5 × 10−3 for CNVs >500 kb) (Fig. 2). This estimate is not significantly different from rates observed in control samples in previous studies with smaller sample sizes (Supplemental Table 7; Supplemental Fig. 8; Sebat et al. 2007; Xu et al. 2008). The de novo CNVs ranged in size from 62 kb to 10 Mb (Table 3). Although our estimate is not significantly different from previous estimates, it is likely to be conservative, as it does not account for CNVs overlapping CNPs, regions for which there is inadequate probe coverage, and regions, such as segmental duplications (SDs), that are often refractory to CNV detection using array-based techniques. If the distribution of CNP is taken as an indicator of how many CNVs are missed due to SDs, it is possible that this estimate may be 15%–30% higher (Kidd et al. 2008; McCarroll et al. 2008; Conrad et al. 2009).
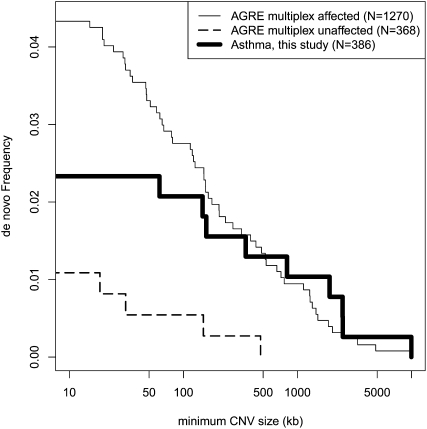
Figure 2.
Observed frequency of de novo CNVs as a function of minimum CNV size.
SDs in both direct and inverted orientation flanked three of the nine de novo CNVs identified, and an additional de novo CNV had SDs adjacent to its distal breakpoint (Table 3). A duplication at chr16:15.4M is flanked by 138 kb of sequence with >99% identity in direct orientation and 176 kb of sequence with >98% identity in inverted orientation. Similarly, two deletions at chr22:17.3M are flanked by 162 kb of sequence with 99% identity in direct orientation and 126 kb of sequence with >96% identity in inverted orientation. The presence of SDs in direct orientation suggests these CNVs were generated via nonallelic homologous recombination (NAHR). In contrast, none of the CNVs observed in the HapMap were flanked at either breakpoint by SDs, although this depletion was not significant compared with the asthma data set (zero of seven vs. three of nine, P = 0.21, two-sided Fisher's exact test) (Table 3; Supplemental Table 3).
De novo CNV mutations and selection
Because we ascertained our CNVs using the same platform and CNV discovery procedure described previously (Itsara et al. 2009), we assume comparable sensitivity and false-negative rates in detecting large CNVs. We therefore estimated the average genome-wide selection coefficient for large CNV deletions and duplications using a simple mutation-selection balance model (Methods; Supplemental Methods). For CNVs >500 kb, μ = (5/772) = 6.5 × 10−3. By using a control set of CNVs (Methods), we estimate the frequency of CNVs >500 kb in the population to be q = 0.04. Assuming a dominant effect of CNVs (h = 1), this yields s = 0.16 (95% confidence interval [CI] 0.02–0.31). The CI on this estimate is large, mainly owing to uncertainty in the CNV mutation rate (Supplemental Methods). A conservative, but more precise, estimate of the mutation rate can be obtained by combining de novo CNV rates from this study and other studies that have detected CNVs using lower-resolution arrays (Sebat et al. 2007; Stefansson et al. 2008). One of these studies examined about 10,000 transmissions, but about 5500 were from parent–child pairs for which no duplications and only a subset of deletions could be detected (Stefansson et al. 2008). This conservative estimate of the mutation rate >500 kb corresponds to a selection coefficient s = 0.09 (95% CI 0.06–0.12) (Supplemental Table 8).
It should be noted that this estimate does not apply to regions of the genome where there is inadequate probe coverage to estimate both mutation rate and allele frequency, including many duplication-rich regions of the genome that remain poorly ascertained on even the best-available SNP platforms (Cooper et al. 2008). In regions where there is adequate probe coverage, this estimate is based off a mutation rate estimate that excludes known sites of CNP and therefore likely represents a lower bound.
AGRE autism analysis
Numerous studies have shown that individuals with neurocognitive disease are enriched for large de novo CNVs (de Vries et al. 2005; Sharp et al. 2006; Sebat et al. 2007; Walsh et al. 2008). This effect is most pronounced for simplex cases compared with familial cases where slight but insignificant increases in the de novo rate have been observed (Sebat et al. 2007) compared with unaffected individuals. We re-examined SNP microarray data generated from one of the largest collections of autism (AGRE [Autism Genetics Resource Exchange]) in an effort to compare de novo rates between affected and unaffected siblings. After accounting for pedigree information, QC, and monozygotic individuals in multiple births, 3896 distinct individuals from the AGRE data set were available for further CNV analysis. From this sample set, it was possible to form 1757 trios, including 1638 multiplex and 119 simplex pedigrees (Table 1). There was no significant difference in the number of CNVs per individual between parents and children (Supplemental Table 9).
Of 10,839 CNV calls examined, 7616 were assigned as inherited (Supplemental Table 10). For 1454 of the 7616 inherited CNVs, a single parental origin could not be assigned. Among the remaining 81%, there was no significant difference in the rate of maternal versus paternal inheritance of CNVs (3103 vs. 3059, P = 0.5838, binomial test).
Overall, the fraction of probands with a CNV >500 kb was 9.9% (233/2352) and 3.7% (88/2352) for CNVs >1 Mb. Compared with a set of Caucasian controls analyzed on the same platform (N = 1370; Methods), this represents a slight but significant elevation in the occurrence of large CNVs (odds ratio [OR] = 1.31, P = 0.03 and OR = 2.09, P = 0.001, respectively, two-tailed Fisher's exact test). We found that large CNVs in the AGRE data set were more likely to affect genes than were CNVs found among controls (1.86-fold enrichment, P = 0.0073, two-sided Wilcoxon rank-sum test); this remained significant after further correcting for CNV size (P = 0.03, two-sided Wilcoxon rank-sum test). These enrichments are strikingly similar to previously observed enrichments (case-control ratios of 1.32 and 1.79 for CNV frequency and gene content, respectively) in a study of rare CNVs and schizophrenia (The International Schizophrenia Consortium 2008). While enrichment in frequency of large CNVs in autism may in part be due to subtle ascertainment differences, our results benefit from having ascertained our CNVs using the same calling algorithm and genotyping platform across all cases and controls.
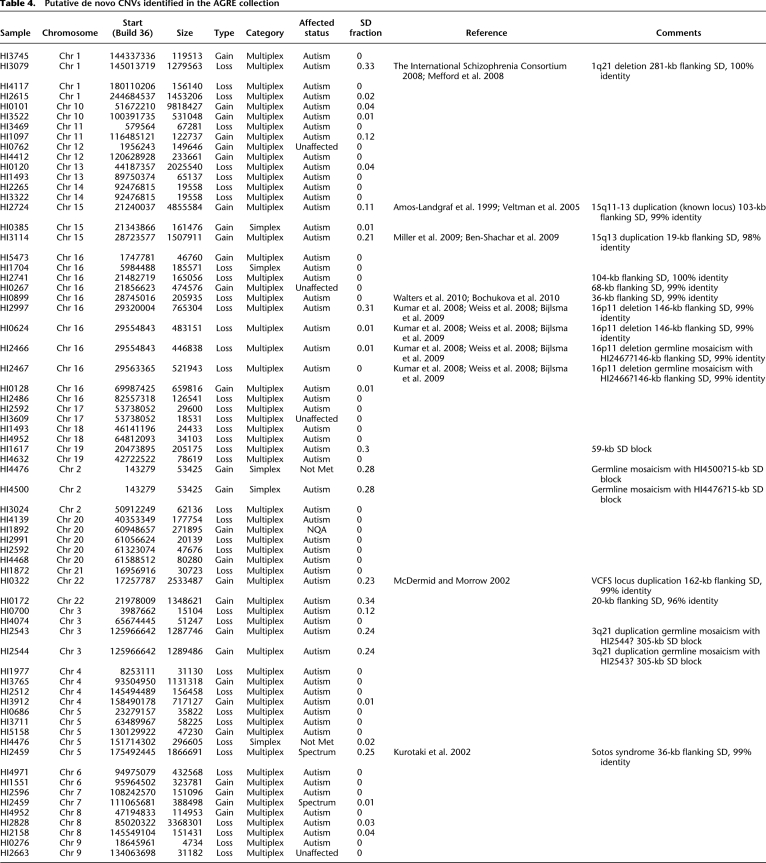
After removing CNVs overlapping known sites of somatic rearrangement, excluding trios that failed familial validation, and excluding CNVs whose phase could not be deduced based on overlap with CNPs, there were 67 putative de novo CNVs (62 in multiplex trios, five in simplex) representing 64 independent events (Tables 2, 4; Supplemental Table 10). In three instances, de novo CNVs were identified in a pair of nonmonozygotic siblings representing potential germline mosaicism and were counted as single events. Several de novo CNVs occur both at loci previously associated with variable phenotypes, including autism (Amos-Landgraf et al. 1999; McDermid and Morrow 2002; Veltman et al. 2005; The International Schizophrenia Consortium 2008; Kumar et al. 2008; Mefford et al. 2008; Weiss et al. 2008; Ben-Shachar et al. 2009; Bijlsma et al. 2009; Miller et al. 2009), as well as sites of recurrent CNV not previously associated with autism (Kurotaki et al. 2002; Bochukova et al. 2010; Walters et al. 2010).
Table 4.
Putative de novo CNVs identified in the AGRE collection
To our knowledge, our analysis represents the largest number of samples in the AGRE collection systematically screened for de novo CNVs (Supplemental Table 11). To assess the performance of our analysis, we compared previously reported de novo CNVs in five analyses (Sebat et al. 2007; Szatmari et al. 2007; Kumar et al. 2008; Weiss et al. 2008; Bucan et al. 2009) of AGRE families to our results (Supplemental Table 12). We identify 59 de novo CNVs previously unreported in these five studies. Excluding six CNVs that have apparently been mislabeled in the Bucan et al. (2009) study and one CNV too small to detect in the Szatmari et al. (2007) study, we were able to identify 18 of 20 previously reported de novo CNVs. Subsequently, we confirmed eight as de novo CNVs, excluded eight as inherited events or mosaic deletions, and could not confirm or exclude the remaining two CNVs due to unavailable parental sample data (Supplemental Fig. 9; Supplemental Table 12).
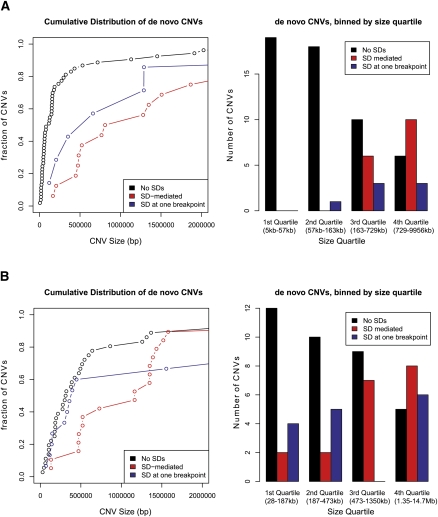
Over one-quarter (17/64) of de novo CNVs in the AGRE collection were flanked by SDs in direct orientation (12/64) or had one of the breakpoints mapping within a cluster of SDs (Table 2). We refer to the latter as SD-associated to distinguish it from events likely created due to NAHR. Combining these results with those from the asthma trios shows a strong trend toward larger de novo events being preferentially associated or mediated by SDs (Fig. 3A). Based on the median size of events (∼165 kbp), we find that larger events are more likely to be mediated (P = 0.001, two-tailed Fisher's exact test) by SDs. To eliminate potential artifacts arising from cell line passage, we repeated the analysis with a set of 70 de novo CNVs derived from blood (Stefansson et al. 2008) and observed a similar trend (Fig. 3B).
Figure 3.
Comparison of de novo CNV size and potential underlying mechanisms. We classified de novo events into three categories: segmental duplication (SD)-mediated (CNV breakpoints were flanked by directly orientated SDs), SD at one breakpoint (a cluster of SDs one side), or no SDs (no SDs were identified). SDs were defined as segments >1 kbp and >90% sequence identity. (A) Using de novo events from the AGRE and asthma trios, the cumulative distribution (scatter plot) of CNV size and the frequency of CNV classes in each CNV size quartile are shown. Both SD-mediated and associated events are significantly enriched for de novo events above the median size (P = 3.1 × 10−7 and 0.018 respectively, two-tailed Fisher's exact test) where they account for 63% (24/38) of the events. (B) The analysis was repeated for a recent analysis of controls obtained from blood DNA (Stefansson et al. 2008). In this study, SD-mediated events were enriched for events above the median CNV size (P = 0.00964, two-tailed Fisher's exact test), while SD-associated events were not (P = 1, two-tailed Fisher's exact test).
A small number (N = 60) of samples in the AGRE collection represent individuals with simplex autism, many of which were previously analyzed by Sebat et al. (2007). After correcting for overlapping sample sets, the rate of de novo CNVs in the study of Sebat et al. (2007) remains significantly elevated. However, the small number of AGRE simplex autism cases in our analysis does not permit statistical distinction between de novo rates observed in the study of Sebat et al. (2007) and unaffected individuals (Supplemental Table 13).
Among the 1638 multiplex autism trios, 1270 had an affected child and 368 had an unaffected child (Table 2). Although cell line artifacts are expected to be enriched in the AGRE collection, we found no significant difference in the rate of de novo CNVs (Fig. 2) between individuals with asthma (nine of 772) and unaffected (four of 736) individuals of the multiplex autism pedigrees (P = 0.27, two-sided Fisher's exact test). Notably, we did observe a dramatic difference in rate of de novo mutation between affected and unaffected siblings from the same autism families. Our analysis indicates that the frequency of de novo CNV carriers is enriched approximately fourfold for affected versus unaffected individuals (P = 1.6 × 10−3, two-sided Fisher's exact test). The statistical significance is even greater when considering only individuals with a strict diagnosis of autism (P = 9.2 × 10-4) (Supplemental Table 14) and is unlikely the result of cell line artifacts since there were no systematic differences in the rate of growth during establishment of cell lines from affected and unaffected individuals (D. Fugman, pers. comm.). Interestingly, this effect remains when separately considering CNVs <500 kb or >500 kb in size (P = 0.03 for both) (Supplemental Table 15). If there is an existing predisposition to autism within a family, an otherwise low-risk de novo CNV may possibly exacerbate and result in phenotypic consequences (Girirajan et al. 2010).
Parental origin of de novo CNVs
By analyzing the B-allele frequency (BAF) in de novo CNVs, we were able to unambiguously determine the parental origin for 47 of 73 de novo CNVs in the asthma, AGRE multiplex autism, and AGRE simplex autism data sets (Table 2; Supplemental Table 16). We identified 21 paternal and 26 maternal de novo events (mean size = 1.25 Mb and 1.19 Mb, respectively) and therefore no evidence for a parent-of-origin preference (P = 0.56). This trend holds irrespective of the study design or the relationship of these events to SDs (Supplemental Table 16). Although the number of CNVs that could be assigned a parental origin was relatively small (N = 47), we have 95% power to detect a bias in the fraction of CNVs arising in one parent versus another of 0.26 or more from the null hypothesis of 0.5.
Discussion
Using Illumina 550K SNP array data generated with peripheral blood DNA, we estimate a genome-wide de novo CNV rate of 1.2 × 10−2 events per transmission per generation (95% CI 5.3 × 10−3–2.2 × 10−2). This estimate is consistent with previous studies' estimates ranging from 0.5%–3% (Supplemental Fig. 8; Sebat et al. 2007; Xu et al. 2008; Conrad et al. 2009). As expected, our estimate falls below that calculated using high-resolution CNV arrays and above that of lower-resolution arrays (Sebat et al. 2007; Conrad et al. 2009). Our estimate has the benefit of being a direct estimate based on approximately twice the number of trios as previous work Although there is less uncertainty in our estimate of the genome-wide CNV mutation rate, it is likely to be conservative as it does not account for regions of the genome not adequately covered by our platform, such as SDs or common sites of CNV as these regions were excluded in our analysis. Conversely, SDs and CNPs cover ∼5%–6%, so that our estimate is largely applicable to ∼94% of the human genome.
The extent to which the exclusion of CNPs has biased our estimate of the de novo CNV mutation rate is difficult to determine. Common CNVs may be of higher frequency by virtue of a combination of an elevated mutation rate and/or reduced purifying selection. Under the assumption that CNPs are under the same degree of purifying selection as de novo CNVs, we have identified here, this bias could be significant (15%–30%) (Kidd et al. 2008; McCarroll et al. 2008; Conrad et al. 2009). However, previous studies of common CNVs have suggested that most diallelic CNPs with a minor allele frequency of >5% are largely ancestral mutations (McCarroll et al. 2008; Conrad et al. 2009) and that many CNVs observed at ∼1% within a population share a single mutational origin (McCarroll et al. 2008). These observations suggest that much of the increased frequency of CNPs is not due to a substantially increased mutation rate but rather relaxed purifying selection compared with rare CNVs.
Whereas previous estimates of de novo point mutation rates range from about 35–90 substitutions per generation (Nachman and Crowell 2000; Kondrashov 2003; Roach et al. 2010), mean CNV sizes of 693 kb and 2 Mb in the AGRE and asthma data sets, respectively, suggest an average of 8–25 kbp de novo CNV sequence per genome. Thus, although de novo CNVs occur at lower overall frequency than point mutations, about 100-fold more base pairs of sequence are affected per generation. A comparison between the chimpanzee and human genomes reported only a fourfold difference in the number of CNV base pairs versus single–base pair substitutions (Cheng et al. 2005; The Chimpanzee Sequencing and Analysis Consortium 2005). We propose that this discrepancy reflects a large difference in the selection pressure operating on new CNV mutations versus de novo SNPs.
Our calculated de novo CNV rate is based on trios from Mexico City for which the child was affected with asthma. Accordingly, there are several sources of potential bias in our de novo CNV rate estimate that may affect its generalizability. First, some of the events we considered as validated de novo CNVs may reflect somatic mosaicism. While the allelic balance in the de novo CNVs detected suggests that they are not mosaic events within blood, we are unable to exclude the possibility that these are somatic events specific to hematopoietic cells. Second, the age of children of our trios at the time of collection was between 5 and 17 yr (Hancock et al. 2009), and neurocognitive disease cannot be definitively excluded. If de novo CNVs cause disease with onset later in life, then the prevalence of de novo CNVs will be biased upward relative to an unaffected cross-section of the population. Finally, if de novo CNVs contribute to the risk of asthma, then our estimate of the CNV mutation rate will be elevated by a factor approximately equal to the average relative risk of asthma in individuals with a de novo CNV. The extent to which de novo CNVs contribute to the risk of asthma is unclear, but our results suggest no enrichment compared with other groups unaffected for neurocognitive disease (Supplemental Table 8).
Two of the de novo CNVs observed in our asthma data set were 22q11 deletions for which the phenotype has previously characterized as associated with Velo-Cardio facial syndrome (VCFS) or DiGeorge syndrome. It is unclear whether individuals with 22q11 deletions are at increased risk for asthma (Staple et al. 2005). Compared with previous prevalence estimates of one in 3900, observing two deletions in 386 represents a significant elevation (P = 0.02, two-sided Fisher's exact test). It is unlikely that these individuals were erroneously ascertained individuals with cardiovascular malformations, as the classification of asthma was based on spirometry. However, one in 3900 is believed to represent a minimal estimate of the prevalence of VCFS, having been based on ascertainment of individuals with cardiac anomalies (Goodship et al. 1998; Botto et al. 2003; Oskarsdottir et al. 2004). In contrast, 22q11 deletions are known to be highly variable in presentation (Bassett et al. 2005; Kobrynski and Sullivan 2007), with reports in some cases of individuals only being identified after diagnosis in their children (McDonald-McGinn et al. 2001), making it unclear whether this represents a significant elevation above the true prevalence of VCFS deletions.
Unlike SNPs or chromosomal aneuploidy events, we find no parent of origin effect, with roughly equal numbers arising from mother and father irrespective of disease study. These results are consistent with a meiotic-based mechanism such as unequal crossover, as opposed to replication-based mechanisms (Lee et al. 2007), as the primary force for driving CNV formation. For example, if replication-based mechanisms were responsible for most de novo CNV formation (Lee et al. 2007), one might expect a paternal bias, as has been observed for point mutations (Crow 2000). One should caution that the number of de novo events in this study is relatively few (47), and the events are in general quite large (median = 150 kbp). Different mutational mechanisms contribute disproportionately to the spectrum of structural variation, and as smaller events are more routinely ascertained, we might expect to discover different biases.
A detailed analysis of the underlying de novo events identified in this study suggest that larger deletion and duplication events are much more likely to be mediated by duplicated sequences. This observation is consistent with previous reports that indicated that the role of NAHR is more pronounced for larger CNV events compared with small events (Tuzun et al. 2005; Korbel et al. 2007; Conrad et al. 2009). Among putative NAHR events, the similarity and length of the SD are extraordinary (average, 98.8% with an average length of 124 kbp). While we find an excess of deletions versus de novo duplications (31 vs. 42) for all events, this trend reverses itself for events in excess of 500 kbp (13 gains vs. nine losses), consistent perhaps with stronger prenatal selection. Ten of 16 de novo events greater than 1 Mbp in size were either bracketed by SDs or had one of their breakpoints mapping within the vicinity of SDs (Fig. 3). The former is consistent with a NAHR of directly orientated duplicated sequences. The latter, however, suggests some other SD-associated mechanism that does not depend directly on sequence similarity but rather is a reflection of the instability conferred by these regions during repair or replication (Lee et al. 2007). The observation of so many SD-associated events may help to explain the phenomenon of primate duplication shadowing whereby lineage-specific duplications were 10 times more likely to occur adjacent to regions of SD (Cheng et al. 2005).
Estimating the CNV mutation rate on the Illumina 550K SNP arrays allows us to compare the frequency of large de novo CNVs to that previously observed in the general population using the same platform and examine the potential effects of selection. A value of s = 0.16 (95% CI 0.02–0.31) suggests that large (>500 kb) CNVs on average are deleterious. Although the CI on this estimate is wide, even the most conservative estimates suggest that s ∼ 0.1 (Supplemental Table 8). However, the relation between negative selection and clinical phenotype remains to be elucidated. For example, the value of s for large CNVs is comparable to previously calculated values for porphyria variegata, lipoid proteinosis, and BRCA1 mutations (Supplemental Table 17; Møorch and Andersen 1941; Crow 1986; Stine and Smith 1990; Pavard and Metcalf 2007). Porphyria variegata has a wide range of manifestations ranging from skin sensitivity to mental retardation, suggesting a broad range of symptoms upon which negative selection may act (Hift et al. 2004). In contrast, negative selection at BRCA1 has been hypothesized to occur only on the fraction of individuals with onset of cancer early enough to affect fertility (Pavard and Metcalf 2007). Finally, although the clinical manifestations of lipoid proteinosis are inherited in an autosomal recessive manner, heterozygotes are believed to be selectively disadvantageous with s = 0.07 under a dominant model of selection (Stine and Smith 1990).
The results presented here are consistent with previously proposed models of neuropsychiatric disease (Zhao et al. 2007; Girirajan et al. 2010). Zhao and colleagues, for example, put forward a model of autism risk in which families fall into two major categories: those in which the overall risk for autism is low, representing the majority of families, and those in which the risk is much higher due to a disease allele with a dominant mode of transmission and sex-dependent penetrance. Under this model, simplex or sporadic autism represents the situation in which autism occurs in a low-risk family due to a spontaneous mutation of high penetrance, whereas multiplex autism occurs due to the inheritance of an existing allele from a mildly affected or asymptomatic parent.
In another model, Girirajan et al. (2010) proposed the necessity in some families for a second mutational hit to lead to severe neuropsychiatric disease. He observed that individuals with childhood developmental delay are enriched approximately fourfold for a rare 520-kb 16p12 deletion. In nearly all cases examined (22/23), the deletion was inherited. Thus, similar to the model proposed by Zhao et al. (2007), 16p12 deletions appear to be an example of inherited predisposition to neuropsychiatric disease with dominant transmission. However, these individuals were more likely to carry a second large (>500 kb) CNV than were matched controls, and clinical features of those with a “second-hit” were typically more severe and recognizable than those with the 16p12 deletion alone. This supports a disease model in which the presence of a 16p12 deletion by itself results in predisposition to disease and, in combination with other risk-conferring variants, can exacerbate neuropsychiatric phenotypes. Thus, for childhood developmental delay (1) there exist rare variants of moderate to high risk that (2) alone or in combination with one another may result in neuropsychiatric disease. In particular, the observed enrichment of second-hit large CNVs suggests that such genetic variants appear to fulfill criteria 1 and 2 and supports the claim that such variants are under purifying selection.
We propose that multiplex autism pedigrees represent a situation where there is an existing inherited predisposition to neuropsychiatric disease, but this alone is not sufficient to cause disease. Secondary insults such as large CNVs as observed in Girirajan et al. (2010) or de novo CNVs as observed in this study are required to manifest as autism. Consequently, the observation of de novo CNVs disproportionately among affected siblings compared with unaffected siblings can be thought of as a depletion of second-hits in the unaffected sib due to this sensitized familial background. The abundance of inherited and de novo CNVs in the general population provides ample opportunity for multiple affected persons to appear within families. We propose that many of these CNVs, by themselves, are not fully penetrant but in combination with other genetic factors provide a molecular etiology for autism and as such are ultimately eliminated by selection. Finally, it is interesting to note that we observe this enrichment for de novo CNVs smaller than 500 kb, suggesting that a broad size range of CNVs, many of which are rare, may act as moderate risk variants for neuropsychiatric diseases.
Methods
SNP microarray and sample collections
For the purposes of CNV detection, samples were required to pass initial QC applied by the Illumina BeadStudio Genotyping Module as well as additional CNV-specific QC filters (Supplemental Methods). Complete trios were screened for de novo CNVs only if the child passed CNV-specific QC filters. In order to estimate a genome-wide de novo CNV mutation rate, we considered SNP microarray data obtained from three different sources: (1) Illumina 1M Duo genotype data from 60 HapMap trios (54 after CNV-specific QC) obtained from 89 CEU and 90 YRI lymphoblastoid cell lines (GEO accession nos. GSE16894 and GSE16896). (2) Cell line–derived DNA from 4271 individuals in multiplex and simplex AGRE (http://www.agre.org) pedigrees genotyped on the Illumina HumanHap550v1 and v3 SNP array originally generated at Children's Hospital of Pennsylvania (Bucan et al. 2009). After validation of familial relationships (see below) and CNV-specific QC, this included 1757 validated trios, of which 1638 and 119 were from multiplex and simplex pedigrees, respectively. (3) The final source was blood DNA from trios in which the child was affected by asthma. Illumina HumanHap550 SNP microarray data were published previously (Hancock et al. 2009). Probands consisted of children aged 5- to 17-yr-old with a mild-to-modest asthma diagnosis given by a pediatric allergist at the allergy referral clinic of a large public pediatric hospital in central Mexico City (Hospital Infantil de México, Federico Gómez). Four-hundred-ninety-eight case–parent triads were genotyped at the University of Washington, Department of Genome Sciences. After initial quality control, 492 complete trios with validated parentage remained in this data set. Three-hundred-eighty-six trios remained after additional CNV-specific QC filters. Note these QC metrics are more stringent for CNVs than for SNP genotype calls. In addition, we generated a CNV call-set from additional control samples. Illumina Hap550 genotyping data used for this purpose consisted of lymphoblastoid cell line DNA from 671 neurologically normal Caucasian controls from a study of Parkinson's disease (dbGaP accession no. phs000089; Simon-Sanchez et al. 2007) and peripheral blood DNA from 699 individuals of European ancestry from the InCHIANTI study of aging (http://www.inchiantistudy.net; Melzer et al. 2008).
CNV discovery
Discovery of de novo CNVs was carried out as an extension of a previously validated method that identifies CNVs from Illumina SNP microarray data (Itsara et al. 2009). Illumina GenomeStudio software was used to generate two summary quantities for each SNP: the LogR ratio, representing the total signal intensity, and the BAF, representing the allelic balance. In order to minimize batch effects, the LogR ratio and BAF for a given sample were generated using genotype clusters defined by the batch in which each sample itself was originally run. Thus, the HapMap sample genotypes and intensities were generated using the default GenomeStudio cluster file (defined by the HapMap itself), the asthma intensities were based off a cluster file generated by the asthma samples themselves, and similarly for the AGRE collection. Due to difficulties normalizing the LogR ratio and BAF in sample collections with both males and females, CNV discovery was restricted to autosomes.
A hidden Markov model (HMM) examines the LogR Ratio and BAF in order to identify duplications, deletions, and homozygous deletion events. Briefly, we specify a four-state HMM corresponding to copy number 0 through 3 based on LogR intensities, transformed into standard normal measurements (Z-scores) over a chromosome, and the square root of a quantity we termed the b-deviation. The b-deviation of a probe was defined as the deviation from the expected BAF given the genotype. For homozygotes, this was defined as the minimum of BAF and 1 − BAF, while for heterozygotes, this was defined as the absolute value of (BAF − 0.5). For failed genotypes or CNV probes, the b-deviation was the minimum value of BAF, 1 − BAF, and the absolute value of (BAF − 0.5). All analyses were performed using human genome assembly Build 36 (hg18) coordinates and were restricted to autosomes. Further details on CNV discovery are available in Supplemental Methods.
De novo CNV identification
Because of relatively high false-negative rates, it is insufficient to use CNV calls from the CNV discovery phase alone to identify candidate de novo events. We estimated that manual curation outside of SDs has high sensitivity (100% and 94.7% for the Illumina 550K and Illumina 1MDuo platforms, respectively) to detect CNVs at a given locus and therefore used it as a screen against inherited CNVs (Supplemental Methods; Supplemental Tables 18, 19). For each CNV identified in a trio child from the discovery phase, SNP array data at the same locus were manually inspected in each member of the corresponding trio. CNVs in each child were then annotated as maternal inheritance, paternal inheritance, unclear inheritance (cases in which either parent could have transmitted a CNV), inheritance from both parents (for homozygous deletions), potential de novo events, or ambiguous. Although not explicitly filtered out, all CNVs flagged as potentially de novo using this technique had >50% of their length outside of SDs. For all data sets, the number of CNVs marked as false-positives were consistent with the previously estimated true-positive rate for our CNV discovery procedure (Itsara et al. 2009). For the AGRE data set, this process was partly automated by marking CNV calls in children with more than 50% mutual overlap with a CNV call in either parent as inherited.
Candidate de novo CNVs were subsequently subjected to three filters: removal of common sites of immune somatic rearrangement or complexity (>50% overlap with immunoglobulin or T-cell receptor loci, or HLA) (Supplemental Table 20), exclusion of sites of common CNPs (McCarroll et al. 2008) because of an inability to unambiguously resolve phase (N = 33), and confirmation of familial relationships. Since the HapMap was from the higher-resolution Illumina 1M Duo platform, candidate de novo CNVs in the HapMap were additionally required to contain less than 50% mutual overlap with any other CNV detected in the HapMap.
To confirm chromosomal transmission and exclude errors in sample labeling and large-scale uniparental disomy, 100 random SNPs were selected on each autosome (2200 SNPs total) and tested for Mendelian inheritance. Parent–child relationships were confirmed by assessing Mendelian inheritance of SNP genotypes (see Supplemental Methods).
Parental origin was determined for each de novo event by analyzing the transmission of the SNP alleles from each parent to the child. In duplications, this was done by using the BAF to genotype heterozygous SNPs as either “AAB” (BAF < 0.4) or “ABB” (BAF > 0.6), thereby determining the duplicated allele. For all CNVs in which informative genotypes existed, SNP inheritance was consistent with inheritance from one parent. De novo CNVs were classified as SD-mediated if flanked by paralogous SDs in direct orientation (Bailey et al. 2001), or as SD-associated if one of two breakpoints mapped within a cluster of SDs (>90% identity; >1 kbp).
Array CGH validation and sensitivity estimates
Validation experiments for de novo events were completed with array CGH using a customized design (NimbleGen 12x135K) targeting de novo events. To determine whether a targeted region of N consecutive probes was validated, we compared the mean signal in the targeted region to mean signals in all windows containing N probes on the array not predicted to be a CNV. A CNV was considered validated if its mean signal corresponded to a P-value < 0.05.
With respect to CNV discovery, the sensitivity of our method in regions with sufficient probe coverage was previously estimated at ∼60% with a true-positive rate >77% (Itsara et al. 2009). Consistent with previous estimates of the true-positive rate, we were able to validate predicted copy number changes for eight of nine CNVs tested.
Frequency of large CNVs in the general population
The frequency of large CNVs in the general population was estimated using Illumina 550K SNP array data from a total of 1370 individuals. This control included SNP genotypes from both lymphoblastoid cell lines (N = 671) and peripheral blood material (N = 699; see above). Previously, we observed, using our CNV discovery technique, that the use of cell line DNA versus peripheral blood DNA is not a major contributor to estimates of CNV burden (Itsara et al. 2009). The fraction of individuals with at least one CNV >500 kb and >1 Mb was 7.7% (106/1370) and 1.8% (25/1370), respectively. These values are similar to previously reported estimates (Itsara et al. 2009). The number of CNVs per haploid genome for CNVs >500 kb was 0.04 (109/[1370 × 2]).
Population genetic analysis
We calculated the selection coefficient based on modifications of the classic mutation-selection balance model (Haldane 1932; Crow 1986) by constructing two models assuming either linked mutations at distinct loci with no recombination or unlinked mutations within a haploid genome (Supplemental Methods). If the limit that the genome-wide mutation rate (μ) and equilibrium frequency of mutation-bearing genomes (q) is small, both models converge to the classical approximation that s = μ/q.
De novo CNV enrichment calculation in AGRE pedigrees
Relative CNV enrichment was calculated within multiplex autism pedigrees using all children, defined as individuals with SNP array data for both parents. In the cases of monozygotic siblings, a single representative was included in the analysis. Individual phenotypes were determined with the AGRE phenotypic database (http://www.agre.org) using the preassigned “Scored Affected Status,” which categorizes affected individuals as autism (meets diagnostic criteria for autism using the ADI-R scoring algorithm), not quite autism (NQA), or broad spectrum. The latter two classifications represent individuals failing to meet the strict criteria for autism and may represent a wide range of phenotypes. Relative CNV enrichment was initially calculated between all affected (autism, NQA, and broad spectrum) and unaffected individuals. Additionally, the enrichment calculation was stratified comparing autism or the combination of broad spectrum and NQA classifications against unaffected individuals.
Acknowledgments
We thank Huiling Li, NIEHS, for expert technical assistance. We thank Grace Chiu at Westat Inc. (Research Triangle Park, NC) and Shuangshuang Dai and John Grovenstein at the National Institute of Environmental Health Sciences for data management. We thank the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families for provided resources. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). We also thank D. Fugman for technical support with the AGRE samples. We thank A. Singleton, L. Ferrucci, and investigators of the InCHIANTI study for sharing control genotype data generated with support from the Intramural Research Program of the National Institute of Aging, National Institutes of Health, Department of Health and Human Services. We also thank T. Brown, S. Girirajan, G.M. Cooper, and P. Green for critical review of the manuscript. This work was supported by a grant from the Simons Foundation (SFARI 137578 to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute. Subject enrollment and Illumina genotyping of the Mexican asthma study were supported by the Intramural Research Program of the National Institute of Environmental Health Sciences (NIEHS; ZIA ES049019 and ZIA ES025045). Subject enrollment was supported in part by the National Council of Science and Technology, Mexico (grant 26206-M).
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE23645.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.107680.110.
References
- Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. 2006. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439: 851–855 [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, Cassidy SB, Driscoll DJ, Rogan PK, Schwartz S, Nicholls RD 1999. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 65: 370–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res 11: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, Husted J, Weksberg R, Caluseriu O, Webb GD, Gatzoulis MA 2005. Clinical features of 78 adults with 22q11 deletion syndrome. Am J Med Genet A 138: 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, Franco LM, Malphrus A, Bottenfield GW, Spence JE, et al. 2009. Microdeletion 15q13.3: A locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet 46: 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, van Haeringen A, Fransen van de Putte DE, Anderlid BM, Lundin J, Lapunzina P, Perez Jurado LA, Delle Chiaie B, et al. 2009. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet 52: 77–87 [DOI] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O'Rahilly S, et al. 2010. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature 463: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto LD, May K, Fernhoff PM, Correa A, Coleman K, Rasmussen SA, Merritt RK, O'Leary LA, Wong LY, Elixson EM, et al. 2003. A population-based study of the 22q11.2 deletion: Phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 112: 101–107 [DOI] [PubMed] [Google Scholar]
- Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, Sonnenblick LI, Alvarez Retuerto AI, Imielinski M, Hadley D, Bradfield JP, et al. 2009. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 5: e1000536 doi: 10.1371/journal.pgen.1000536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, Khaitovich P, Graves T, Osoegawa K, Church D, DeJong P, Wilson RK, Paabo S, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437: 88–93 [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87 [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. 2009. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA 2008. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 40: 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF 1986. Basic concepts in population, quantitative, and evolutionary genetics. W.H. Freeman, New York [Google Scholar]
- Crow JF 2000. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet 1: 40–47 [DOI] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, Janssen IM, Reijmersdal S, Nillesen WM, Huys EH, Leeuw N, et al. 2005. Diagnostic genome profiling in mental retardation. Am J Hum Genet 77: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R III, Abel KJ, Manning A, Panhuysen C, Farrer LA 2005. Complement factor H polymorphism and age-related macular degeneration. Science 308: 421–424 [DOI] [PubMed] [Google Scholar]
- Fellermann K, Stange DE, Schaeffeler E, Schmalzl H, Wehkamp J, Bevins CL, Reinisch W, Teml A, Schwab M, Lichter P, et al. 2006. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet 79: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, et al. 2010. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet 42: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodship J, Cross I, LiLing J, Wren C 1998. A population study of chromosome 22q11 deletions in infancy. Arch Dis Child 79: 348–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS 1932. The causes of evolution. Longmans, Green and Co, London [Google Scholar]
- Hancock DB, Romieu I, Shi M, Sienra-Monge JJ, Wu H, Chiu GY, Li H, del Rio-Navarro BE, Willis-Owen SA, Weiss ST, et al. 2009. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in Mexican children. PLoS Genet 5: e1000623 doi: 10.1371/journal.pgen.1000623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hift RJ, Meissner D, Meissner PN 2004. A systematic study of the clinical and biochemical expression of variegate porphyria in a large South African family. Br J Dermatol 151: 465–471 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium 2005. A haplotype map of the human genome. Nature 437: 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium 2008. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455: 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, et al. 2009. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet 84: 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrynski LJ, Sullivan KE 2007. Velocardiofacial syndrome, DiGeorge syndrome: The chromosome 22q11.2 deletion syndromes. Lancet 370: 1443–1452 [DOI] [PubMed] [Google Scholar]
- Kondrashov AS 2003. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat 21: 12–27 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH Jr, Dobyns WB, et al. 2008. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet 17: 628–638 [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, et al. 2002. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 30: 365–366 [DOI] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lupski JR 2007. Genomic rearrangements and sporadic disease. Nat Genet 39: S43–S47 [DOI] [PubMed] [Google Scholar]
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, et al. 1991. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66: 219–232 [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. 2008. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet 40: 1166–1174 [DOI] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE 2002. Genomic disorders on 22q11. Am J Hum Genet 70: 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Tonnesen MK, Laufer-Cahana A, Finucane B, Driscoll DA, Emanuel BS, Zackai EH 2001. Phenotype of the 22q11.2 deletion in individuals identified through an affected relative: Cast a wide FISHing net! Genet Med 3: 23–29 [DOI] [PubMed] [Google Scholar]
- Mefford H.C, Sharp A.J, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney V.K, Crolla J.A, Baralle D, et al. 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med.359: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, et al. 2008. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet 4: e1000072 doi: 10.1371/journal.pgen.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Shen Y, Weiss LA, Korn J, Anselm I, Bridgemohan C, Cox GF, Dickinson H, Gentile J, Harris DJ, et al. 2009. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J Med Genet 46: 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møorch ET, Andersen H 1941. Chondrodystrophic dwarfs in Denmark. E. Munksgaard, Copenhagen [Google Scholar]
- Nachman MW, Crowell SL 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsdottir S, Vujic M, Fasth A 2004. Incidence and prevalence of the 22q11 deletion syndrome: A population-based study in western Sweden. Arch Dis Child 89: 148–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavard S, Metcalf CJ 2007. Negative selection on BRCA1 susceptibility alleles sheds light on the population genetics of late-onset diseases and aging theory. PLoS ONE 2: e1206 doi: 10.1371/journal.pone.0001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, et al. 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328: 636–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. 2007. Strong association of de novo copy number mutations with autism. Science 316: 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. 2006. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 38: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, et al. 2007. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet 16: 1–14 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM 2008. Novel microdeletion syndromes detected by chromosome microarrays. Hum Genet 124: 1–17 [DOI] [PubMed] [Google Scholar]
- Staple L, Andrews T, McDonald-McGinn D, Zackai E, Sullivan KE 2005. Allergies in patients with chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) and patients with chronic granulomatous disease. Pediatr Allergy Immunol 16: 226–230 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. 2008. Large recurrent microdeletions associated with schizophrenia. Nature 455: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine OC, Smith KD 1990. The estimation of selection coefficients in Afrikaners: Huntington disease, porphyria variegata, and lipoid proteinosis. Am J Hum Genet 46: 452–458 [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, et al. 2007. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet 39: 319–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, Beck S, Hurles ME 2008. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat Genet 40: 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- van Ommen GJ 2005. Frequency of new copy number variation in humans. Nat Genet 37: 333–334 [DOI] [PubMed] [Google Scholar]
- Veltman MW, Craig EE, Bolton PF 2005. Autism spectrum disorders in Prader-Willi and Angelman syndromes: A systematic review. Psychiatr Genet 15: 243–254 [DOI] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. 2008. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320: 539–543 [DOI] [PubMed] [Google Scholar]
- Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, Falchi M, Chen F, Andrieux J, Lobbens S, et al. 2010. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature 463: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. 2008. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358: 667–675 [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M 2008. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet 40: 880–885 [DOI] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, et al. 2007. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci 104: 12831–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]