Abstract
Approximately 98% of mammalian DNA is noncoding, yet we understand relatively little about the function of this enigmatic portion of the genome. The cis-regulatory elements that control gene expression reside in noncoding regions and can be identified by mapping the binding sites of tissue-specific transcription factors. Cone-rod homeobox (CRX) is a key transcription factor in photoreceptor differentiation and survival, but its in vivo targets are largely unknown. Here, we used chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) on CRX to identify thousands of cis-regulatory regions around photoreceptor genes in adult mouse retina. CRX directly regulates downstream photoreceptor transcription factors and their target genes via a network of spatially distributed regulatory elements around each locus. CRX-bound regions act in a synergistic fashion to activate transcription and contain multiple CRX binding sites which interact in a spacing- and orientation-dependent manner to fine-tune transcript levels. CRX ChIP-seq was also performed on Nrl−/− retinas, which represent an enriched source of cone photoreceptors. Comparison with the wild-type ChIP-seq data set identified numerous rod- and cone-specific CRX-bound regions as well as many shared elements. Thus, CRX combinatorially orchestrates the transcriptional networks of both rods and cones by coordinating the expression of photoreceptor genes including most retinal disease genes. In addition, this study pinpoints thousands of noncoding regions of relevance to both Mendelian and complex retinal disease.
Photoreceptors are the first point of contact between our nervous system and the outside world, serving to transform light energy into visual signals in the retina (Rodieck 1998). Over 200 Mendelian loci have been implicated in inherited retinal disease (RetNet database, http://www.sph.uth.tmc.edu/Retnet/), and the majority of the causative genes that have been identified are specifically expressed or enriched in photoreceptors (Blackshaw et al. 2001). Defects in photoreceptor genes result in dysfunction and death of photoreceptors, often leading to blindness (Rattner et al. 1999). Despite the high correlation between gene expression and disease vulnerability, there is currently no systems-level understanding of how transcriptional regulation is globally coordinated in the retina. Defining the architecture of the mammalian photoreceptor transcription network is therefore a major objective of vision science research.
The structure of a cell type–specific transcriptional regulatory network is determined by the transcription factors which the cell expresses, their target genes, and the cis-regulatory elements which mediate interaction between the two. A range of different transcription factors has been shown to contribute to photoreceptor gene regulation with a particularly prominent role attributed to CRX, NRL, and NR2E3 (Furukawa et al. 1999; Akhmedov et al. 2000; Mears et al. 2001). Although significant progress has been made in the computational identification of putative photoreceptor regulatory elements (Qian et al. 2005; Hsiau et al. 2007), our knowledge of the cis-regulatory regions relevant to photoreceptor gene expression remains incomplete.
During retinal development, photoreceptor cell fate is established by the expression of the homeodomain transcription factor, OTX2, in retinal progenitor cells (Nishida et al. 2003). This transcription factor then activates expression of another homeodomain transcription factor, CRX, in presumptive rods and cones (Chen et al. 1997; Furukawa et al. 1997). CRX expression is subsequently maintained in both photoreceptor cell types into adulthood. Mutations in human CRX are associated with several retinal diseases including cone-rod dystrophy and a severe form of blindness in newborns known as Leber's congenital amaurosis (Freund et al. 1997, 1998; Jacobson et al. 1998; Sohocki et al. 1998). CRX has been shown to influence the expression of many genes in the retina and is critical for normal photoreceptor differentiation (Furukawa et al. 1999; Livesey et al. 2000; Blackshaw et al. 2001; Hsiau et al. 2007). Crx mutant mice fail to form outer segments, the photosensitive organelles of photoreceptors, and lack detectable photoreceptor function (Furukawa et al. 1999).
NRL is a leucine zipper transcription factor which is expressed specifically in rod photoreceptors (Swaroop et al. 1992; Liu et al. 1996; Swain et al. 2001). Mutations in human NRL cause autosomal dominant retinitis pigmentosa, and mice mutant for Nrl show an en masse conversion of presumptive rod photoreceptors into cones (Bessant et al. 1999; Mears et al. 2001; DeAngelis et al. 2002; Daniele et al. 2005). This latter finding suggests that Nrl acts as an endogenous photoreceptor cell fate switch, and a variety of morphological, electrophysiological, and gene expression studies have confirmed the cone-like nature of the photoreceptors in the Nrl−/− retina (Mears et al. 2001; Daniele et al. 2005; Hsiau et al. 2007).
Other transcription factors such as NR2E3, RAX, NEUROD1, THRB, RXRG, RORA, PRDM1, and ESRRB also play important roles in regulating photoreceptor gene expression (Ng et al. 2001; Pennesi et al. 2003; Corbo and Cepko 2005; Roberts et al. 2005; Cheng et al. 2006; Fujieda et al. 2009; Brzezinski et al. 2010; Onishi et al. 2010). Furthermore, additional uncharacterized transcription factors are likely to have a role in this process (Hsiau et al. 2007). Clearly, the photoreceptor transcriptional network is complex and contains several major hubs as well as numerous minor ones.
Despite the recognized role of CRX in photoreceptor gene regulation, the vast majority of its in vivo binding sites are unknown. In the present study we have generated a comprehensive genomic map of CRX binding sites in the adult mouse retina using CRX chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq). This map pinpoints the noncoding regions of the genome relevant to photoreceptor gene expression and establishes CRX as a global regulator of the rod and cone transcriptional networks.
Results
Genome-wide distribution of CRX binding sites in rod photoreceptors
To define the genomic targets of CRX, we carried out CRX ChIP-seq on 8-wk-old wild-type mouse retinas. A total of ∼4.3 × 106 high-quality sequence reads were mapped to the genome for each of two CRX replicates and ∼3.7 × 106 for an IgG control yielding a total of 5595 replicated read clusters (Supplemental Table S1), henceforth referred to as CRX-bound regions (CBRs). Chromatin immunoprecipitation of selected CBRs performed on Crx−/− retinas failed to show enrichment, indicating that the antibody is specific to CRX (Supplemental Fig. S1). In the mouse retina, CRX is highly expressed in both rod and cone photoreceptors and at considerably lower levels in a subset of bipolar cells (Fig. 1A). Rod photoreceptors constitute >70% of all cells in the mouse retina with cones and bipolar cells comprising ∼2% and ∼7%, respectively (Jeon et al. 1998). This suggests that the vast majority of sequence reads derived from CRX ChIP-seq on whole wild-type retinas will correspond to CRX binding in rods.
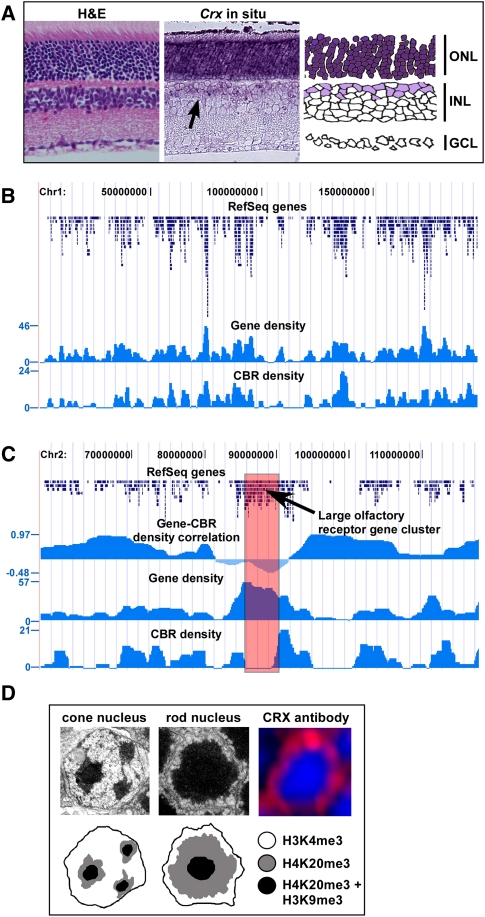
Figure 1.
Genomic distribution of CRX-bound regions in rod photoreceptors. (A) Hematoxylin and eosin (H&E)–stained section of adult retina and in situ hybridization on adult retina using a probe against Crx. There is strong, uniform staining for Crx throughout the outer nuclear layer (ONL; dark purple) which is composed of the cell bodies of both rod and cone photoreceptors. In addition, there is fainter staining in a subset of cells in the inner nuclear layer (INL) that represent bipolar cells. GCL, Ganglion cell layer. (B) Graph of gene density and CBR density across mouse chromosome 1, showing a strong correlation between the two. (C) Graph of gene density, CBR density, and the correlation between the two for a portion of mouse chromosome 2. In the central portion of the graph, there is a region of poor correlation between gene density and CBR density, which represents a large cluster of olfactory receptor genes. (D) Electron micrographs of a cone and rod nucleus along with antibody staining for CRX in a rod nucleus. In the antibody staining, the nuclei are counterstained with DAPI which highlights the heterochromatin. The bottom tier of the figure depicts schematics of the cone and rod nuclei, indicating the expected pattern of a marker for gene-rich euchromatin (H3K4me3) and two markers for gene-poor heterochromatin (H4K20me3 and H3K9me3). These patterns of chromatin markers are based on a prior study (Solovei et al. 2009).
CBR density closely parallels gene density over the entire genome (r = 0.66; Pearson's correlation coefficient) (Fig. 1B), but several regions show poor correlation and correspond to large clusters of olfactory receptor genes on mouse chromosomes 2, 7, and 9 (Fig. 1C; data not shown). The rod photoreceptors of nocturnal mammals such as the mouse have a nuclear architecture which is unique among mammalian cell types (Solovei et al. 2009). The central region of the rod nucleus is occupied by heterochromatin which contains gene-poor, lowly expressed regions of the genome (Fig. 1D). In contrast, the gene-rich, actively transcribed regions of the genome are restricted to a shell of euchromatin immediately subjacent to the nuclear membrane (Fig. 1D). It appears that CRX protein is largely restricted to the euchromatic portion of the rod nucleus (Fig. 1D). This finding may explain the correlation between CBR density and gene density across the genome since the heterochromatic, gene-poor regions of the genome are largely inaccessible to CRX protein.
Phylogenetically conserved CRX-bound regions show high GC content and increased predicted nucleosome binding
Upon analyzing the sequence structure of the CBRs (Supplemental Tables S2, S3), we found that the single most overrepresented motif corresponds very closely to the previously characterized binding preference of CRX (Fig. 2A,B; Lee et al. 2010). To determine the distribution of CRX binding sites in CBRs, we evaluated 1 kb of genomic sequence centered on the 5595 replicated CBRs. We found a prominent peak of CRX binding sites in the center of the CBRs (Fig. 2C) which corresponded to a peak of strong phylogenetic conservation (Fig. 2D). The presence of strong phylogenetic conservation over CBRs suggests that these regions could be under selective pressure and therefore may be functionally important (Visel et al. 2007).
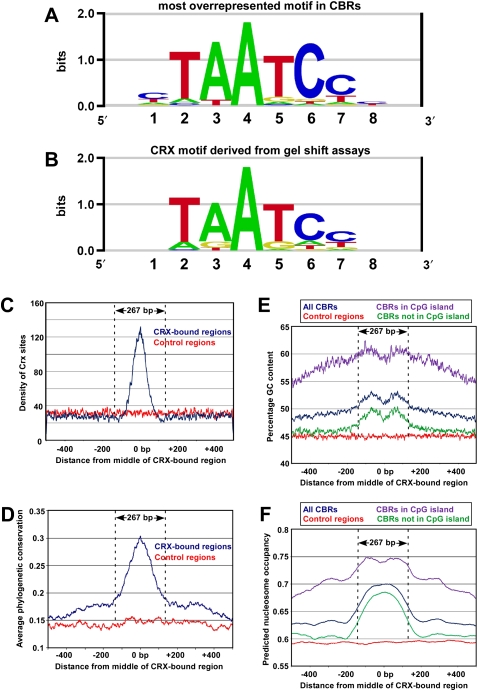
Figure 2.
Sequence analysis of CRX-bound regions. (A) Sequence logo representing the single most highly overrepresented motif found in 10,212 CBRs derived from wild-type retina. (B) Sequence logo of the DNA-binding preference of in vitro synthesized CRX protein as determined by quantitative relative affinity gel shift assays (Lee et al. 2010). (C) The distribution of CRX binding sites across a 1-kb region centered on all replicated CBRs (blue curve) and a set of control sequences (red curve). The y-axis indicates the number of CRX sites per nucleotide that have an affinity ≥0.05 of the affinity of a consensus CRX site. The average size of the CBRs (267 bp) is indicated. (D) The average phylogenetic conservation across all replicated CBRs (blue curve) and a set of control sequences (red curve). The y-axis indicates the average phastCons score per nucleotide (Siepel et al. 2005). (E) Percentage GC content across all replicated CBRs (blue curve) and a set of control sequences (red curve). Also shown is the percentage GC content for all replicated CBRs that did (purple curve) or did not (green curve) overlap with CpG islands. (F) Predicted nucleosome occupancy based on a prior study (Kaplan et al. 2009), across all replicated CBRs (blue curve) and a set of control sequences (red curve). Also shown is the predicted nucleosome occupancy for all replicated CBRs that did (purple curve) or did not (green curve) overlap with CpG islands.
We also found an elevated GC content across the entire region (Fig. 2E), which was only partially attributable to overlap with CpG islands (cf. purple and green curves in Fig. 2E). Since primary DNA sequence can influence nucleosome placement (Tillo and Hughes 2009), we hypothesized that the GC-rich peak within CBRs might represent a nucleosome positioning signal. We therefore determined the predicted nucleosome occupancy over all replicated CBRs using a previously published set of predictions for nucleosome positioning across the mouse genome (Kaplan et al. 2009). Compared with control sequences (red curve in Fig. 2F), there was a peak of increased predicted nucleosome occupancy centered on the middle of the CBRs (blue curve in Fig. 2F), suggesting that CBRs contain signals within their primary DNA sequence that favor nucleosome placement.
CRX-bound regions cluster around photoreceptor genes
CBRs tended to cluster within and around photoreceptor genes (Supplemental Fig. S2; Supplemental Table S3). For example, the Gnat1 locus which encodes the alpha-subunit of rod transducin, shows four discrete CBRs: at −0.6 kb upstream of the transcription start site (TSS), immediately upstream of the TSS, and within the first and eighth introns (Fig. 3A). Expression of Gnat1 is markedly reduced or absent in Crx and Nrl mutant retinas, respectively (Hsiau et al. 2007), and the CBR immediately upstream of its TSS contains CRX and NRL binding sites which have been shown to be required for promoter activity (Fig. 3B; Lee et al. 2010). Thus, CRX ChIP-seq accurately detects known and novel cis-regulatory elements around photoreceptor genes.
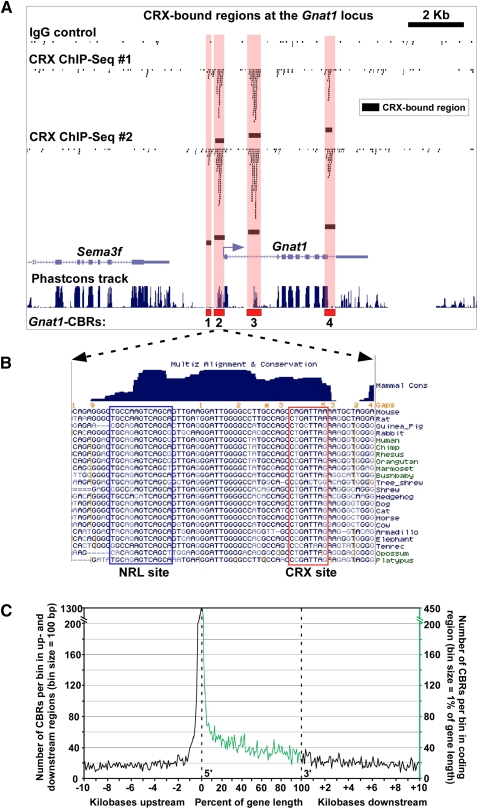
Figure 3.
Distribution of CRX-bound regions around photoreceptor genes. (A) Pattern of CRX-bound regions around Gnat1 that encodes rod alpha-transducin, a component of the phototransduction cascade. Sequence reads derived from two ChIP-seq replicates using an anti-CRX antibody (“CRX ChIP-seq #1” and “CRX ChIP-seq #2”) or an IgG control (“IgG control”) are shown along with the corresponding CBRs. Also shown is the “Phastcons track,” which indicates the pattern of phlyogenetic conservation across the region (Siepel et al. 2005). In this and subsequent figures, CBRs are numbered from 5′ to 3′ with respect to the transcription start site of the gene with which they are associated. (B) Sequence-level view of a portion of Gnat1-CBR2. Note the presence of phylogenetically conserved CRX and NRL binding sites within this region. Additional conserved motifs are also evident, but their binding factors are currently unknown. (C) Distribution of CBRs around mouse genes. This graph shows the density of CBRs over the length of all mouse genes, as well as in the first 10 kb upstream of and downstream from all genes. Note that the location of CBRs within genes is given as percentage of gene length.
Next, we sought to evaluate the distribution of CBRs around gene loci in a genome-wide fashion; 52.9% of CBRs occurred in intergenic regions, 46.2% fell within a single gene, and 0.9% overlapped more than one gene. CBRs showed a pronounced tendency to aggregate around the TSS of genes (Fig. 3C). Under the assumption that CBRs represent cis-regulatory regions controlling the expression of individual genes, we devised a simple algorithm for assigning CBRs to genes on a genome-wide scale: If a CBR falls within a gene, it is assigned to that gene; otherwise, it is assigned to the gene whose TSS is nearest. In this fashion we assigned 10,212 CBRs (including both replicated CBRs as well as those occurring in only a single ChIP-seq replicate) to a total of 6272 genes (representing 22.6% of all genes; Supplemental Table S4).
If the CBR assignments are correct, we should see a marked enrichment around photoreceptor genes, many of which will be dysregulated in the Crx−/− retina. We therefore determined whether CBRs were preferentially assigned to CRX-dysregulated genes. We found that 67% (329/492) of CRX-dysregulated genes had at least one CBR assigned to them which represents a highly significant enrichment compared with all genes (P = 3.62 × 10−99, hypergeometric distribution). Upon separating CRX-dysregulated genes into downregulated and upregulated genes, we found that 81% (242/299) of CRX-downregulated genes had at least one CBR assigned to them (P = 1.15 × 10−102, hypergeometric distribution). In contrast, only 45% (87/193) of CRX-upregulated genes had at least one CBR assigned to them (P = 2.51 × 10−12, hypergeometric distribution). Finally, out of 1289 genes that were dysregulated in Crx, Nrl, or Nr2e3 mutant retinas, 58% (752/1289) had at least one CBR assigned to them (P = 1.40 × 10−177, hypergeometric distribution). These results suggest that the vast majority of genes that are downregulated in the Crx mutant are directly regulated by CRX. In contrast, it appears that only about half of CRX-upregulated genes are directly regulated by CRX.
CRX-bound regions represent photoreceptor-specific cis-regulatory elements
To test whether CBRs represent active cis-regulatory elements we compared them with a list of 33 previously published photoreceptor cis-regulatory regions. We found that 90.9% (30/33) of these regions corresponded to CBRs (Supplemental Table S5). Interestingly, two of the three regions that did not contain CBRs regulate expression of the cone-specific genes, cone arrestin 3 (Arr3) and blue cone opsin 1 (Opn1sw) (Chen et al. 1994; Chiu and Nathans 1994; Zhu et al. 2002). This fact suggests that the sequence reads corresponding to CRX binding in cones may be below the detection threshold of this assay. On the other hand, CBRs were detected in the vicinity of a number of other cone-specific genes including Opn1mw, Gnat2, Gnb3, Pde6c, Pde6h, and Gngt2 (Supplemental Table S3). This latter finding raises the alternative possibility that some of the cis-regulatory regions of cone genes may be bound by CRX in rods but remain inactive.
We then correlated computationally predicted photoreceptor cis-regulatory elements (Hsiau et al. 2007) with the occurrence of CBRs. Of the 100 CBRs with the greatest number of sequence reads, 48 corresponded to predicted cis-regulatory elements (Supplemental Fig. S3). The computational predictions generally correlated well with the CBR data set (examples in Supplemental Fig. S3B–D), but many CBRs were identified which did not correspond to computationally predicted regions (Supplemental Fig. S3; data not shown), a finding that underscores the greater sensitivity of ChIP-seq for detecting in vivo regulatory elements.
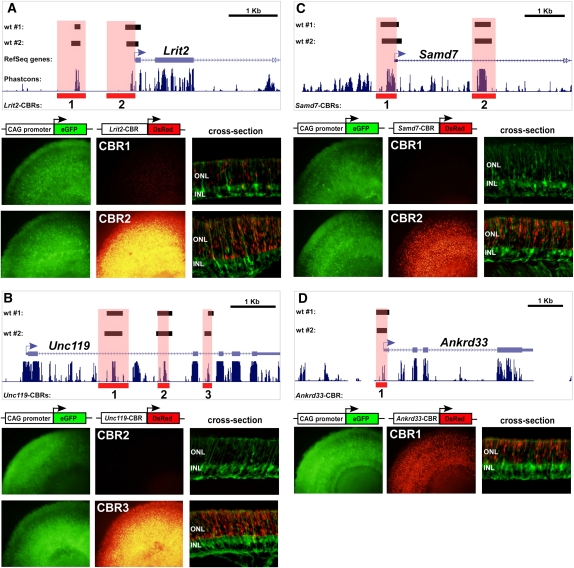
Next, we systematically examined the cis-regulatory activity of CBRs around selected photoreceptor loci. A total of 27 CBRs around 13 different genes was chosen either on the basis of their known or inferred photoreceptor expression pattern or the pattern of CBRs around the locus (Supplemental Table S6). Regions spanning individual CBRs were tested for promoter activity by electroporation as CBR-reporter fusions into mouse retina. Fifty-two percent (14/27) of all individual CBRs tested drove detectable expression in photoreceptors with at least one positive CBR being detected around 92% (12/13) of all loci examined (Fig. 4; Supplemental Fig. S4). Individual CBRs varied widely in their expression strength, but there was no correlation between activity and the position of the CBR relative to the TSS. For example, Lrit2-CBR2 located in the immediate upstream promoter region of Lrit2 drove strong photoreceptor-specific expression whereas Lrit2-CBR1, located ∼1.2 kb upstream of the TSS of the gene, drove much weaker expression (Fig. 4A). In contrast, Unc119-CBR3 drove strong expression in photoreceptors whereas Unc119-CBR2, which was closer to the TSS, did not (Fig. 4B). The number of sequence reads within a CBR was also not predictive of cis-regulatory activity. Whereas Samd7-CBR1 and Samd7-CBR2 had comparable numbers of sequence reads, only Samd7-CBR2 showed activity in this assay (Fig. 4C).
Figure 4.
CRX-bound regions are photoreceptor-specific cis-regulatory elements. (A) The CBRs around a novel photoreceptor-enriched gene, Lrit2, act as photoreceptor-specific cis-regulatory elements. There are two CBRs within the first 2 kb upstream of the transcription start site that were bound in both CRX ChIP-seq replicates from wild-type retinas (“wt #1” and “wt #2”). PCR products encompassing these CBRs (highlighted in light red) were cloned into a DsRed reporter construct and co-electroporated along with a ubiquitously expressing CAG promoter into explanted P0 mouse retinas. The retinas were grown for 8 d and then imaged in both red and green channels in flat-mount and as cross-sections. All flat-mount images in this figure were exposed for the same length of time to permit comparison of the strength of expression. (B) CBRs around a known photoreceptor gene, Unc119, which, when mutated in humans, results in cone-rod dystrophy. Unc119-CBR1 was shown previously to drive strong photoreceptor-specific expression in electroporated retinas (Hsiau et al. 2007). Unc119-CBR3 also drives strong photoreceptor expression, whereas Unc119-CBR2 does not. (C) CBRs around another novel photoreceptor-enriched gene, Samd7. Samd7-CBR2 drives strong, photoreceptor-specific expression, whereas Samd7-CBR1 does not. (D) CBR around a photoreceptor-enriched gene, Ankrd33, which has recently been shown to inhibit the DNA-binding activity of CRX (Sanuki et al. 2010). Only a single CBR was found in the vicinity of Ankrd33. It shows strong photoreceptor-specific cis-regulatory activity.
A subset of the tested CBRs was also subjected to CRX ChIP-qPCR and luciferase assays. Although this subset included Samd7-CBR1 and Slc24a1-CBR1, which failed to drive detectable expression in photoreceptors (Fig. 4C; Supplemental Fig. S4F), all CBRs showed CRX binding in vivo and were able to drive higher levels of luciferase expression in tissue culture cells when cotransfected with a CRX-expressing plasmid (Supplemental Fig. S5). Taken together, these findings demonstrate that not all CBRs are able to autonomously drive photoreceptor expression, even though they are bound by CRX in vivo.
Photoreceptor cis-regulatory elements act in a combinatorial fashion
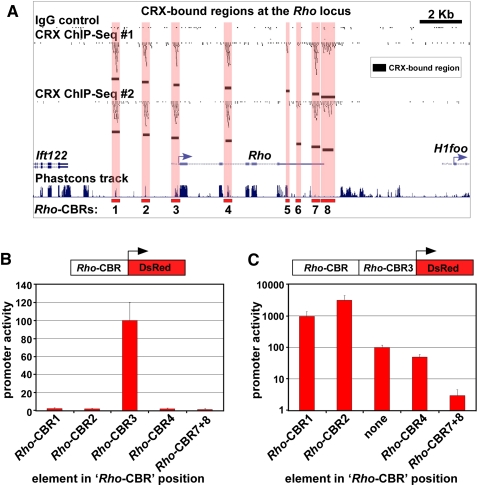
In order to test the idea that multiple CBRs around photoreceptor genes may act in a combinatorial fashion, we analyzed the rod-specific rhodopsin (Rho) locus in detail. The Rho promoter region has been extensively studied over many years (Morabito et al. 1991; Zack et al. 1991; Yu et al. 1993; DesJardin and Hauswirth 1996) and possesses two previously identified cis-regulatory elements (Zack et al. 1991; Nie et al. 1996): a promoter region (Rho-CBR3 in Fig. 5A) and an enhancer further upstream (Rho-CBR2 in Fig. 5A). In addition to these two known elements, we found six novel CBRs in the immediate vicinity of Rho (Fig. 5A). We tested Rho-CBR1, Rho-CBR2, Rho-CBR3, Rho-CBR4, and Rho-CBR7 + 8 for their ability to drive expression in rods when fused to a minimal basal promoter (Fig. 5B). As previously shown in transgenic mice (Nie et al. 1996), Rho-CBR3 was able to drive strong rod expression whereas Rho-CBR2 was not. Strikingly, none of the other Rho-CBRs we tested showed any activity in this assay. Next, we cloned Rho-CBR1, Rho-CBR2, Rho-CBR4, and Rho-CBR7 + 8 upstream of a more extended proximal promoter region (Rho-CBR3) and tested whether they could enhance the expression driven by this region. Quantification of expression in electroporated retinas showed that Rho-CBR1 and Rho-CBR2 strongly enhanced expression, ∼10- and 31-fold over Rho-CBR3 alone, respectively (Fig. 5C). In contrast, fusion of Rho-CBR4 and Rho-CBR7 + 8 upstream of Rho-CBR3 resulted in ∼2- and 33-fold decreases in expression compared with Rho-CBR3 alone, respectively (Fig. 5C). These findings suggest that Rho-CBRs have enhancer or silencer activity which requires interaction with an extended proximal promoter region. In addition, our findings suggest that multiple CBRs around Rho and other photoreceptor genes may act in a combinatorial fashion to fine-tune overall transcriptional output.
Figure 5.
CRX-bound regions act in a combinatorial fashion to drive photoreceptor gene expression. (A) Distribution of CBRs around the rhodopsin (Rho) gene, which encodes rod opsin, the primary light-sensing molecule of rod photoreceptors. (B) Quantitative analysis of Rho-CBR cis-regulatory activity in electroporated retinas. The indicated Rho-CBRs were cloned into a DsRed reporter and electroporated into P0 mouse retinas along with a Rho-CBR3-eGFP loading control. After 8 d in culture, the retinas were imaged in flat-mount, and promoter activity was quantified by measuring fluorescence (see Methods for details). Shown is the mean ± standard deviation of three replicate electroporations. All values are normalized to that of Rho-CBR3, which is set equal to 100. (C) Quantitative analysis of Rho-CBR cis-regulatory activity when cloned upstream of the Rho proximal promoter region (Rho-CBR3). Shown is the mean ± SD of three replicate electroporations. All values are normalized to that of Rho-CBR3 alone, which is set equal to 100. Note that the y-axis is on a log scale.
CRX-bound regions are enriched for pairs of transcription factor binding sites
To identify additional binding sites that might interact with CRX sites within CBRs, we analyzed overrepresentation of pairs of sites within 50 bp of each other. We found that the top 20 most highly ranked motif pairs could be categorized into five classes (Supplemental Table S7). Strikingly, CRX–CRX pairs represented the two most highly ranked motifs suggesting that multiple clustered CRX binding sites may represent an important aspect of CBR architecture.
In addition to CRX–CRX pairs, we found overrepresentation of CRX–nuclear receptor binding site pairs, CRX–E-box pairs, CRX–NRL pairs, and pairs consisting of a CRX site along with a variety of GC-rich motifs (Supplemental Table S7). These findings accord with the well-known role of nuclear receptors such as NR2E3 and E-box binding transcription factors such as NEUROD1 in regulating rod gene expression (Pennesi et al. 2003; Chen et al. 2005). In addition, the present findings corroborate the importance of CRX–NRL interactions in photoreceptor gene regulation on a genome-wide scale (Mitton et al. 2000; Hsiau et al. 2007).
CRX binding sites control transcriptional output in a spacing- and orientation-dependent fashion
In order to elucidate the functional role of CRX–CRX motifs within CBRs, we examined the patterning of CRX–CRX pairs in more detail. Pairs of CRX sites can have one of three possible relative orientations: tandemly repeated, head-to-head, or tail-to-tail (Supplemental Fig. S6A–C). We tallied the number of CRX–CRX pairs between 8 and 100 bp apart within the central region of the CBRs and found that pairs of sites in all three orientations show a decreasing abundance at greater intersite spacing (Supplemental Fig. S6A–C). In addition, we found a strong preference for tandemly repeated sites between 8 and 11 bp apart, suggesting that CBRs favor the presence of pairs of CRX sites approximately one helical turn apart and in the same orientation.
To test the function of closely spaced CRX sites within promoters, we created a series of synthetic cis-regulatory elements and assayed their activity in electroporated retinas. A minimal basal promoter including the first 35 bp upstream of the TATA box of the bovine Rho gene and containing a single CRX site fails to drive any expression in electroporated mouse retinas (Supplemental Fig. S6D). Addition of a synthetic 50-bp DNA fragment upstream of this minimal promoter containing a second CRX site spaced 32 bp upstream of the first CRX site also fails to drive expression (Supplemental Fig. S6E). However, further addition of a third CRX site 10 bp upstream of the second site results in robust photoreceptor-specific expression (Supplemental Fig. S6F). A construct containing two CRX sites and an NRL site drives similar levels of photoreceptor-specific expression (Supplemental Fig. S6G). Thus, these experiments demonstrate a synergistic interaction between closely spaced CRX sites as well as closely spaced CRX and NRL sites.
Next, we evaluated the spacing and orientation dependence of this CRX–CRX interaction. We created a series of synthetic promoters in which we progressively increased the spacing between CRX sites 2 and 3, without otherwise altering the intervening sequences, and quantified their activity in electroporated retinas. We found that as the intersite spacing increases, the promoter activity rapidly decreases (Supplemental Fig. S6I). A similar result was observed when the orientation of CRX site 3 was flipped (Supplemental Fig. S6K). Thus, CRX sites act synergistically to drive photoreceptor-specific expression and show greater activity with short intersite spacings and a tandem orientation. These results directly mirror the situation observed in endogenous photoreceptor cis-regulatory elements (CBRs) and demonstrate that the combinatorial cis-regulatory logic of photoreceptors is fundamentally similar to that observed in a wide range of cell types and organisms (Makeev et al. 2003; Davidson 2006; Rothbacher et al. 2007; Gertz et al. 2009; Zinzen et al. 2009; Ravasi et al. 2010).
The gene networks of rods and cones contain both shared and cell type–specific CRX-bound regions
Cones represent only a small fraction of the cells in the mouse retina, and the absence of CBRs within the known regulatory regions of cone genes such as Opn1sw and Arr3 suggest that few of the sequence reads in our CRX ChIP-seq analysis of wild-type retinas derive from cones. In order to map CRX binding sites in cones, we took advantage of the Nrl−/− retina which shows an en masse conversion of rods into cones and therefore represents an enriched source of this otherwise rare cell type (Daniele et al. 2005). Changes in rod- and cone-specific gene expression in the Nrl−/− retina compared with wild type are summarized in Figure 6A–D. We performed CRX ChIP-seq on 8-wk-old Nrl−/− retinas in two biological replicates along with IgG controls and identified a total of 8039 and 4076 CBRs (Supplemental Tables S2, S8). Despite the lower number of CBRs in the second replicate, the overall number of discrete CBRs between both replicates (9661) is comparable to the overall number obtained in the analysis of wild-type retinas (10,212).
Figure 6.
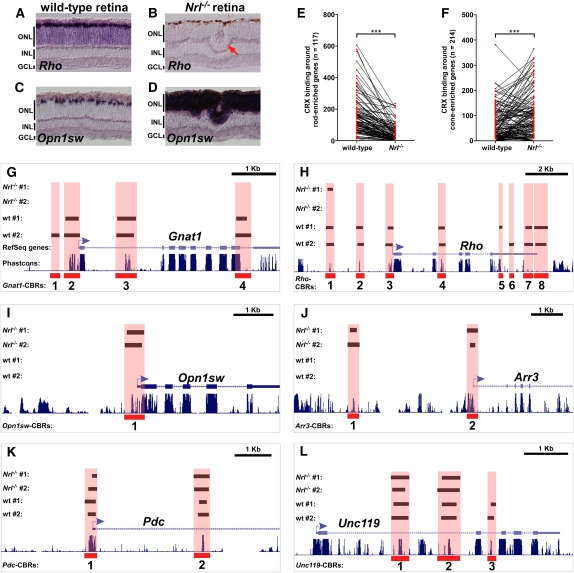
Rods and cones have both shared and cell type–specific CRX-bound regions. (A–D) In situ hybridization pattern of rod-specific rhodopsin (Rho) on wild-type (A) and Nrl−/− (B) retinas, and cone-specific blue opsin (Opn1sw) on wild-type (C) and Nrl−/− (D) retinas. In the wild-type retina, Rho is expressed in the majority of cells in the ONL, whereas Opn1sw is only expressed in a small subset of cells at the outer edge of the ONL. The Nrl−/− retina shows the converse pattern: Rho is completely absent, whereas Opn1sw is strongly expressed throughout the entire ONL. Rosette formation in common in the ONL of Nrl−/− retinas (red arrow in B). (E) CRX binding around rod-enriched genes in wild-type and Nrl−/− retinas. Each pair of red dots connected by a black line represents a single rod-enriched gene. The y-axis indicates the number of sequence reads within all CBRs assigned to that gene. There is a marked decrease in the number of assigned sequence reads for most rod genes in the Nrl−/− retina relative to wild-type. ***P < 0.0001, paired Student's t-test. (F) CRX binding around cone-enriched genes in wild-type and Nrl−/− retinas. In this case, there is an overall increase in CRX binding around cone genes in Nrl−/− retinas compared with wild-type. ***P < 0.0001, paired Student's t-test. Gnat1 (G) and Rho (H), both rod-specific genes, show a near absence of CBRs in the Nrl−/− retina. Opn1sw (I) and Arr3 (J), both cone-specific genes, show prominent CBRs in the Nrl−/− retina but not in wild-type. Pdc (K) and Unc119 (L) are expressed at similar levels in both rods and cones. They show similar levels of CRX binding in both wild-type and Nrl−/− retinas.
Given the conversion of rods into cones in the Nrl−/− retina, we hypothesized that the Nrl mutant would show an increase in CRX binding around cone genes and a corresponding decrease around rod genes. To test this idea, we quantified the extent of CRX binding around rod- and cone-enriched genes in both wild-type and Nrl−/− retinas. We found that rod gene loci showed a significant decrease in CRX binding in the Nrl mutant relative to the wild type (Fig. 6E). Rod-specific genes such as Rho and Gnat1 demonstrated an almost total loss of CRX binding in their vicinity as shown by the disappearance of nearly all of the CBRs present around these loci in wild-type retinas (Fig. 6G,H). In contrast, cone genes showed an overall increase in CRX binding in nearby CBRs (Fig. 6F). Interestingly, novel CBRs that were not present in the wild-type data set appeared in the promoter regions of cone genes such as Opn1sw and Arr3 in the Nrl−/− ChIP-seq data set (Fig. 6I,J). Despite these differences, there was a strong genome-wide correlation between the distribution of CBRs in wild-type and Nrl−/− retinas (r = 0.72, Pearson's correlation coefficient), indicating that the two cell types have very similar overall patterns of CRX binding. This fact is underscored by the observation that photoreceptor genes that show coexpression in rods and cones often showed similar levels of CRX binding in wild-type and Nrl−/− retinas (Fig. 6K,L).
CRX directly regulates photoreceptor transcription factors
Next we evaluated CRX's role in regulating transcription factors in the photoreceptor gene network. Numerous CBRs corresponding to previously identified cis-regulatory regions were found around the Crx locus itself (Supplemental Fig. S7A,B). In order to assess the combinatorial nature of transcriptional regulation at the Crx locus, we tested the activity of a composite cis-regulatory element containing portions of both upstream (Crx-CBR4) and downstream (Crx-CBR6) regulatory elements. This construct drove very strong expression in both photoreceptors and a subset of bipolar cells (Supplemental Fig. S7C,D; cf. Fig. 1A). Thus, the endogenous levels and pattern of Crx expression require multiple cis-regulatory regions spread over tens of kilobases around the locus. These results confirm prior suggestions that CRX directly regulates its own expression (Furukawa et al. 2002) and are reminiscent of the autoregulation observed at many other transcription factor loci (Bateman 1998; Crews and Pearson 2009).
CRX also directly regulates the rod determination pathway in a multitiered fashion (Supplemental Fig. S7A,E–G). Multiple CBRs were found around all three components of the transcriptional cascade leading to rod cell fate: Rorb, Nrl, and Nr2e3 (Supplemental Fig. S7E–G). In the case of the Nr2e3 and Nrl loci, multiple CBRs overlap with previously identified cis-regulatory regions, confirming that these CBRs likely represent bona fide cis-regulatory elements (Akimoto et al. 2006; Hsiau et al. 2007; Oh et al. 2008). In addition, CRX binding was found at nearly all other well-characterized photoreceptor transcription factor loci including Rax, Neurod1, Thrb, Rxrg, Rora, Esrrb, Mef2c, and Prdm1 (Supplemental Table S3). These findings suggest a pervasive role for CRX in directly regulating other transcription factors in photoreceptors and thus confirm its status as a global regulator in this cell type.
CRX controls the majority of retinal disease genes
In order to evaluate CRX binding around loci implicated in retinal disease, we identified the mouse orthologs of 125 human retinal disease genes (Supplemental Table S9). We found that 70.4% (88/125) of these genes had associated CBRs in wild-type or Nrl−/− retinas (Supplemental Table S9), suggesting extensive transcriptional regulation of retinal disease loci by CRX. Considering only those subsets of genes with retina-enriched or retina-specific patterns of expression, the percentage of CBR-associated genes increases to 95.3% (61/64) and 97.8% (46/47), respectively. This finding suggests that nearly all previously identified retinal disease genes with retina-restricted expression are likely CRX targets.
We therefore hypothesized that our data set of CBRs could help to pinpoint novel retinal disease genes. All conserved mouse CBRs were accordingly mapped to the orthologous human genomic regions and an alignment with intervals containing uncloned retinal disease genes was performed (Supplemental Table S10). With this approach we were able to reduce the total number of 4958 candidate genes present in the 31 mapped retinal disease intervals to 724 high-priority genes associated with CBRs. In some cases, this marked reduction in the number of candidate genes resulted in the identification of only one or a few candidates within a given disease gene interval (Supplemental Table S10). In addition, if more than one CBR-associated gene fell within a candidate disease region, it was possible to further prioritize the candidates based on the extent of CRX binding as measured by the number of ChIP-seq sequence reads assigned to the individual genes (Supplemental Table S10). Thus, CBR clustering may serve as a useful tool for identifying human retinal disease genes within mapped genomic intervals.
Discussion
Using ChIP-seq technology, we have generated a comprehensive map of the cis-regulatory regions of mammalian photoreceptors and established CRX as a global regulator of the photoreceptor transcriptional network.
CBRs represent photoreceptor cis-regulatory elements which act in a combinatorial fashion to regulate gene expression. Pioneering studies in Drosophila and other model organisms have shown that the complex expression patterns of many embryonically expressed genes are attributable to the action of multiple, spatially discrete cis-regulatory elements which individually mediate expression in a subpart of a gene's overall expression domain (Small et al. 1993; Davidson 2006). In addition, recent work has identified “shadow enhancers” around many early Drosophila genes that drive expression in a spatiotemporal pattern similar to the originally defined “primary” enhancer at that locus (Hong et al. 2008). We find that mouse photoreceptor cis-regulatory regions are spatially arrayed in an analogous fashion. Even genes in the terminal tier of differentiation that are specific to rod photoreceptors (such as Rho and Gnat1) have multipartite, distributed cis-regulatory regions. Instead of mediating different aspects of spatial expression, these regions appear to modulate quantitative levels of expression in the same cell type. From an evolutionary perspective, the multiplicity of CBRs around these loci may act as a failsafe mechanism to ensure that precise and reliable levels of expression are maintained in the steady state. Indeed, a recent report suggested that Drosophila shadow enhancers might serve to quantitatively fine-tune levels of expression within defined spatial domains and thereby provide robustness to environmental perturbations (Frankel et al. 2010). Despite the multipartite nature of photoreceptor cis-regulatory regions, the present study demonstrates that the logic of photoreceptor gene regulation is fundamentally simple. A single transcription factor plays a pervasive role in regulating hundreds if not thousands of genes via structurally simple cis-regulatory elements consisting of closely clustered binding sites. Thus, CRX represents a classic example of a terminal selector gene (Hobert 2008).
In this study, only about half of the CBRs tested for cis-regulatory activity were able to drive detectable expression in photoreceptors by themselves. Yet, a number of these “nonexpressors” could potently modulate the levels of expression driven by an adjacent CBR. A prior study in the mouse tested phylogenetically conserved noncoding regions for cis-regulatory activity via promoter-reporter transgenes and found that 45% of the tested regions possessed enhancer activity (Pennacchio et al. 2006). In that study, it was suggested that nonexpressing regions might possess enhancer activity at time points in development other than the one assayed. Another possible explanation for the lack of expression is that many phylogenetically conserved noncoding regions could represent enhancers which require cooperation with other noncoding regions to show expression. Thus, many functional cis-regulatory elements are likely to escape detection via simple promoter-reporter fusion assays in which a single genomic region is fused to a basal promoter.
The concept that cis-regulatory elements consist of clusters of transcription factor binding sites is well established (Davidson 2006), but the detailed grammar rules that govern the intersite spacing and orientation of individual binding sites within these elements is not well understood in any system. We previously reported that pairs of CRX and NRL sites within 40 bp of each other are associated with high-level expression in photoreceptors (Hsiau et al. 2007). We have now shown that close pairing of CRX sites is another important feature of CBRs and that there is a particular preference for CRX sites approximately one helical turn apart and in the same orientation. The importance of intersite spacing has been shown previously for the Drosophila homeodomain transcription factor, bicoid, which has a DNA-binding preference almost identical to that of CRX (Hanes and Brent 1991; Hanes et al. 1994). Interestingly, a similar helical periodicity of bicoid binding sites was also previously reported (Makeev et al. 2003). Despite these similarities, it appears that CBRs tolerate a relatively wide range of spacings and orientations between CRX sites, and thus the observed spacing and orientation preference is not absolute, a finding also observed with other transcription factors (Makeev et al. 2003; Chiang et al. 2006).
CBRs have an elevated GC content which may reflect the presence of a nucleosome positioning signal. One possible explanation for the presence of predicted nucleosome binding sites centered directly over CBRs is that they may serve to silence the activity of these elements in nonphotoreceptor cell types (Tillo et al. 2010). The presence of a distributed nucleosome positioning signal superimposed on cell type–specific transcription factor binding sites suggests that individual nucleotides within a cis-regulatory element may be under dual evolutionary pressures to maintain adequate binding of a particular transcription factor as well as to favor nucleosome placement.
The distribution of CBRs across the genome suggests that CRX potentially regulates the expression of thousands of genes in photoreceptors. Although many of the direct targets we identified in this study were previously shown to be dysregulated in the Crx−/− retina (Livesey et al. 2000; Hsiau et al. 2007), a large proportion showed no evidence of dysregulation. One possible explanation for this discrepancy is that other transcription factors may compensate to a variable extent for the loss of CRX. One candidate for this compensatory activity is OTX2 which is expressed in adult photoreceptors at lower levels than CRX and which has a nearly identical DNA-binding preference to that of CRX (Nishida et al. 2003; Chatelain et al. 2006). Although a significant fraction of all genes (22.6%) had at least one CBR in their vicinity, many of these appear to have only modest amounts of CRX binding as reflected by low numbers of sequence reads. It is therefore possible that CRX may bind somewhat promiscuously to many regions within that portion of the genome to which it has physical access, and that much of this low-level binding may not be functional. Similarly, widespread, putatively nonfunctional binding of transcription factors has been previously reported in Drosophila (Li et al. 2008).
CRX's high degree of network connectivity accounts for the severity of the human and mouse phenotypes when it is mutated. In addition, the pervasive regulation of known retinal disease genes by CRX strongly suggests that additional disease loci are likely to be found among its many newly discovered target genes. A key feature of the present data set is that it pinpoints many of the noncoding regions relevant to both Mendelian as well as complex diseases affecting photoreceptors. There remain many pedigrees affected by retinal disease for which the causative gene has not been identified. It is possible that a subset of these families actually have mutations in the noncoding regions of known retinal disease genes. Knowledge of the noncoding cis-regulatory elements at those loci will now permit targeted resequencing to screen for such mutations.
In addition, despite recent advances in identifying the genetic loci contributing to multifactorial retinal diseases such as age-related macular degeneration (Swaroop et al. 2009), we still have an incomplete understanding of why only a subset of patients with predisposing genetic alterations develop the disease. Given the importance of photoreceptors in this disease process, it is likely that genetic variation in both the coding and noncoding portions of the genome relevant to photoreceptor function could contribute to susceptibility. Thus, the present data set will dramatically reduce the total “sequence space” that must be searched to identify relevant mutations.
Methods
Mouse husbandry
Adult CD1, C57BL/6, Crx−/− (Furukawa et al. 1999), and Nrl−/− (Mears et al. 2001) mice were maintained in an air-conditioned environment on a 12-h light–dark schedule at 20°C –22°C and had free access to food and water. The health of the animals was regularly monitored. All animal procedures were approved by the University of Regensburg animal rights committee and complied with the German Law on Animal Protection and the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals, 1999. Animals were also housed at Washington University in St. Louis, and all studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act and were approved by the Washington University in St. Louis Institutional Animal Care and Use Committee (approval no. 20080058).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously (Langmann et al. 2008) with minor modifications. In brief, for each ChIP, six 8-wk-old retinas were dissected in 1× phosphate-buffered saline (PBS), pooled, minced, and treated with 1% formaldehyde for 15 min at room temperature. The reaction was stopped by incubation with 0.125 M glycine. Cells were lysed in 0.5% NP-40. The nuclear pellet was lysed with 1% SDS and 0.5% EmpigenBB and homogenized by sonication, 15 × 10 sec, at 50% amplitude (SONICS Vibracell VCX400 sonicator). Immunoprecipitations were performed overnight at 4°C with 2.5 μg of anti-CRX antibody (Santa Cruz, sc-30150X) or anti-IgG antibody (Upstate, 12-370) bound to protein A Sepharose beads. After washing and elution steps, cross-links were reversed at 65°C overnight. The immunoprecipitated DNA was purified using QIAquick purification columns (Qiagen).
High-throughput sequencing
Sequencing was carried out using the 1G Illumina Genome Analyzer (Solexa). Two independent lanes were sequenced from independent biological replicates of CRX-ChIP and IgG-ChIP DNA derived from wild-type (C57BL/6) and Nrl−/− retinas. CRX-bound or IgG-precipitated DNA (10 ng) was size-fractionated and purified by SDS-PAGE to obtain 100–200-bp fragments. To prepare DNA fragments for adapter ligation, a single adenosine was added to the 3′ end of the blunt phosphorylated DNA using Klenow fragment (3′ to 5′ exo minus). Adapters were added and DNA was subjected to 18 cycles of PCR for enrichment. Each library was validated using an Agilent 2100 Bioanalyzer and was then sequenced on the Illumina cluster station and 1G analyzer. High-quality 36-bp tags were mapped to the mouse genome (NCBI Build 37) using the Genomatix (http://www.genomatix.de/) mapping algorithm.
Clustering of sequence reads and identification of CRX-bound regions
The distribution of reads in the CRX-ChIP and IgG-ChIP data sets was determined by counting the number of reads in a 100-bp sliding window using the Genomatix NGS Analyzer (Sultan et al. 2008). Assuming a Poisson distribution, the threshold applied by the clustering algorithm was eight reads per 100 bp. Nonspecific enrichments detected in the IgG control data were subtracted from the local enrichments (clusters) representing genomic regions bound by CRX protein. Fundamentally similar peak-calling results were obtained with the QuEST algorithm using moderate stringency parameters (Supplemental Table S11; Valouev et al. 2008). High confidence ChIP-seq sequence reads derived from wild-type and Nrl−/− retinas are publicly available at the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) as series record GSE20012.
CBRs in the two ChIP-seq replicates which overlapped by one or more nucleotides were defined as “double-hit” CBRs. 73.5% of the wild-type replicate #1 CBRs overlapped with replicate #2 CBRs, and 68.7% of wild-type replicate #2 CBRs showed overlap with those from replicate #1. The extent of the double-hit CBR was defined as the 5′-most and 3′-most extent of the individual overlapping CBRs. In some cases, two or more CBRs from one replicate overlapped one CBR from the other replicate. In such cases, the entire overlapping cluster of CBRs was combined together as a single “double-hit” CBR. Those CBRs that did not overlap with any CBR from the other replicate were defined as “single-hit” CBRs.
Sequence analysis of CRX-bound regions
De novo motif discovery in CBRs was performed with the Genomatix CoreSearch database and the resulting sequence logo was constructed using enoLOGOS (Workman et al. 2005). All further sequence analyses were performed on 1-kb regions of genomic DNA centered on the middle nucleotide of all double-hit CBRs. A control set of 1-kb sequences was obtained by adding 100 kb to the start and end coordinates of CBRs. CRX binding sites were identified within CBRs as previously described using a cutoff affinity value of ≥0.05 of the affinity of CRX's consensus binding site (Lee et al. 2010). The number of identified CRX sites was tallied per nucleotide position within the 1-kb region. Phylogenetic conservation within CBRs was determined by averaging the phastCons score at each nucleotide (Siepel et al. 2005). GC content within these regions was determined by averaging the GC content over a 5-bp sliding window moving 1 nt at a time. If a region overlapped a CpG island by at least 1 nt (as defined by the UCSC Genome Browser, http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=151155442&c=chr15&g=cpgIslandExt), it was defined as being “in” a CpG island. Nucleosome occupancy around CBRs was calculated using mouse genome nucleosome occupancy predictions (Kaplan et al. 2009), downloaded from the Segal lab website (http://genie.weizmann.ac.il/software/nucleo_genomes.html). CBRs were also scanned for pairs of transcription factor binding sites using Genomatix RegionMiner and position weight matrices from the MatInspector database. Z-scores and other pertinent data were calculated as described (Ho Sui et al. 2005).
Defining a comprehensive set of mouse genes
A comprehensive set of mouse genes was defined by merging the RefSeq gene set with additional genes and transcription units specified by features from the Affymetrix GeneChip Mouse Genome 430 2.0 microarray. Any Affymetrix feature that overlapped (either partially or completely) with a RefSeq gene was assigned to that gene. If an Affymetrix feature overlapped with more than one RefSeq gene on the same strand, the Affymetrix feature was assigned to that gene with which it shared maximal overlap. Any Affymetrix feature which showed no overlap with a RefSeq gene on the same strand was defined as a “gene” and named according to that Affymetrix feature. Prior to this assignment procedure, all Affymetrix features and all RefSeq genes that mapped to more than one location in the genome were removed from the analysis. A subset of RefSeq genes had no corresponding Affymetrix features. A total of 27,735 mouse genes were defined in this manner (Supplemental Table S4).
Determining the distribution of CRX-bound regions along chromosomes
CBR density along chromosomes was determined by tallying the number of CBRs that fell within a 1-Mb sliding window, moving 100 kb at a time. The middle nucleotide of each CBR was used to define its location. Gene density was calculated in the same fashion using the mouse gene set defined above. The gene-CBR density correlation was determined in Excel by calculating the Pearson's correlation coefficient between 10 successive gene density and CBR density values in a 10-Mb sliding window, stepping 100 kb at a time. Gene density, CBR density, and gene–CBR density correlations were displayed graphically using UCSC Genome Browser custom tracks (Kuhn et al. 2009).
Determining the distribution of CRX-bound regions around genes
The middle nucleotide of 10,212 double-hit and single-hit CBRs from wild-type retinas was mapped relative to genes (as defined above) across the genome. For those CBRs that fell within a single gene, its position within the gene was normalized to gene length by dividing the distance of the CBR from the TSS by the length of the gene. The number of CBRs that fell into each of 100 equal-sized bins along the length of the gene was tallied. Next, the position of the middle nucleotide of each intergenic CBR within the first 10 kb upstream of or downstream from all mouse genes was determined. The number of CBRs that fell into each of 100 bins (each 100 bp in length) within both the upstream and downstream regions was counted. If a particular CBR was within 10 kb upstream of one gene and 10 kb downstream from another gene, both positions were recorded.
Automated assignment of CRX-bound regions to genes and assignment validation
All double-hit and single-hit CBRs were assigned to one of 27,735 mouse genes as defined above. Any CBR whose middle nucleotide fell within a single gene was assigned to that gene. All other CBRs were assigned to the gene with the nearest transcription start site. These CBR-to-gene assignments were validated by comparing the list of genes with assigned CBRs to lists of genes dysregulated in Crx−/−, Nrl−/−, or Nr2e3−/− retinas. Dysregulation was determined based on previously published data from Affymetrix GeneChip Mouse Genome 430 2.0 microarrays (Hsiau et al. 2007). The values for individual Affymetrix features from the Crx−/−, Nrl−/−, or Nr2e3−/− microarray experiments were compared with the corresponding wild-type control value, and the Affymetrix feature was determined to be dysregulated if the value in either wild-type or mutant was ≥400 and the mutant value was greater than or equal to twofold changed relative to the control (i.e., either up- or downregulated). When all three data sets were combined, there were a total of 1289 dysregulated genes, with a number of genes being dysregulated in more than one mutant background. Enrichment of CBR-associated genes within the mutant gene sets was determined using the hypergeometric distribution.
Computational analysis of CRX binding site pairs
CRX sites with a predicted affinity ≥0.05 of the CRX consensus sequence within the central 100 bp of all double hit CBRs were identified. Then, the position and orientation of all other CRX sites between 8 and 100 bp from an identified site were tallied. A similar analysis was performed on a control set of sequences.
Rod- and cone-enriched gene sets
Rod- and cone-enriched gene sets were derived from a previously published study (Corbo et al. 2007). Any Affymetrix features which mapped to more than one location in the genome or which did not show an Affymetrix score of at least 400 in either the wild-type or Nrl−/− retina data sets were removed from the analysis. A total of 214 and 117 cone and rod genes, respectively, were analyzed.
Identification of human retinal disease gene candidates
Genomic coordinates defining the critical regions of all 31 mapped but uncloned human retinal disease loci were retrieved from RetNet (http://www.sph.uth.tmc.edu/retnet) and publications cited therein. The relevant marker sequences were mapped to the current human genome annotation hg19 using the UCSC Genome Browser to obtain the genomic coordinates. The R/Bioconductor library biomaRt (http://cran.r-project.org/) (Durinck et al. 2005) was used to obtain a list of all annotated genes for each locus interval and their unique mouse orthologs. All mouse genes containing CBRs (Supplemental Table S4) were matched with this list to define human candidate disease genes based on CBRs around the corresponding mouse orthologs.
Quantitative real-time PCR
qPCR was performed on an ABI 7900HT Fast Real-Time PCR System using SYBR Green. The fold enrichment of each target site was calculated as 2 to the power of the cycle threshold (Ct) difference between IgG- and CRX-ChIP samples. Successful CRX-mediated chromatin enrichment from C57BL/6 retinas was verified by qPCR using primers specific for the proximal promoter of the mouse retinoschisin gene (Rs1-A: 5′-AATTAGGGGCCCACATCTTC-3′, Rs1-B: 5′-GTTTAGCAAGGGAGGTGCTG-3′). Successful CRX-mediated chromatin enrichment from Nrl−/− retinas was validated by qPCR using primers specific for the proximal promoter of the cone genes, Gnat2 and Gnb3: (Gnat2-A F: 5′-CAGGGAACAGAGACTGCAGAG-3′, Gnat2-B R: 5′-CTGCCAACCAACTGACTTGA-3′; Gnb3-A F: 5′-AACCATGCTTCCTCGTTGAG-3′, Gnb3-B R: 5′-CAACTAGGATCAGGCCCAAG-3′). Additional qPCR primers used for verification of CRX-ChIP enrichment are listed in Supplemental Table S12.
Luciferase reporter assays
Luciferase reporters were based on Promega plasmid vectors pGL4.10 for regions immediately upstream of the TSS (i.e., promoter regions) and pGL3 for regions further from the TSS (i.e., enhancer regions). Four promoter reporters (Wdr17-CBR1, Slc24a1-CBR1, Samd7-CBR1, Lrit2-CBR2) and three enhancer reporters (Samd7-CBR2, Lrit2-CBR1, Ric8b-CBR2) were generated using PCR primers listed in Supplemental Table S12. HEK cells in 12-well plates were transfected with 0.2 μg of luciferase vectors and 0.4 μg of CRX-expression vector or pcDNA3.1/V5 control vector, respectively, using TransIT-LT1 (Mirus). Luciferase activity was determined as described previously (Langmann et al. 2008).
Immunohistochemistry, RNA in situ hybridization, and electron microscopy
Retinal tissue from a 25-wk-old CD-1 mouse was fixed in 4% paraformaldehyde for 1 h at room temperature and washed three times in 1× PBS. The lens was left in place during fixation and removed during the final wash step. Tissue was then embedded in 4% agarose in 1× PBS and 50 μm vibratome sections were collected. Sections were blocked with normal goat serum (NGS; 2% goat serum and 0.5% Triton X-100 in 1× PBS) for 1 h at room temperature. Rabbit polyclonal CRX H-120 X (sc-30150 X, Santa Cruz) was diluted in NGS (1:500) and incubated with sections at 4°C overnight. The primary antibody was then removed and the sections were washed in PBS prior to incubation in goat anti-rabbit biotinylated secondary (1:500 in NGS) (BA-1000, Vector Laboratories) for 1 h at room temperature. Following three PBS washes, sections were incubated in ABC solution (Elite ABC kit standard, Vector Laboratories) for 30 min at room temperature, washed again in 1× PBS, and incubated in tyramide solution for 10 min at room temperature. The tyramide solution contained 0.8% Cy3 conjugated tyramide (TSA cyanine 3, Perkin Elmer) in amplification diluent (0.1 M borate at pH 8.5, 0.003% H2O2). After washing in 1× PBS, sections were DAPI-counterstained (1:4000) for 5 min at room temperature. Following two 1× PBS washes, sections were mounted with gelvatol and coverslipped. Images were captured using a BX61WI microscope (Olympus) equipped with a DSU spinning disc and an ORCA-ER CCD camera (Hamamatsu).
In situ hybridization on C57BL/6 and Nrl−/− tissue sections derived from 4- to 9-wk-old mice was performed as previously described (Chen and Cepko 2000; Hsiau et al. 2007). The probes for Crx, Rho, and Opn1sw were also described previously (Corbo et al. 2007). Electron microscopy was performed as described previously (Corbo and Cepko 2005).
Construction of CBR-reporter fusion constructs
PCR primers were designed using Primer3 (Rozen and Skaletsky 2000) and are given in Supplemental Table S6. Two different starting vectors were used for the construction of CBR-reporter fusion constructs: Rho-basal and no-basal. The Rho-basal vector contains nucleotides −36 to +79 around the TATA box of bovine Rho cloned upstream of DsRed (Hsiau et al. 2007). CBRs were cloned in a polylinker upstream of this minimal Rho promoter region. This vector was used for cloning all CBRs that resided at some distance for a gene's endogenous promoter region. The second vector, no-basal, consists of a polylinker immediately upstream of DsRed without a basal promoter. This vector was used for cloning any CBR that was localized immediately upstream of a TSS. Further details on these vectors were already published (Hsiau et al. 2007). Constructs that contained more than one CBR were created by first cloning the promoter-proximal CBR into the no-basal vector. The second CBR was then cloned immediately upstream of the first CBR. All restriction enzyme sites used for cloning CBRs into reporter vectors are given in Supplemental Table S6.
Engineering synthetic cis-regulatory elements
Synthetic cis-regulatory elements including all CRX spacing constructs as well as “Nrl × 1; Crx × 2” (Supplemental Fig. S6G) were generated in the following manner. For the CRX spacing constructs, a random 50-bp sequence was generated in silico with the requirement that it possess no CRX or NRL sites and that it approximate the GC content of the mouse genome. Substitutions were then created in this sequence to generate high-affinity CRX sites with the required spacing and orientation. Two complementary oligonucleotides containing this sequence were synthesized and additional nucleotides were added to the ends so that, after kinasing and annealing, the oligonucleotides would form a double-stranded DNA molecule with an XbaI half-site at the 5′ end and an EcoRI half-site at the 3′ end. This DNA molecule was then cloned into the Rho-basal vector that had been digested with XbaI and EcoRI to generate the final synthetic reporter construct. “Nrl × 1; Crx × 2” was generated in a similar fashion using another “backbone” DNA sequence generated in silico. The oligonucleotides used to generate these constructs are listed in Supplemental Table S12. Note that CRX sites are highlighted in red and NRL sites in blue.
Retinal electroporation and explant culture
In vitro electroporation and explant culture of retinas were performed as described (Hsiau et al. 2007). In brief, eyes were removed from decapitated P0 CD1 pups in a sterile fashion, and the retinas were isolated from the sclera and surrounding tissue leaving the lens in place. The retinas were then subjected to electroporation and grown for 8 d in explants culture. Retinas were subsequently harvested, fixed, and imaged in both flat-mount and section exactly as described previously (Hsiau et al. 2007).
Quantification of cis-regulatory activity in explanted retinas
Quantification of cis-regulatory activity was performed on flat-mounted retinas as described previously (Lee et al. 2010). For analysis of Rho-CBRs, individual constructs were coelectroporated with a loading control, Rho-CBR3-eGFP, into explanted P0 mouse retinas, grown for 8 d and then imaged. Analysis of synthetic CRX spacing constructs was performed in the same fashion except that the synthetic construct “Crx × 3” (Supplemental Fig. S6F) fused to eGFP was used as the loading control. All subsequent image analysis steps were as described (Lee et al. 2010).
Acknowledgments
We thank C. Cepko for helping establish this collaboration between the Corbo and Langmann labs and for providing Crx−/− mice. We thank A. Swaroop for sharing Nrl−/− mice and for providing a Crx expression plasmid. We thank J. Lieb and D. Tillo for their advice on nucleosome positioning as well as for sharing unpublished results. We also thank N. Williams and C. Odonkor for their participation in the early phases of the CRX spacing analysis. We thank D. Ibberson and R. Carmouche from the GeneCore sequencing team and Bernward Klocke from Genomatix for their excellent support. Finally, we thank C. Micchelli, H. Stöhr, Y. Walczak, and all the members of the Corbo and Langmann labs for advice and input. This project was funded by the NIH (EY018826), The American Health Assistance Foundation, the German Research Foundation (LA1203/7-4), and Pro Retina.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE20012.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109405.110.
References
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, et al. 2000. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci 97: 5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, et al. 2006. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci 103: 3890–3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E 1998. Autoregulation of eukaryotic transcription factors. Prog Nucleic Acid Res Mol Biol 60: 133–168 [DOI] [PubMed] [Google Scholar]
- Bessant DA, Payne AM, Mitton KP, Wang QL, Swain PK, Plant C, Bird AC, Zack DJ, Swaroop A, Bhattacharya SS 1999. A mutation in NRL is associated with autosomal dominant retinitis pigmentosa. Nat Genet 21: 355–356 [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Fraioli RE, Furukawa T, Cepko CL 2001. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell 107: 579–589 [DOI] [PubMed] [Google Scholar]
- Brzezinski JA IV, Lamba DA, Reh TA 2010. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 137: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain G, Fossat N, Brun G, Lamonerie T 2006. Molecular dissection reveals decreased activity and not dominant negative effect in human OTX2 mutants. J Mol Med 84: 604–615 [DOI] [PubMed] [Google Scholar]
- Chen CM, Cepko CL 2000. Expression of Chx10 and Chx10-1 in the developing chicken retina. Mech Dev 90: 293–297 [DOI] [PubMed] [Google Scholar]
- Chen J, Tucker CL, Woodford B, Szel A, Lem J, Gianella-Borradori A, Simon MI, Bogenmann E 1994. The human blue opsin promoter directs transgene expression in short-wave cones and bipolar cells in the mouse retina. Proc Natl Acad Sci 91: 2611–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ 1997. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19: 1017–1030 [DOI] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J 2005. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci 25: 118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A 2006. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet 15: 2588–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Nix DA, Shultzaberger RK, Gasch AP, Eisen MB 2006. Flexible promoter architecture requirements for coactivator recruitment. BMC Mol Biol 7: 16 doi: 10.1186/1471-2199-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu MI, Nathans J 1994. A sequence upstream of the mouse blue visual pigment gene directs blue cone-specific transgene expression in mouse retinas. Vis Neurosci 11: 773–780 [DOI] [PubMed] [Google Scholar]
- Corbo JC, Cepko CL 2005. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet 1: e11 doi: 10.1371/journal.pgen.0010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Myers CA, Lawrence KA, Jadhav AP, Cepko CL 2007. A typology of photoreceptor gene expression patterns in the mouse. Proc Natl Acad Sci 104: 12069–12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST, Pearson JC 2009. Transcriptional autoregulation in development. Curr Biol 19: R241–R246 doi: 10.1016/j.cub.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN Jr 2005. Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci 46: 2156–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH 2006. The regulatory genome: Gene regulatory networks in development and evolution. Academic Press, London, UK [Google Scholar]
- DeAngelis MM, Grimsby JL, Sandberg MA, Berson EL, Dryja TP 2002. Novel mutations in the NRL gene and associated clinical findings in patients with dominant retinitis pigmentosa. Arch Ophthalmol 120: 369–375 [DOI] [PubMed] [Google Scholar]
- DesJardin LE, Hauswirth WW 1996. Developmentally important DNA elements within the bovine opsin upstream region. Invest Ophthalmol Vis Sci 37: 154–165 [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W 2005. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21: 3439–3440 [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. 1997. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91: 543–553 [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM 1998. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet 18: 311–312 [DOI] [PubMed] [Google Scholar]
- Fujieda H, Bremner R, Mears AJ, Sasaki H 2009. Retinoic acid receptor-related orphan receptor alpha regulates a subset of cone genes during mouse retinal development. J Neurochem 108: 91–101 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Cepko CL 1997. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91: 531–541 [DOI] [PubMed] [Google Scholar]
- Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL 1999. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet 23: 466–470 [DOI] [PubMed] [Google Scholar]
- Furukawa A, Koike C, Lippincott P, Cepko CL, Furukawa T 2002. The mouse Crx 5′-upstream transgene sequence directs cell-specific and developmentally regulated expression in retinal photoreceptor cells. J Neurosci 22: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA 2009. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457: 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes SD, Brent R 1991. A genetic model for interaction of the homeodomain recognition helix with DNA. Science 251: 426–430 [DOI] [PubMed] [Google Scholar]
- Hanes SD, Riddihough G, Ish-Horowicz D, Brent R 1994. Specific DNA recognition and intersite spacing are critical for action of the bicoid morphogen. Mol Cell Biol 14: 3364–3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW 2005. oPOSSUM: Identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res 33: 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O 2008. Regulatory logic of neuronal diversity: Terminal selector genes and selector motifs. Proc Natl Acad Sci 105: 20067–20071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC 2007. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE 2: e643 doi: 10.1371/journal.pone.0000643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Huang Y, Hanna DB, Freund CL, Affatigato LM, Carr RE, Zack DJ, Stone EM, McInnes RR 1998. Retinal degenerations with truncation mutations in the cone-rod homeobox (CRX) gene. Invest Ophthalmol Vis Sci 39: 2417–2426 [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH 1998. The major cell populations of the mouse retina. J Neurosci 18: 8936–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 doi: 10.1093/nar/gkn875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T, Lai CC, Weigelt K, Tam BM, Warneke-Wittstock R, Moritz OL, Weber BH 2008. CRX controls retinal expression of the X-linked juvenile retinoschisis (RS1) gene. Nucleic Acids Res 36: 6523–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Myers C, Williams N, Abdulaziz M, Corbo JC 2010. Quantitative fine-tuning of photoreceptor cis-regulatory elements through affinity modulation of transcription factor binding sites. Gene Ther (in press) doi: 10.1038/gt.2010.77 [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A 1996. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene 12: 207–211 [PubMed] [Google Scholar]
- Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL 2000. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol 10: 301–310 [DOI] [PubMed] [Google Scholar]
- Makeev VJ, Lifanov AP, Nazina AG, Papatsenko DA 2003. Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res 31: 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A 2001. Nrl is required for rod photoreceptor development. Nat Genet 29: 447–452 [DOI] [PubMed] [Google Scholar]
- Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A 2000. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem 275: 29794–29799 [DOI] [PubMed] [Google Scholar]
- Morabito MA, Yu X, Barnstable CJ 1991. Characterization of developmentally regulated and retina-specific nuclear protein binding to a site in the upstream region of the rat opsin gene. J Biol Chem 266: 9667–9672 [PubMed] [Google Scholar]
- Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D 2001. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet 27: 94–98 [DOI] [PubMed] [Google Scholar]
- Nie Z, Chen S, Kumar R, Zack D 1996. RER, an evolutionarily conserved sequence upstream of the rhodopsin gene, has enhancer activity. J Biol Chem 271: 2667–2675 [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T 2003. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci 6: 1255–1263 [DOI] [PubMed] [Google Scholar]
- Oh EC, Cheng H, Hao H, Jia L, Khan NW, Swaroop A 2008. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res 1236: 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi A, Peng GH, Poth EM, Lee DA, Chen J, Alexis U, de Melo J, Chen S, Blackshaw S 2010. The orphan nuclear hormone receptor ERRbeta controls rod photoreceptor survival. Proc Natl Acad Sci 107: 11579–11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Pennesi ME, Cho JH, Yang Z, Wu SH, Zhang J, Wu SM, Tsai MJ 2003. BETA2/NeuroD1 null mice: A new model for transcription factor-dependent photoreceptor degeneration. J Neurosci 23: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Esumi N, Chen Y, Wang Q, Chowers I, Zack DJ 2005. Identification of regulatory targets of tissue-specific transcription factors: Application to retina-specific gene regulation. Nucleic Acids Res 33: 3479–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner A, Sun H, Nathans J 1999. Molecular genetics of human retinal disease. Annu Rev Genet 33: 89–131 [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MR, Hendrickson A, McGuire CR, Reh TA 2005. Retinoid X receptor γ is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci 46: 2897–2904 [DOI] [PubMed] [Google Scholar]
- Rodieck RW 1998. The first steps in seeing. Sinauer, Sunderland, MA [Google Scholar]
- Rothbacher U, Bertrand V, Lamy C, Lemaire P 2007. A combinatorial code of maternal GATA, Ets and β-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development 134: 4023–4032 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sanuki R, Omori Y, Koike C, Sato S, Furukawa T 2010. Panky, a novel photoreceptor-specific ankyrin repeat protein, is a transcriptional cofactor that suppresses CRX-regulated photoreceptor genes. FEBS Lett 584: 753–758 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Arnosti DN, Levine M 1993. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119: 762–772 [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP 1998. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet 63: 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368 [DOI] [PubMed] [Google Scholar]
- Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. 2008. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321: 956–960 [DOI] [PubMed] [Google Scholar]
- Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A 2001. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem 276: 36824–36830 [DOI] [PubMed] [Google Scholar]
- Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N 1992. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci 89: 266–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaroop A, Chew EY, Rickman CB, Abecasis GR 2009. Unraveling a multifactorial late-onset disease: From genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet 10: 19–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Hughes TR 2009. G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442 doi: 10.1186/1471-2105-10-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE 5: e9129 doi: 10.1371/journal.pone.0009129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Bristow J, Pennacchio LA 2007. Enhancer identification through comparative genomics. Semin Cell Dev Biol 18: 140–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV 2005. enoLOGOS: A versatile web tool for energy normalized sequence logos. Nucleic Acids Res 33: W389–W392 doi: 10.1093/nar/gki439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chung M, Morabito MA, Barnstable CJ 1993. Shared nuclear protein binding sites in the upstream region of the rat opsin gene. Biochem Biophys Res Commun 191: 76–82 [DOI] [PubMed] [Google Scholar]
- Zack DJ, Bennett J, Wang Y, Davenport C, Klaunberg B, Gearhart J, Nathans J 1991. Unusual topography of bovine rhodopsin promoter-lacZ fusion gene expression in transgenic mouse retinas. Neuron 6: 187–199 [DOI] [PubMed] [Google Scholar]
- Zhu X, Ma B, Babu S, Murage J, Knox BE, Craft CM 2002. Mouse cone arrestin gene characterization: Promoter targets expression to cone photoreceptors. FEBS Lett 524: 116–122 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]