Abstract
Here, we fully characterize the genomes of 14 Plasmodium falciparum patient isolates taken recently from the Iquitos region using genome scanning, a microarray-based technique that delineates the majority of single-base changes, indels, and copy number variants distinguishing the coding regions of two clones. We show that the parasite population in the Peruvian Amazon bears a limited number of genotypes and low recombination frequencies. Despite the essentially clonal nature of some isolates, we see high frequencies of mutations in subtelomeric highly variable genes and internal var genes, indicating mutations arising during self-mating or mitotic replication. The data also reveal that one or two meioses separate different isolates, showing that P. falciparum clones isolated from different individuals in defined geographical regions could be useful in linkage analyses or quantitative trait locus studies. Through pairwise comparisons of different isolates we discovered point mutations in the apicoplast genome that are close to known mutations that confer clindamycin resistance in other species, but which were hitherto unknown in malaria parasites. Subsequent drug sensitivity testing revealed over 100-fold increase of clindamycin EC50 in strains harboring one of these mutations. This evidence of clindamycin-resistant parasites in the Amazon suggests that a shift should be made in health policy away from quinine + clindamycin therapy for malaria in pregnant women and infants, and that the development of new lincosamide antibiotics for malaria should be reconsidered.
The World Health Organization (WHO) campaign to eradicate malaria in the 1950s and 1960s was initially largely successful in decreasing the burden of malaria. Drug failure eventually led to a resurgence in the number of malaria cases in the 1990s and caused vast numbers of deaths that could have been avoided through a better appreciation of the prevalence of drug-resistant malaria and more informed choices of first-line drugs (Greenwood et al. 2008). While malaria deaths are now likely to decline, primarily because of the introduction of artemisinin-based combination therapy (ACT) as well as increased insecticide spraying and the use of bed nets (Greenwood et al. 2008), this may only be temporary. Indeed, there has been a general increase in the parasite clearance times in ACT-treated Plasmodium falciparum malaria cases from near the Thai–Cambodian border, suggesting that case numbers may soon begin increasing (Dondorp et al. 2009).
Remarkably, although artemisinin is used on tens of millions of individuals annually, we have little idea about how it acts or which genes contribute to resistance, confounding the community's ability to monitor the spread of resistance using molecular markers and to deploy new therapies (Eastman and Fidock 2009). The fact that the genes involved in artemisinin resistance are not known is due to a variety of problems, including the fact that in vivo resistance has not been replicated in vitro (Dondorp et al. 2009). Additionally, the association of phenotypes with genotypes in P. falciparum is hampered by the difficulties in performing complementation studies due to extremely low transfection efficiencies and the fact that laboratory crosses of drug-sensitive and drug-resistant P. falciparum cannot be easily performed. Thus, it took more than 40 yr between the identification of chloroquine resistance in the field (Harinasuta et al. 1965) and confirmation that resistance is due to mutations in the chloroquine resistance transporter (pfcrt, MAL7P1.27) (Wellems et al. 1990; Fidock et al. 2000; Sidhu et al. 2002). While the recombinant progeny from one of the three extant crosses (Walliker et al. 1987; Wellems et al. 1990; Hayton et al. 2008) have most famously been used to map chloroquine resistance (Wellems et al. 1990), they have been used to map loci contributing to a wide variety of phenotypes that distinguish parental clones. For example, they have already been scored for a variety of different phenotypes that are related to drug sensitivity, including antifolate sensitivity (Wang et al. 2004b), quinine sensitivity (Ferdig et al. 2004), expression levels (Gonzales et al. 2008), and plasmodial surface ion channels (Alkhalil et al. 2009), but they could be scored for any phenotype that quantitatively distinguishes the parental strains Dd2 and HB3, such as propensity to mutate in the laboratory (Rathod et al. 1997). However, there are a limited number of phenotypes that distinguish these two laboratory strains that were derived from patients 40 yr ago. While more crosses would provide valuable data for many researchers interested in parasite genetics, there are ethical considerations associated with using primates in research. An alternative to creating new recombinant progeny is to find existing recombinant isolates that arose from recent meioses occurring in humans. Such parasites might be identified by performing full-genome analyses on parasites from a limited geographical area and could provide the malaria community with an unprecedented number of parasites differing in a variety of phenotypes for use in linkage or quantitative trait locus (QTL) studies.
One attractive group of parasites for full genome investigation is from the Peruvian Amazon. Due to the low transmission rates it is expected that parasites isolated from an individual will be from a single clone infection. In addition, malaria was eradicated in the 1960s in this region but re-emerged with epidemics in the early 1990s (Aramburu Guarda et al. 1999; Branch et al. 2005), suggesting that the genomes might contain signatures of selective sweeps (Wootton et al. 2002; Roper et al. 2004). At first, malaria in this region was treated with chloroquine (first-line), sulfadoxine-pyrimethamine (second-line), and quinine with clindamycin or tetracycline (third-line) (Aramburu Guarda et al. 1999), but the emergence of resistance resulted in widespread chloroquine and antifolate treatment failure (Durand et al. 2007). Today, malaria remains hypoendemic with low levels of seasonal transmission of P. falciparum and P. vivax parasites in the region surrounding Iquitos, Peru (Roshanravan et al. 2003). Previous studies of parasites in the region describe only one or two independent haplotypes for important drug-resistance genes, suggesting a limited number of founders for this population (Bacon et al. 2009) and suggest that we might find recombinant progeny in this region.
In this study, we performed genome scanning on 14 P. falciparum patient isolates from a limited geographical region. We show that the parasite population in the Peruvian Amazon is very closely related, with combinations of only two to four different genotypes for drug resistance genes, suggesting at most four parental haplotypes. Furthermore, some parasites taken from different patients who presented with symptoms were essentially clones of one another, while others were recent meiotic siblings that could be useful in linkage studies or eQTL analyses. Unexpectedly, genome scanning also revealed uncharacterized mutations in the apicoplast 23S rRNA that distinguished some Peruvian strains from the reference strain, 3D7. Because one of these mutations had been previously associated with lincosamide antibiotic resistance in the chloroplast and many bacterial species (Vester and Douthwaite 2001), sensitivity testing was performed revealing that the parasites harboring the mutation had indeed become resistant to clindamycin, a drug used in combination with quinine to treat pregnant women and infants for malaria in Peru. These are the first documented cases of resistance to this class of drugs in malaria and suggest that the use of lincosamide drugs in treating malaria should be reconsidered.
Results
Initial genotyping shows a limited number of parental haplotypes
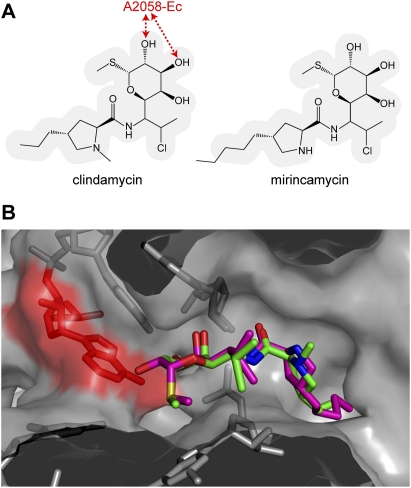
An initial motivation for conducting this study was to examine field isolates to determine whether the same types of genomic changes typically found in laboratory strains of P. falciparum would be discovered. To this end, patient isolates of P. falciparum were obtained with informed consent from the region surrounding Iquitos, Peru during the peak malaria season (February through July) of 2006 (Supplemental File 1). The isolates were taken into short-term in vitro culture for periods of weeks and subjected to genotyping of known resistance genes by PCR. As indicated by increased sulfadoxine-pyrimethamine sensitivity in Peru (Durand et al. 2007), we detected wild-type pfdhps (dihydropteroate synthetase, PF08_0095) and single and triple mutant alleles (NCNI and ICNL, respectively) of pfdhfr-ts (bifunctional dihydrofolate reductase-thymidylate synthase, PFD0830w) by conventional genotyping, as well as the K76T mutation that confers resistance to chloroquine resistance mutation (Sidhu et al. 2002). Altogether, there were four genotypes for pfmdr1 (multidrug resistance protein, PFE1150w), three for pfdhps, three for pfdhfr-ts, and two for pfcrt, suggesting four parental haplotypes in the Peruvian population.
Gene copy number analysis reveals only widespread subtelomeric variants
Because drug resistance can also be conferred by copy number variations (CNVs), the isolates were next examined by microarray. Genomic DNA from 14 isolates was analyzed on high-density full-genome tiling microarrays (Table 1; Dharia et al. 2009). First, 10–15 μg of whole genomic DNA from parasites grown in leukocyte-depleted red cells from each Peruvian isolate was fragmented with DNase I, end-labeled, and hybridized to a tiling microarray that covers ∼90% of coding regions, which comprise 53% of the genome and 60% of noncoding regions. The array contains 4.8 million, mostly 25-mer, single-stranded probes that are perfect matches to the reference 3D7 genome and are spaced, on average, 2–3 nt apart on alternating strands. Only probes that map to a single location in the genome were used for these analyses. Although there are fewer probes to repetitive regions, each nucleotide in the P. falciparum coding genome generally can be probed nine to 13 times on the microarray.
Table 1.
Patient isolates and genotypes
Genomic DNA from 14 patient-derived P. falciparum isolates was applied to the microarray from throughout the 2006 malarious season from various healthcare centers surrounding Iquitos, Peru. Standard genotyping was used to determine the drug resistance alleles for pfdhfr-ts (N51I, C59R, S108T/N, I164L, *=C50R), pfdhps (A437G, K540E, A581G), pfmdr1 (N86Y, D142G, Y184F, S1034C, N1042D, D1246Y), and pfcrt (C72S, V73, M74I, N75E, K76T, H97Q). The apicoplast 23S (api 23S) rRNA genotype was determined by microarray analysis combined with PCR and direct sequencing. Mutations are underscored.
The advantage of using microarrays is that they can easily detect copy number variations and deletions. Recent reports indicate CNVs can be a way for malaria parasites to provide dosage compensation for a protein whose activity is either inhibited by a drug or whose expression otherwise increases resistance (Kidgell et al. 2006; Sidhu et al. 2006; Nair et al. 2008). We therefore first tested to see whether the Peruvian strains would also harbor CNVs by examining the genome-wide hybridization patterns. As described previously (Kidgell et al. 2006; Dharia et al. 2009), we calculated the log2 ratios for the average intensity of the probes corresponding to each gene in the genome versus a 3D7 reference hybridization and systematically looked in all isolates for sets of more than three consecutive genes with increased log2 ratios above 0.5. We did not see large-scale amplifications surrounding the pfmdr1 locus, which have previously been observed in laboratory strains (Kidgell et al. 2006). This was expected as there were point mutations in pfmdr1, and while amplification of wild-type pfmdr1 is associated with mefloquine resistance (Price et al. 2004), there have been no reports of amplification of mutant pfmdr1. Surprisingly, we did not see amplification around the pfgch1 (GTP cyclohydrolase I, PFL1155w) locus in response to antifolate use despite the historical use of this treatment in Peru (Kidgell et al. 2006; Nair et al. 2008). This may be related to the re-emergence of antifolate-sensitive alleles of pfdhfr-ts and pfdhps, or it may be related to the lack of the quadruple point mutation pfdhfr-ts in Peru (Bacon et al. 2009). Although single-gene events may have been missed, we only saw evidence of widespread multigene CNVs in the Peruvian isolates in subtelomeric regions like the deletions described below (Supplemental File 2), indicating that they appear to be less common in field isolates than in laboratory strains.
Deletions in diagnostic genes may confound P. falciparum diagnoses
The MOID algorithm was also used as previously described to identify regions that were likely to be deleted in the Peruvian strains (Dharia et al. 2009). We found deleted regions covering 20–25 kb in subtelomeric regions, including the apparent deletion of important diagnostic loci like histidine-rich proteins II and III (HRP2, MAL7P1.231 and HRP3, MAL13P1.480, respectively) (Supplemental File 2; Supplemental Fig. S1; Wongsrichanalai et al. 2007; Martin et al. 2009). Because many malaria diagnostic tests that distinguish between P. falciparum and P. vivax are based on immune recognition between commercial antibodies and these proteins (Wongsrichanalai et al. 2007), it is likely that false-negative readings could be obtained. These data suggest that the use of HRP2- or HRP3-based diagnostic tests in this region should be reconsidered.
Gene polymorphism frequencies revealed expected levels of variation in the population
The microarray design used here can also be used to detect single nucleotide polymorphisms (SNPs). We previously used this design to detect over 90% of SNPs probed by the microarray that distinguished different strains of P. falciparum with a 10% false-positive rate (Dharia et al. 2009). In order to globally detect polymorphism frequencies for different gene families, we ran our polymorphism detection algorithm for each isolate with the 3D7 reference hybridization (Supplemental File 3). We saw an expected high frequency of polymorphisms in genes annotated “antigenic variation” (GO:0020033), with 60.2% average polymorphic probe frequency across all 14 isolates and a low frequency of polymorphisms in genes annotated “metabolic process” (GO:0008152), with 2.6% average polymorphic probe frequency across all 14 isolates. This distribution is similar to frequencies detected in the analysis of a variety of different laboratory strains (Kidgell et al. 2006). In the Peruvian population, important possible vaccine candidates, including the circumsporozoite protein (PFC0210c), apical membrane antigen 1 (PF11_0344), merozoite surface protein 1 (PFI1475w), thrombospondin-related anonymous protein (PF13_0201), and CelTOS (PFL0800c), all ranked amongst the top 10% most variable of all genes in the genome including subtelomeric genes, highlighting the difficulty in finding antigens that will provide universal protection (Winzeler 2008).
Microarray analysis demonstrates limited diversity
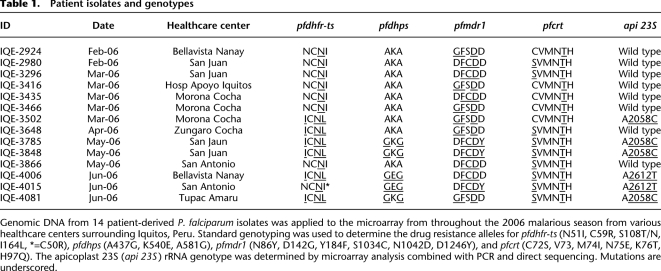
Our next goal was to determine whether or not there might be haplotypes in linkage disequilibrium in the population that might reveal novel drug resistance loci. We thus first performed pairwise analysis on the patient isolates by first computationally identifying all polymorphisms that distinguished each isolate of the pair. We then plotted 100-kb blocks that appeared to share the same genotype as a function of genome position. (Fig. 1A–I). Unexpectedly, pairwise comparisons of isolate hybridization intensities revealed that many isolates shared the same genotype for large portions of chromosomes or were virtually “clones” of one another. The low endemicity, low rates of multiple infections, and the spread of “clonal” parasite genotypes suggest high rates of self-mating as previously described in Venezula (Urdaneta et al. 2001). However, with “clonal” genotypes isolated over a period of months there was a great amount of diversity in subtelomeric regions, including var, rifin, and stevor genes, and internal var gene clusters suggesting high levels of subtelomeric mutations (Fig. 1D,E) introduced by self-mating or mitotic recombination that may be influenced by immune selection given that antigenic genes often reside in these regions. Cloning and sequencing of a member of the var gene family (PFD1015c; Supplemental Fig. S2) revealed different sequence patterns between “clonal” isolates, thus confirming the expected differences. While the lack of diversity could be due to array insensitivity, especially as other comparative genome hybridization approaches have shown that only a fraction of SNPs (12%–24%) are typically detected using arrays complementary to the P. falciparum genome (Tan et al. 2009), this seems unlikely. First, Tan et al. (2009) also showed that sensitivity is inversely proportional to probe length, and thus it is difficult to compare the performance of 25-mer arrays, such as those used here, to 45–65-mer arrays. In addition, probe density is also a factor, as there are 10× more probes on the array used here than those used in the Tan et al. (2009) study. Indeed, previous work has shown that up to 90% of the existing variation in unique coding regions can be detected using our array (Dharia et al. 2009), and this array has been used to successfully discover newly emerged SNPS in the P. falciparum genome (Rottman et al. 2010).
Figure 1.
Pairwise comparison of isolates reveal progeny of a natural cross. (A) Log2 ratios of probe intensities plotted along chromosome 14 for the hybridization of genomic DNA from isolates IQE2924 and IQE3502 display polymorphisms, indicated by upward and downward spikes for the left half of the chromosome. The normal distribution of log2 ratios around 0 for the right half of the chromosome indicate that these two isolates share the same DNA sequence for this half of the chromosome. (B) Log of P-value for polymorphism detection plotted on chromosome 14 shows polymorphisms predicted only for half of the chromosome. The blue lines indicate polymorphisms in IQE2924 using IQE3502 as the reference, and the green indicates polymorphisms in IQE3502 using IQE2924 as the reference. The horizontal dashed line indicates a P-value cutoff of 1 × 10−5. (C) Blocks of 100 kb of sequence that displayed no polymorphisms by pairwise comparison of isolates were drawn (colored blue or red in panels C–H), and chromosome 14 is displayed. (D,E) Essentially “clonal” parasites, IQE2924 and IQE3416 (D) and IQE3848 and IQE3785 (E) show very few polymorphisms for the core chromosomal region. (F,G) Other pairwise comparisons showed parasites that shared the same genotype blocks for approximately half of the core chromosomal regions. IQE2924 and IQE3502 shared ∼60%, while IQE3848 and IQE3502 shared ∼40%. (H) Overlaying the genotype blocks comparing IQE3502 to IQE2924 and IQE3848 revealed that the latter two genotypes could explain the genotype of IQE3502, indicating that it was the progeny of a natural cross between these two genotypes. Magenta regions did not contain any polymorphisms between the parental and progeny genotypes, and thus are of uncertain assignment. (I) Analysis of inheritance of organellar genomes revealed that IQE3502 inherited the IQE3848 genotype, suggesting that this was the maternal genotype, while IQE2924 was the paternal.
Searching for isolate pairs that could define the entire genotype of a third isolate, we discovered that isolate IQE3502 was the product of a cross between parasites sharing the IQE2924/3416/3466 and IQE3848/3785 genotypes (Fig. 1F–H). In the progeny, we detected between 13 definitive and 19 possible recombination breakpoints in this cross that corresponds to a recombination distance of 12–18 kb per centimorgan, correlating well with the reported value of ∼15 kb per centimorgan (Su et al. 2007). Due to the maternal inheritance of the nonnuclear mitochondrial and apicoplast genomes (Creasey et al. 1994; Okamoto et al. 2009), analysis of the parental and offspring genotypes demonstrated that the maternal genotype was IQE3848/3785 and the paternal genotype was IQE2924/3416/3466 (Fig. 1I). The pairwise comparison data, as well as the fact that there are at most four different drug resistance alleles for each gene examined, indicated that P. falciparum parasites in the area surrounding Iquitos are likely the descendents of only four multidrug-resistant parental clones.
The Peruvian isolates have two mitochondrial and four apicoplast genotypes
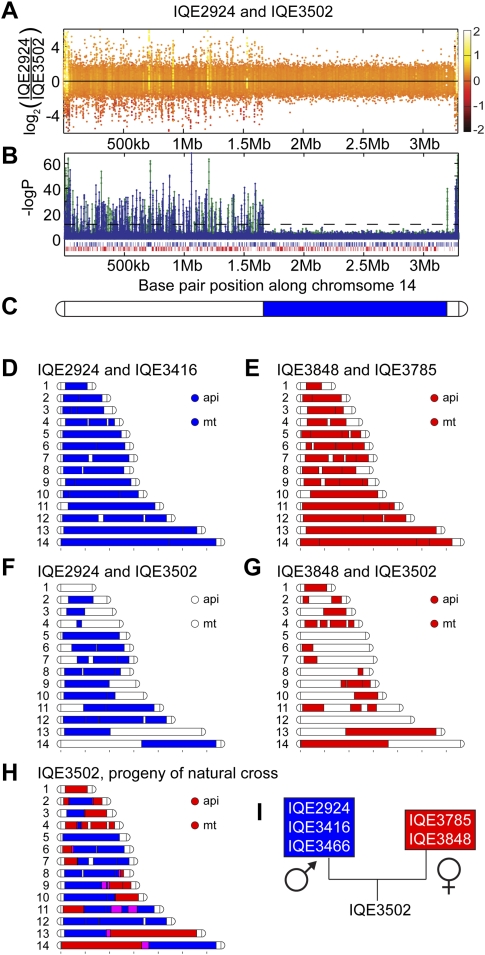
The extensive mitochondrial genotyping carried out to determine the age of P. falciparum demonstrates a single mitochondrial genotype (haplotype 5) in Peruvian parasite populations in 1999 (Joy et al. 2003). We confirmed that our microarray could be used for limited mitochondrial genotyping by validating Dd2 and HB3 hybridizations (Dharia et al. 2009) with data from Joy et al. (2003). Our samples from 2006 indicate the presence of two mitochondrial haplotypes in Peru by microarray with mutations near base pair positions 1690 (Fig. 2Ad), 4807, and 4947 (Fig. 2Ae; Supplemental File 1), but alleles may have been missed due to a lack of unique probes in repetitive regions of the mitochondrial genome.
Figure 2.
Apicoplast polymorphisms correlated with clindamycin resistance. (A) Polymorphism detection was performed for the organellar genomes (api, apicoplast; mt, mitochondrion) versus the reference 3D7. IQE3502 and IQE4006 represent the mutations we saw in the Peruvian parasites. PCR amplification and sequencing revealed these polymorphisms to be point mutations: A4210C (a) and A4743T (b) in PFC10_API0010, a gene located in the inverted repeat region of the apicoplast genome (IRA, IRB). Mutation T16659C (c) was a synonymous SNP in apicoplast gene PFC10_API0060. The mitochondrion polymorphisms were predicted to be near base pair positions 1690 (d), 4807 (data not shown), and 4947 (e). (B) The peptidyl transferase domain of the 23S rRNA secondary structure was modeled based on E. coli. Mutations of residues (blue letters) were implicated in bacteria and algal chloroplasts resistance to lincosamides. The red circles indicate residues that were mutated in our patient isolates. While A2612U was not correlated with resistance, A2058C was correlated with greater than 100-fold increase in clindamycin in vitro EC50.
Little work has been done to look at mutations in the apicoplast genome in P. falciparum, an essential chloroplast-like organelle thought to be of algal origin (Dahl and Rosenthal 2008). Due to the high AT content (85.75%) of the plastid genome, we only have unique probes to 55.9% of the 34-kb apicoplast genome. Like the chloroplast genome, the circular apicoplast genome has inverted repeat regions of length ∼5 kb, containing ribosomal RNA and tRNA genes. Because of the probable copy correction of the inverted repeats like in the chloroplast (Myers et al. 1982), we considered probes that map to the inverted repeats as unique. Upon hybridization and analysis of the Peruvian isolates, we determined the presence of four different apicoplast haplotypes characterized by combinations of two mutations in the 23S rRNA gene (PFC10_API0010) and one polymorphism in an apicoplast-encoded molecular chaperone/AAA+ gene (PFC10_API0060) (Supplemental File 1). PCR amplification and sequencing revealed these polymorphisms to be point mutations: A4210C (Fig. 2Aa) and A4743T (Fig. 2Ab) in PFC10_API0010 and T16659C (synonymous SNP) in PFC10_API0060 (Fig. 2Ac).
Clindamycin resistance is predicted by a point mutation in the apicoplast-encoded 23S rRNA
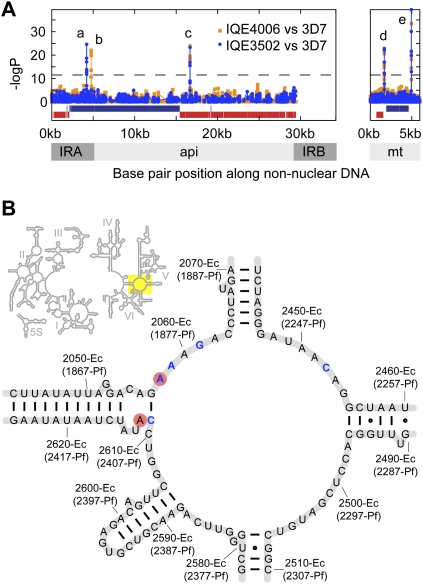
The polymorphisms A4210C and A4743T correspond to A1875C-Pf and A2409U-Pf in the 23S rRNA (positions A2058-Ec and C2612-Ec, respectively, Ec denoting Escherichia coli and Pf denoting P. falciparum numbering) (Fig. 2B, based on E. coli secondary structure) (Cannone et al. 2002). The A2058C-Ec mutation is in the same location as a described mutation in the peptidyl transferase cavity of the 23S ribosomal subunit of bacteria and algal chloroplasts that confers resistance to macrolides and lincosamides (lincomycin and clindamycin) (Harris et al. 1989; Vester and Douthwaite 2001). Clindamycin, like several antibiotics, can effectively treat malaria, presumably through the inhibition of processes in the apicoplast. Antimalarials that are thought to act against the apicoplast display a “delayed-death” phenotype, where parasite development is inhibited during the second cycle of erythrocyte invasion (Dahl and Rosenthal 2008). Complete parasite resistance to clindamycin has not been reported, but in vitro studies have suggested that mutations in the apicoplast genome confer resistance to the macrolide antibiotic azithromycin (Sidhu et al. 2007).
To measure the conservation of this RNA gene in Plasmodium species, we used publicly available reads from genome sequencing projects for P. falciparum isolates and other Plasmodium species to assemble the apicoplast-encoded sequence. In fact, all of the worldwide P. falciparum isolates that we could assemble had 100% identity for the entire 3′ 23S rRNA (Africa: D6, FCR3, 106-1, R033, SEN_34_04; South East Asia: D10, FCB, FCC-2, K1, VS-1; India: IGH-CR14, RAJ116; Central America: HB3, SANTA_LUCIA; South America: 7G8; Uncertain origin: 3D7). Not only is the sequence highly conserved within P. falciparum, the regions surrounding the mutations, including every residue displayed in Figure 2B, are 100% conserved across all species for which we could assemble the RNA gene (P. vivax, P. ovale, P. reichenowi, P. knowlesi, P. berghei) (Supplemental Files 4, 5).
Crystal structures of the Haloarcula marismortui and Deinococcus radiodurans large subunit ribosomal RNA (LSU) with clindamycin bound demonstrate that A2058-Ec is an important hydrogen-bond partner (Schlunzen et al. 2001; Tu et al. 2005). Specifically, Tu and colleagues suggest that the desolvation of the N2 of a guanine at position 2058-Ec accounts for more than a 10,000-fold decrease in affinity to macrolides and lincosamides, and thus, the A2058G-Ec mutation confers resistance in eubacteria (Tu et al. 2005). Similarly, we hypothesized that an A2058C-Ec mutation would result in an analogous desolvation effect for one of the three hydrogen-bonding groups in cystosine and, thus, would result in clindamycin resistance in P. falciparum. We conducted docking studies of clindamycin and a related lincosamide proposed antimalarial, mirincamycin (Olliaro and Wells 2009), to a P. falciparum homology model of the 23S peptidyl transferase domain based on the D. radiodurans structure (Supplemental Fig. S3; Schlunzen et al. 2001). We observed that both drugs bind in the same orientation, interacting similarly with A2058-Ec (Fig. 3A,B), and thus the A2058C-Ec mutation should greatly decrease the affinity of both compounds. In contrast to A2058-Ec, the other mutation we observed, A2612-Ec is distant from the clindamycin-binding pocket, suggesting that Peruvian parasites harboring the A2612U-Ec mutation would not be resistant to either clindamycin or mirincamycin.
Figure 3.
Structures of clindamycin and mirincamycin and the predicted interactions with 23S rRNA peptidyl transferase cavity. (A) Previous crystallography studies have shown A2058-Ec (red) is important for making contacts with clindamycin (Schlunzen et al. 2001), and the similarity of chemical structure of clindamycin and mirincamycin suggested that cross-resistance may occur. (B) Lowest energy dockings of clindamycin (green) and mirincamycin (magenta) to a homology model of the P. falciparum peptidyl transferase domain demonstrated that both molecules interact with the 23S rRNA in the same way, and thus, A2058C-Ec (red) was predicted to confer resistance to both lincosamide drugs.
In vitro drug susceptibility tests reveal clindamycin resistance
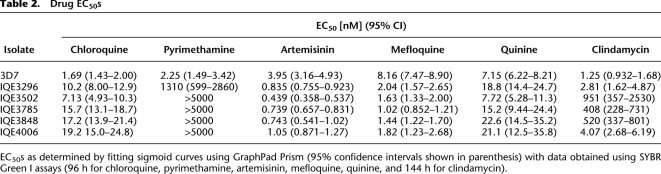
Although not all strains could be revived for drug-sensitivity testing, we were able to determine the EC50 for clindamycin using SYBR Green I assays with 144-h incubations for the reference strain 3D7 and for one isolate that had wild-type 23S rRNA and four that carried the A2058C-Ec or A2612U-Ec (Table 2). Treatment of parasites with clindamycin is thought to result in a “delayed-death” phenotype due to the loss of the apicoplast in the next generation after administration of the drug (Ramya et al. 2007). The effect of the drug is therefore only measurable after the second generation of parasites, and a longer incubation period is needed. As predicted, we discovered that the A2058C-Ec mutation was correlated with clindamycin resistance, while the A2612T-Ec mutation was not. Although the mutations in the apicoplast rRNA may have arisen in response to other antibiotics that are used to treat bacterial infections in the region, Peruvian P. falciparum isolates with A2058C-Ec were clindamycin resistant with greater than a 100-fold increase in EC50.
Table 2.
Drug EC50s
EC50s as determined by fitting sigmoid curves using GraphPad Prism (95% confidence intervals shown in parenthesis) with data obtained using SYBR Green I assays (96 h for chloroquine, pyrimethamine, artemisinin, mefloquine, quinine, and 144 h for clindamycin).
Unfortunately, since there is no method for allelic replacement of apicoplast rRNA genes to confirm that the A2058C-Ec mutation confers clindamycin resistance in P. falciparum, it is formally possible that clindamycin resistance may be, in part, due to another mutation in the genome. In order to identify portions of the genome that may confer resistance, we assumed resistance was inherited through shared haplotype blocks. Thus, by comparing the resistant isolates to each other and to the sensitive isolates, we were able to exclude 83.4% of the parasite nuclear genome (Supplemental Table S1). While the remaining 16.6% of the genome (Supplemental File 6) does encode rRNA methylases, efflux proteins, or inactivating enzymes similar to those described in bacterial clindamycin resistance (Roberts 2008), there is a multitude of evidence that the 2058-Ec mutation confers some level of clindamycin resistance in these isolates. First, there is the evidence from E. coli, Brachyspira hyodysenteriae, Helicobacter pylori, Mycoplasma pneumoniae, Propionibacteria, Streptococcus pneumoniae, Streptomyces ambofaciens, and the Chlamydomonas reinhardtii chloroplast that a mutation at position A2058-Ec to G, C, or U confers licosamide resistance (Vester and Douthwaite 2001). Second, there is the evidence from the clindamycin-bound crystal structures of the 23S rRNA from H. marismortui and D. radiodurans discussed above (Schlunzen et al. 2001; Tu et al. 2005). Third, treatment with clindamycin results in a “delayed-death” phenotype consistent with inhibition of apicoplast function, suggesting that the apicoplast genome is a likely place to look for resistance-conferring mutations. Fourth, clindamycin-resistant malaria has not been reported before and none of the available sequences for the Plasmodium apicoplast have this mutation (Supplemental File 5). Finally, the related Apicomplexan parasite Toxoplasma gondii was selected for resistance to clindamycin and had acquired a mutation at 2061-Ec in the same conserved region of the peptidyl transferase domain of the apicoplast-encoded rRNA (Camps et al. 2002). Although additional testing and genotyping of additional strains will be needed to absolutely rule out a chromosomal contribution, it seems highly likely that clindamycin resistance is caused by this mutation.
The K76T mutation gives low levels of chloroquine resistance in Peruvian strains
While testing for clindamycin resistance we also examined the isolates' sensitivites to other drugs using the SYBR green proliferation assay that we routinely use in drug discovery (Wu et al. 2009), which gives accurate estimates of the EC50s (Table 2). Substantial pyrimethamine resistance was confirmed as predicted by the single-mutant pfdhfr-ts allele and to a greater extent by the triple-mutant pfdhfr-ts allele. Those isolates whose EC50s were above 5 μM all contained the pfdhfr-ts quadruple mutant. Three Peruvian isolates showed slightly decreased susceptibility to quinine, consistent with the point mutation N1042D in pfmdr1 conferring quinine resistance (Sidhu et al. 2005). As predicted, all of the Peruvian isolates had greater than 10-fold increase in chloroquine EC50 compared with the chloroquine-sensitive reference 3D7 consistent with genotyping, indicating that all Peruvian isolates had the hallmark K76T mutation in pfcrt (Sidhu et al. 2002). Nevertheless, the measured EC50s of 10–20 nM does not approach that of the well-characterized strain Dd2 (typically 179 nM in our hands), which also bears the K76T allele, suggesting that additional genetic determinants contained in laboratory strains are important for levels of chloroquine sensitivity. Likewise, all Peruvian isolates tested were also more mefloquine- and artemisinin-sensitive than 3D7. This may be related to the fact that these isolates contain different pfmdr1 alleles from 3D7 (Sidhu et al. 2005), or it may be due to the presence of alleles in other pumps or transporters that have been implicated in malaria parasite drug responses, including the P. falciparum sodium hydrogen exchanger, pfnhe (Ferdig et al. 2004; Bennett et al. 2007), pfatpase6 (Eckstein-Ludwig et al. 2003; Jambou et al. 2005; Uhlemann et al. 2005), pfmdr2 (Rosenberg et al. 2006), or the multidrug resistance-associated protein (Raj et al. 2009).
Discussion
Our findings that the Peruvian parasite population is highly related and that there is an apparent low frequency of meiotic recombination is not entirely surprising. Previous population studies in areas of low endemicity in the Americas showed limited genetic diversity and high self-mating rates, although they were based on very few markers (Ariey et al. 1999; Haddad et al. 1999). Peru was also expected to have a parasite population with limited diversity, and indeed, genotyping of major vaccine candidates showed less diversity than populations in Africa (Chenet et al. 2008). Additionally, a study of the malaria drug-resistance genes using 167 P. falciparum samples collected between 1999 and 2007 has found only a limited number (two to five) of haplotypes in this region (Bacon et al. 2009). While this highly structured population may not be ideal for looking for genomic regions in linkage disequilibrium (Volkman et al. 2007), strains collected from the regions could be used in linkage analyses, and in QTL studies. The hybridization data collected here constitutes a set of genetic markers that could be useful in further analyses.
While our findings that parasite strains are separated by a limited number of meioses may only apply to regions of low endemicity, previous population studies in Africa, where recombination frequencies are expected be much higher, have mostly relied on small numbers of markers to understand how drug-resistant parasites spread and determine rates of sexual recombination between different genotypes. For example, microsatellite genotyping at 12 loci of worldwide isolates suggested that the genetic structure is linked to epidemiology with low heterozygosity in areas where transmission is low (Anderson et al. 2000). A study of the breakdown of linkage disequilibrium over a 5-kb region of the genome demonstrated that meiotic recombination is high in P. falciparum, and the apparent “clonality” of parasites is probably due to inbreeding in areas with low endemicity (Conway et al. 1999). More recent approaches using sequencing (Jeffares et al. 2007; Volkman et al. 2007) or SNP chips (Neafsey et al. 2008; Mu et al. 2010) have identified recombination rates and regions under selection with much greater precision. Similarly, our whole-genome approach may allow for a more comprehensive view of genetic structure and recombination in local, and more dispersed parasite populations in regions of high endemicity in Africa by classifying haplotypes for each core chromosomal gene in an isolate.
Additionally, our study has several implications for the diagnosis and treatment of malaria. First, the study documents the inherent genetic variability and long-term instability that is found in many subtelomeric genes. While the isolates used in these studies were all taken into short-term in vitro culture for a period of weeks in order to isolate sufficient quantities of genomic material, it is unlikely that this instability arose in vitro, since multiple clonal isolates shared the same patterns of variability in subtelomeric regions, and it has also subsequently been confirmed in uncultured isolates (Gamboa et al. 2010). This variability may contribute to the failure of rapid diagnostic tests that are directed toward antigens such as the histidine-rich protein II, encoded by a subtelomeric region that appears to be deleted in a subpopulation of Peruvian P. falciparum. This deletion has been previously described (Pieroni et al. 1998; Bell et al. 2005; Wongsrichanalai et al. 2007; Martin et al. 2009), and the presence of this deletion in the Amazon suggests that widespread use of malaria rapid diagnostic tests that depend on HRP2 will result in missed diagnoses.
Second, we show that a mutation in the apicoplast 23S rRNA correlates with a 100-fold increase in EC50 of clindamycin. Because it is relatively difficult to test for clindamycin sensitivity in vitro, active monitoring for the 23S rRNA mutation may be advisable in regions where lincosamide antibiotics are used to treat malaria. Our analysis of available sequence data suggests that mutations in the 23S rRNA are uncommon, and clindamycin resistance is not widespread. However, the parasite isolates used to draw these conclusions were, in many cases, obtained more than 10 yr ago. On the other hand, a previous study in which PCR amplification of a 595-bp fragment of the apicoplast RNA gene from Indian, Pakistani, Laotian, and Singaporean blood spots in 1997 (Tan et al. 1997) was conducted, showed evidence of several mutations near the A2058-Ec region, suggesting that clindamycin resistance may exist in Asia. Indeed, there are reports in South East Asia of pregnant women with recurrent P. falciparum infection despite treatment with quinine or artesunate (plus or minus clindamycin) (Hall et al. 1975). Although no genotyping is available, Burkhardt et al. (2007) recently showed that some individuals infected with Amazonian parasites from Brazil take much longer to clear infections after clindamycin treatment than individuals infected with African parasites. Further studies, especially in South America, will be needed to assess how prevalent the mutation is and from whence it came, especially as recent clinical trials of fosmidomycin + clindamycin show its potential as an efficacious combination therapy (Borrmann et al. 2004; Olliaro and Wells 2009).
Third, a new lincosamide drug, mirincamycin, is in preclinical development, being prepared by the Medicines for Malaria Venture for Investigational New Drug status (Olliaro and Wells 2009) and has been shown to have good in vitro activity against P. falciparum isolates from Gabon (Held et al. 2010). While this drug is promising, because it has been shown to have synergistic effects with primaquine for clearing liver-stage parasites in animal models (Schmidt 1985), there is a concern for cross-resistance with clindamycin. Comparing the structure of mirincamycin to clindamycin reveals that these two molecules share the moiety implicated for making contacts with the 23S rRNA at the peptidyl transferase cavity, as demonstrated by the crystal structure of clindamycin bound to bacterial ribosomal LSU (Schlunzen et al. 2001; Tu et al. 2005). Thus, both clindamycin and mirincamycin may make contacts with residue A2058-Ec (Fig. 3A,B) suggesting that there may be cross-resistance to mirincamycin in the Amazon region with the A2058C-Ec mutation. In light of this evidence that mirincamycin resistance may already exist, the development of this and future lincosamide antimalarial drugs should be reconsidered.
Conclusions
The methods described here are useful for broadly looking at population structure, and our data show how novel insights can be gained through full genome approaches. In some cases it may be possible to determine whether the population has been structured through immune selection on specific antigens. However, because microarrays offer a lower resolution view, analyses of specific antigens are still best performed through sequencing or conventional genotyping. Furthermore, these methods can also be used to discover new alleles involved in resistance. Our finding that alleles associated with clindamycin resistance exist in South America highlights the importance of developing drugs with novel mechanisms of action and presents new molecular strategies for monitoring clindamycin resistance in South America and also in the rest of the world.
Methods
Study site and collection of malaria patient-derived isolates
Briefly, the Department of Loreto comprises almost one-fourth of the landmass of Peru and has an ecosystem characteristic of the Amazon lowlands (Aramburu Guarda et al. 1999). The rural population of Loreto, estimated at ∼474,000 inhabitants, is clustered in towns and villages located in the Amazon tributary system. The city of Iquitos, the largest urban center in Loreto with a population of ∼345,000, is located 120 m above sea level, at the juncture of the Ucayali and Napo Rivers, forming the Amazon River proper, and is accessible only by air or river. Iquitos has a tropical climate, with a mean temperature of 27.5°C, a mean annual rainfall of 4 m, and a mean annual precipitation of 2.7 m. The climate in this area is typical of the Amazon basin, with a rainy season from November to May and a dry season from June through October. The peak transmission rate for P. falciparum occurs during the rainy season (Aramburu Guarda et al. 1999). The population is predominantly mixed Spanish and American Indian (mestizo). Household economies are sustained by agriculture, fishing, or tourism. The 2006 study samples came from Iquitos.
The 2006 malaria isolates were collected as part of an ongoing febrile surveillance protocol (approval no. NMRCD.2000.0006 from the Naval Medical Research Center Detachment, Lima, Peru) from patients with symptoms typical of malaria who visited clinics located in the jungle city of Iquitos. Once they had been diagnosed with malaria by microscopy, blood was spotted onto filter paper for characterization of SNPs and microsatellites. Venous blood was collected into 3-mL EDTA tubes for laboratory studies from febrile patients when P. falciparum malaria was diagnosed by microscopic examination. A total of 600 μL of blood was taken from each tube under sterile conditions in a laminar-flow hood for drug susceptibility testing. One milliliter was used for the cryopreservation of parasites. Of the remainder, some was spotted onto Whatman 3M chromatography paper (Whatman, Inc.) for PCR; some was used to prepare thick and thin smears for parasite confirmation and quantification; and the rest was frozen at –80°C. Microsatellite testing showed a very low frequency of polyclonal infections (<1%) (Bacon et al. 2009) in the region, and samples from polyclonal infections were not used in these studies.
DNA methods
3D7 (MRA-151) was obtained from the Malaria Research and Reference Reagent Resource Center (MR4; American Type Culture Collection). 3D7 and Peruvian P. falciparum patient isolates were propagated in human erythrocytes as previously described (Trager and Jenson 1978) with medium for 3D7 containing 5% human serum and 1.2% GIBCO AlbuMAX II (Invitrogen), while the medium for patient isolates contained 10% human serum. Genomic DNA was isolated by standard phenol-chloroform extraction. Fifteen micrograms of genomic DNA from each isolate and 2.5 ng each of Bio B, Bio C, Bio D, and Cre Affymetrix control plasmids (Affymetrix Inc.) were fragmented with DNase I and end-labeled with biotin (Winzeler et al. 1998). The samples were hybridized to the microarrays overnight at 45°C in Affymetrix buffers, washed, and scanned using a modified protocol with wash temperatures of 23°C to account for the high AT content of P. falciparum (Supplemental File 7: Dharia et al. 2009).
Microarray analysis
Briefly, for polymorphism detection, using a sliding window of three overlapping probes, we scanned for sets of probes that had significantly lower hybridization when compared with the reference indicative of a polymorphism as determined by Z-test using a P-value cutoff of 1 × 10−5 and a reference 3D7 hybridization. Gene copy number variation scanning was performed by calculating the log2 ratio of the mean intensity for probes to a gene divided by the mean intensity of the probes for a reference 3D7 hybridization as previously described (Dharia et al. 2009).
Pairwise comparisons were performed with microarray hybridizations of 14 patient isolates using one isolate as a reference for another, thus producing 182 comparisons. Polymorphisms were called using P-value cutoffs of 1 × 10−5, and we defined isogenic blocks as all 100-kb windows that had no polymorphisms between a pair of isolates. Next, we identified the progeny of a natural cross by scanning through all possible combinations of three isolates. Specifically, we looked for two isolates that each, separately, were ∼50% isogenic to a third isolate, but in combination were isogenic for >90% of the third isolate. To detect polymorphisms in the organellar apicoplast and mitochondrial genomes, the Peruvian isolates were compared with 3D7 reference hybridizations using a P-value cutoff of 1 × 10−5.
EC50 determination
96h EC50 assays were performed for chloroquine, pyrimethamine, mefloquine, artemisinin, and quinine using a 384-well format. A prolonged incubation period was required, as the Peruvian isolates grew at slower rates than standard laboratory strains, and a 10- to 20-fold difference in signal between control DMSO treated and high-dose drug-treated wells was necessary. Parasites of each Peruvian isolate in media containing 50% human serum and 50% GIBCO AlbuMAX II (Invitrogen) were dispensed into the plates, and compounds were dispensed using a PinTool with a 50 nL head, so that the final volume of each well was 50 μL. The final hematocrit was 2.5% with a 0.075% parasitemia, and the final DMSO concentration was 0.1%. The assay plates were incubated at 37°C for 96 h in airtight incubation units which were gassed daily with 93% nitrogen, 4% carbon dioxide, and 3% oxygen. A total of 10 μL of detection reagent, 10× SYBR Green I (Invitrogen; supplied in 10,000× concentration) in lysis buffer (20 mM Tris-HCl, 5 mM EDTA, 0.16% Saponin w/v, 1.6% Triton X-100 v/v), was dispensed into each well. The plates were incubated at room temp for 24 h. The fluorescence intensity from the plate was read using an Analyst GT multimode reader (Molecular Devices); the reader required a 505 dichroic mirror with 485-nm excitation and 530-nm emission settings, and the plate was read from the bottom.
For the longer 144h EC50 assays with clindamycin (“delayed-death”), a modified protocol was required. The 144-h time point was chosen by taking into account the slower growth rate, on average, of the Peruvian isolates compared with standard laboratory strains. From a 10-mM compound stock, a 1:5 dilution with PBS was made. Then, 1:3 serial dilutions were performed in a 1:5 dilution solution of DMSO and PBS across a 96-well compound plate. A 150-μL suspension of each Peruvian isolate was aliquoted into a separate Costar 96-well round bottom plate (Corning Inc.) in medium that contained human serum with no AlbuMAX. The final hematocrit was 2.5% with 0.15% parasitemia. Using a matrix pipette, 0.75 μL of compound from the compound plate was transferred into the assay plate. The final DMSO concentration was 0.1%. The assay plates were incubated at 37°C for 144 h in airtight incubation units as described above. Parasite growth was monitored by taking blood smears from untreated wells. The medium was removed with a multichannel pipette and 150 μL of medium containing no human serum, or AlbuMAX was added to each well. The contents of the wells were transferred to a Costar 96-well white clear bottom plate (Corning Inc.). A total of 30 μL of 10× SYBR Green I detection reagent described above was dispensed into each well. The plates were incubated at room temp for 24 h. The fluorescence intensity from the bottom of the plate was read using an Analyst GT multimode reader (Molecular Devices).
Assembly of 23S rRNA coding sequence from other isolates
We downloaded all P. falciparum, P. vivax SalI, P. ovale Nigeria 1 CDC, P. reichenowi, P. knowlesi, and P. berghei ANKA reads from NCBI TraceDB (ftp://ftp.ncbi.nih.gov/pub/TraceDB/) (downloaded Feb 2009), BLASTed the reads for each isolate against PFC10_API0010 sequence, trimmed reads with Lucy to remove poor-quality data (Chou and Holmes 2001), assembled the reads de novo with Minimus (Sommer et al. 2007), aligned with nucmer (Kurtz et al. 2004), followed by visual inspection and manual assembly.
Modeling of 23S rRNA and docking
A P. falciparum homology model of the peptidyl transferase domain of the 23S rRNA based on the crystal structure 1JZX (Schlunzen et al. 2001) was used to perform clindamycin and mirincamycin docking. The nucleotides of the peptidyl transferase domain (Fig. 2B) of D. radiodurans 23S rRNA crystal structure (1JZX) were mutated to P. falciparum nucleotides using ModeRNA (M Rother, K Rother, T Puton, JM Bujnicki, in prep.). The homology model was solvated in TIP3P truncated octahedron box, neutralized by the addition of sodium ions, and subjected to minimization. All minimizations were performed using the AMBER 9 software package (Case et al. 2006) with the Wang et al. (2004a) force field. Clindamycin and mirincamycin were docked to the model using AutoDock Vina (Trott and Olson 2009) with the option exhaustiveness set to 100 after PDBQT files of the ligands and receptor were prepared using AutoDockTools (Morris et al. 2009). The lowest energy docking confirmations were visualized, and images were generated using MacPyMol (DeLano Scientific, LLC).
Apicoplast mutation verification
Primers were designed to amplify 400–500 bp of apicoplast genomic DNA with the predicted mutation near the center (Supplemental Table S2). Independent PCR products were sequenced by ABI sequencing (Applied Biosystems Inc.).
var gene variability verification
Primers were designed to amplify a 300-bp segment of PFD1015c (Supplemental Table S2). PCR products from genomic DNA from isolates or 3D7 were cloned into TOPO TA cloning vector (Invitrogen), and four clones per sample were sequenced by ABI sequencing (Applied Biosystems, Inc.). Protein sequences were aligned with ClustalW.
Acknowledgments
We thank C. McNamara and S.J. Westenberger for helpful discussions. D.J.B. and T.J.K. are military service members, and this work was prepared as part of their official duties. E.A.W. was supported by grants from the WM Keck Foundation and the National Institutes of Health (R01AI059472). Malaria work at GNF is partially supported by grants from the Medicines for Malaria Venture and the Wellcome Trust. The views expressed in this article are those of the authors and do not necessarily reflect the official policy of the Department of the Navy, the Department of Defense, the Department of Health and Human Services, or the U.S. government. The study protocol was approved by the Naval Medical Research Center Institutional Review Board in compliance with all applicable Federal regulations governing the protection of human subjects.
Author contributions: N.V.D. conceived, designed, and performed experiments and data analysis and wrote the manuscript. D.P., S.E.R.B., G.E.G., C.L., T.J.K., C.S., and V.S. conceived, designed, and performed experiments. B.B. performed homology modeling. D.J.B. and E.A.W. conceived, designed, and directed the project and wrote the manuscript.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE22861 and are also at http://www.scripps.edu/cb/winzeler/resources/pf_peru.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.105163.110.
References
- Alkhalil A, Pillai AD, Bokhari AA, Vaidya AB, Desai SA 2009. Complex inheritance of the plasmodial surface anion channel in a Plasmodium falciparum genetic cross. Mol Microbiol 72: 459–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, et al. 2000. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 17: 1467–1482 [DOI] [PubMed] [Google Scholar]
- Aramburu Guarda J, Ramal Asayag C, Witzig R 1999. Malaria reemergence in the Peruvian Amazon region. Emerg Infect Dis 5: 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F, Chalvet W, Hommel D, Peneau C, Hulin A, Mercereau-Puijalon O, Duchemin JB, Sarthou JL, Reynes JM, Fandeur T 1999. Plasmodium falciparum parasites in French Guiana: Limited genetic diversity and high selfing rate. Am J Trop Med Hyg 61: 978–985 [DOI] [PubMed] [Google Scholar]
- Bacon DJ, McCollum AM, Griffing SM, Salas C, Soberon V, Santolalla M, Haley R, Tsukayama P, Lucas C, Escalante AA, et al. 2009. Dynamics of malaria drug resistance patterns in the Amazon basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob Agents Chemother 53: 2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DR, Wilson DW, Martin LB 2005. False-positive results of a Plasmodium falciparum histidine-rich protein 2-detecting malaria rapid diagnostic test due to high sensitivity in a community with fluctuating low parasite density. Am J Trop Med Hyg 73: 199–203 [PubMed] [Google Scholar]
- Bennett TN, Patel J, Ferdig MT, Roepe PD 2007. Plasmodium falciparum Na+/H+ exchanger activity and quinine resistance. Mol Biochem Parasitol 153: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann S, Issifou S, Esser G, Adegnika AA, Ramharter M, Matsiegui PB, Oyakhirome S, Mawili-Mboumba DP, Missinou MA, Kun JF, et al. 2004. Fosmidomycin-clindamycin for the treatment of Plasmodium falciparum malaria. J Infect Dis 190: 1534–1540 [DOI] [PubMed] [Google Scholar]
- Branch O, Casapia WM, Gamboa DV, Hernandez JN, Alava FF, Roncal N, Alvarez E, Perez EJ, Gotuzzo E 2005. Clustered local transmission and asymptomatic Plasmodium falciparum and Plasmodium vivax malaria infections in a recently emerged, hypoendemic Peruvian Amazon community. Malar J 4: 27 doi: 10.1186/1475-2875-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt D, Wiesner J, Stoesser N, Ramharter M, Uhlemann AC, Issifou S, Jomaa H, Krishna S, Kremsner PG, Borrmann S 2007. Delayed parasite elimination in human infections treated with clindamycin parallels ‘delayed death’ of Plasmodium falciparum in vitro. Int J Parasitol 37: 777–785 [DOI] [PubMed] [Google Scholar]
- Camps M, Arrizabalaga G, Boothroyd J 2002. An rRNA mutation identifies the apicoplast as the target for clindamycin in Toxoplasma gondii. Mol Microbiol 43: 1309–1318 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2 doi: 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, et al. 2006. Amber 9. University of California, San Francisco, CA [Google Scholar]
- Chenet SM, Branch OH, Escalante AA, Lucas CM, Bacon DJ 2008. Genetic diversity of vaccine candidate antigens in Plasmodium falciparum isolates from the Amazon basin of Peru. Malar J 7: 93 doi: 10.1186/1475-2875-7-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HH, Holmes MH 2001. DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1104 [DOI] [PubMed] [Google Scholar]
- Conway DJ, Roper C, Oduola AM, Arnot DE, Kremsner PG, Grobusch MP, Curtis CF, Greenwood BM 1999. High recombination rate in natural populations of Plasmodium falciparum. Proc Natl Acad Sci 96: 4506–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey A, Mendis K, Carlton J, Williamson D, Wilson I, Carter R 1994. Maternal inheritance of extrachromosomal DNA in malaria parasites. Mol Biochem Parasitol 65: 95–98 [DOI] [PubMed] [Google Scholar]
- Dahl EL, Rosenthal PJ 2008. Apicoplast translation, transcription and genome replication: Targets for antimalarial antibiotics. Trends Parasitol 24: 279–284 [DOI] [PubMed] [Google Scholar]
- Dharia NV, Sidhu AB, Cassera MB, Westenberger SJ, Bopp SE, Eastman RT, Plouffe D, Batalov S, Park DJ, Volkman SK, et al. 2009. Use of high-density tiling microarrays to globally identify mutations and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol 10: R21 doi: 10.1186/gb-2009-10-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361: 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S, Marquino W, Cabezas C, Utz G, Fiestas V, Cairo J, Puray M, Lucas C, Salas C, Gutierrez S, et al. 2007. Unusual pattern of Plasmodium falciparum drug resistance in the northwestern Peruvian Amazon region. Am J Trop Med Hyg 76: 614–618 [PubMed] [Google Scholar]
- Eastman RT, Fidock DA 2009. Artemisinin-based combination therapies: A vital tool in efforts to eliminate malaria. Nat Rev Microbiol 7: 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein-Ludwig U, Webb RJ, Van Goethem ID, East JM, Lee AG, Kimura M, O'Neill PM, Bray PG, Ward SA, Krishna S 2003. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424: 957–961 [DOI] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE 2004. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol 52: 985–997 [DOI] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, et al. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, Incardona S, Perkins M, Bell D, McCarthy J, et al. 2010. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: Implications for malaria rapid diagnostic tests. PLoS ONE 5: e8091 doi: 10.1371/journal.pone.0008091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT 2008. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol 6: e238 doi: 10.1371/journal.pbio.0060238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood BM, Fidock DA, Kyle DE, Kappe SH, Alonso PL, Collins FH, Duffy PE 2008. Malaria: Progress, perils, and prospects for eradication. J Clin Invest 118: 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D, Snounou G, Mattei D, Enamorado IG, Figueroa J, Stahl S, Berzins K 1999. Limited genetic diversity of Plasmodium falciparum in field isolates from Honduras. Am J Trop Med Hyg 60: 30–34 [DOI] [PubMed] [Google Scholar]
- Hall AP, Doberstyn EB, Nanokorn A, Sonkom P 1975. Falciparum malaria semi-resistant to clindamycin. Br Med J 2: 12–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harinasuta T, Suntharasamai P, Viravan C 1965. Chloroquine-resistant falciparum malaria in Thailand. Lancet 2: 657–660 [DOI] [PubMed] [Google Scholar]
- Harris EH, Burkhart BD, Gillham NW, Boynton JE 1989. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: Correlation of genetic and physical maps of the chloroplast genome. Genetics 123: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, et al. 2008. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe 4: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held J, Westerman R, Kremsner PG, Mordmuller B 2010. In vitro activity of mirincamycin (U24729A) against Plasmodium falciparum isolates from Gabon. Antimicrob Agents Chemother 54: 540–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambou R, Legrand E, Niang M, Khim N, Lim P, Volney B, Ekala MT, Bouchier C, Esterre P, Fandeur T, et al. 2005. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet 366: 1960–1963 [DOI] [PubMed] [Google Scholar]
- Jeffares DC, Pain A, Berry A, Cox AV, Stalker J, Ingle CE, Thomas A, Quail MA, Siebenthall K, Uhlemann AC, et al. 2007. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet 39: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy DA, Feng X, Mu J, Furuya T, Chotivanich K, Krettli AU, Ho M, Wang A, White NJ, Suh E, et al. 2003. Early origin and recent expansion of Plasmodium falciparum. Science 300: 318–321 [DOI] [PubMed] [Google Scholar]
- Kidgell C, Volkman SK, Daily J, Borevitz JO, Plouffe D, Zhou Y, Johnson JR, Le Roch K, Sarr O, Ndir O, et al. 2006. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog 2: e57 doi: 10.1271/journal.ppat.0020057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12 doi: 10.1185/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SK, Rajasekariah GH, Awinda G, Waitumbi J, Kifude C 2009. Unified parasite lactate dehydrogenase and histidine-rich protein ELISA for quantification of Plasmodium falciparum. Am J Trop Med Hyg 80: 516–522 [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson A.J 2009. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, Chotivanich K, Wilairatana P, Krudsood S, White NJ, et al. 2010. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet 42: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Grant DM, Rabert DK, Harris EH, Boynton JE, Gillham NW 1982. Mutants of Chlamydomonas reinhardtii with physical alterations in their chloroplast DNA. Plasmid 7: 133–151 [DOI] [PubMed] [Google Scholar]
- Nair S, Miller B, Barends M, Jaidee A, Patel J, Mayxay M, Newton P, Nosten F, Ferdig MT, Anderson TJ 2008. Adaptive copy number evolution in malaria parasites. PLoS Genet 4: e1000243 doi: 10.1371/journal.pgen.1000243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Schaffner SF, Volkman SK, Park D, Montgomery P, Milner DA Jr, Lukens A, Rosen D, Daniels R, Houde N, et al. 2008. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol 9: R171 doi: 10.1186/gb-2008-0-12-4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Spurck TP, Goodman CD, McFadden GI 2009. Apicoplast and mitochondrion in gametocytogenesis of Plasmodium falciparum. Eukaryot Cell 8: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliaro P, Wells TN 2009. The global portfolio of new antimalarial medicines under development. Clin Pharmacol Ther 85: 584–595 [DOI] [PubMed] [Google Scholar]
- Pieroni P, Mills CD, Ohrt C, Harrington MA, Kain KC 1998. Comparison of the ParaSight-F test and the ICT Malaria Pf test with the polymerase chain reaction for the diagnosis of Plasmodium falciparum malaria in travellers. Trans R Soc Trop Med Hyg 92: 166–169 [DOI] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, et al. 2004. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 364: 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj DK, Mu J, Jiang H, Kabat J, Singh S, Sullivan M, Fay MP, McCutchan TF, Su XZ 2009. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem 284: 7687–7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramya TN, Mishra S, Karmodiya K, Surolia N, Surolia A 2007. Inhibitors of nonhousekeeping functions of the apicoplast defy delayed death in Plasmodium falciparum. Antimicrob Agents Chemother 51: 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod PK, McErlean T, Lee PC 1997. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci 94: 9389–9393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett 282: 147–159 [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305: 1124 doi: 10.1126/science.1098876 [DOI] [PubMed] [Google Scholar]
- Rosenberg E, Litus I, Schwarzfuchs N, Sinay R, Schlesinger P, Golenser J, Baumeister S, Lingelbach K, Pollack Y 2006. pfmdr2 confers heavy metal resistance to Plasmodium falciparum. J Biol Chem 281: 27039–27045 [DOI] [PubMed] [Google Scholar]
- Roshanravan B, Kari E, Gilman RH, Cabrera L, Lee E, Metcalfe J, Calderon M, Lescano AG, Montenegro SH, Calampa C, et al. 2003. Endemic malaria in the Peruvian Amazon region of Iquitos. Am J Trop Med Hyg 69: 45–52 [PubMed] [Google Scholar]
- Rottman M, McNamara C, Yeung BKS, Lee MCS, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, et al. 2010. Spiroindolones, a potent compound class for the treatment of Malaria. Science 329: 1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821 [DOI] [PubMed] [Google Scholar]
- Schmidt LH 1985. Enhancement of the curative activity of primaquine by concomitant administration of mirincamycin. Antimicrob Agents Chemother 27: 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Verdier-Pinard D, Fidock DA 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298: 210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Valderramos SG, Fidock DA 2005. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol 57: 913–926 [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA 2006. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis 194: 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Sun Q, Nkrumah LJ, Dunne MW, Sacchettini JC, Fidock DA 2007. In vitro efficacy, resistance selection, and structural modeling studies implicate the malarial parasite apicoplast as the target of azithromycin. J Biol Chem 282: 2494–2504 [DOI] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL, Pop M 2007. Minimus: A fast, lightweight genome assembler. BMC Bioinformatics 8: 64 doi: 10.1186/1471-2105-8-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Hayton K, Wellems TE 2007. Genetic linkage and association analyses for trait mapping in Plasmodium falciparum. Nat Rev Genet 8: 497–506 [DOI] [PubMed] [Google Scholar]
- Tan TM, Nelson JS, Ng HC, Ting RC, Kara UA 1997. Direct PCR amplification and sequence analysis of extrachromosomal Plasmodium DNA from dried blood spots. Acta Trop 68: 105–114 [DOI] [PubMed] [Google Scholar]
- Tan JC, Patel JJ, Tan A, Blain JC, Albert TJ, Lobo NF, Ferdig MT 2009. Optimizing comparative genomic hybridization probes for genotyping and SNP detection in Plasmodium falciparum. Genomics 93: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jenson JB 1978. Cultivation of malarial parasites. Nature 273: 621–622 [DOI] [PubMed] [Google Scholar]
- Trott O, Olson AJ 2009. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore PB, Steitz TA 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121: 257–270 [DOI] [PubMed] [Google Scholar]
- Uhlemann AC, Cameron A, Eckstein-Ludwig U, Fischbarg J, Iserovich P, Zuniga FA, East M, Lee A, Brady L, Haynes RK, et al. 2005. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat Struct Mol Biol 12: 628–629 [DOI] [PubMed] [Google Scholar]
- Urdaneta L, Lal A, Barnabe C, Oury B, Goldman I, Ayala FJ, Tibayrenc M 2001. Evidence for clonal propagation in natural isolates of Plasmodium falciparum from Venezuela. Proc Natl Acad Sci 98: 6725–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B, Douthwaite S 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 45: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA Jr, Daily JP, Sarr O, Ndiaye D, Ndir O, et al. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet 39: 113–119 [DOI] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R 1987. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science 236: 1661–1666 [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA 2004a. Development and testing of a general amber force field. J Comput Chem 25: 1157–1174 [DOI] [PubMed] [Google Scholar]
- Wang P, Nirmalan N, Wang Q, Sims PF, Hyde JE 2004b. Genetic and metabolic analysis of folate salvage in the human malaria parasite Plasmodium falciparum. Mol Biochem Parasitol 135: 77–87 [DOI] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ 1990. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature 345: 253–255 [DOI] [PubMed] [Google Scholar]
- Winzeler EA 2008. Malaria research in the post-genomic era. Nature 455: 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, McCullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ, et al. 1998. Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197 [DOI] [PubMed] [Google Scholar]
- Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH 2007. A review of malaria diagnostic tools: Microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77 (Suppl): 119–127 [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418: 320–323 [DOI] [PubMed] [Google Scholar]
- Wu T, Nagle A, Sakata T, Henson K, Borboa R, Chen Z, Kuhen K, Plouffe D, Winzeler E, Adrian F, et al. 2009. Cell-based optimization of novel benzamides as potential antimalarial leads. Bioorg Med Chem Lett 19: 6970–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]