Abstract
Genomes encode multiple signals, raising the question of how these different codes are organized along the linear genome sequence. Within protein-coding regions, the redundancy of the genetic code can, in principle, allow for the overlapping encoding of signals in addition to the amino acid sequence, but it is not known to what extent genomes exploit this potential and, if so, for what purpose. Here, we systematically explore whether protein-coding regions accommodate overlapping codes, by comparing the number of occurrences of each possible short sequence within the protein-coding regions of over 700 species from viruses to plants, to the same number in randomizations that preserve amino acid sequence and codon bias. We find that coding regions across all phyla encode additional information, with bacteria carrying more information than eukaryotes. The detailed signals consist of both known and potentially novel codes, including position-dependent secondary RNA structure, bacteria-specific depletion of transcription and translation initiation signals, and eukaryote-specific enrichment of microRNA target sites. Our results suggest that genomes may have evolved to encode extensive overlapping information within protein-coding regions.
The requirement to organize the multitude of signals encoded by a genome into a linear DNA sequence imposes complex constraints on the way in which these signals can be encoded. For example, the region of the mRNA transcript that encodes an amino acid sequence inevitably overlaps with the RNA secondary structure signals that the transcript encodes as a whole. In addition, since transcription initiates upstream of coding regions, coding regions may be constrained to preferentially avoid signals for transcription initiation in order to minimize aberrant transcription. Thus, unlike prose, in which each letter participates in exactly one word, the same genomic sequence may in some cases be required to simultaneously encode multiple types of signals and avoid other signals. One way in which the same sequence can comply with such constraints is if codes have a degree of redundancy, namely, if a given code can encode the exact same information in multiple distinct ways. The genetic code exhibits such redundancy since the same amino acid sequence can be encoded by many different DNA sequences. A recent study even suggested that the genetic code is nearly optimal for accommodating additional information (Itzkovitz and Alon 2007).
Several studies highlighted measures of information content that are unique to the protein-coding sequence, with the goal of detecting genes in genomic DNA (Fickett and Tung 1992; Burge and Karlin 1998; Stormo 2000; Green et al. 2003). Studies that focused on the phenomenon of overlapping codes demonstrated that certain codes are enriched or depleted from coding regions of specific organisms, including depletion of microsatellite repeats from coding sequences in Escherichia coli, Saccharomyces cerevisiae, and Caenorhabditis elegans (Ackermann and Chao 2006); restriction enzyme target sites in bacteria and archaea (Gelfand and Koonin 1997); and enrichment for microRNA targets in human (Forman et al. 2008) and for RNA secondary structure (Katz and Burge 2003). A striking example of the use of overlapping information within coding sequences is the encoding of overlapping genes on the same strand or on opposite strands, which is prominent in viruses (for review, see Normark et al. 1983). However, a systematic study, across diverse phyla, of the identity and extent to which protein-coding sequences harbor additional overlapping codes is still lacking.
Results
Markov chain Monte Carlo randomization of protein-coding regions
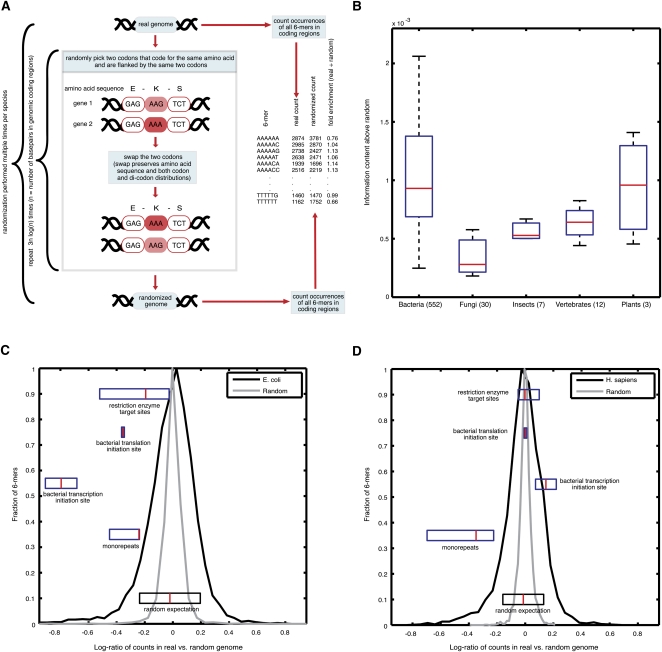
To measure the extent to which the protein-coding regions of a given genome carry additional information beyond the amino acid sequence, we broadly defined information in terms of the distribution of short sequences (Burge et al. 1992) of length 6 bp or 7 bp. We chose these k-mer lengths as they are the longest sequences for which we could obtain ample statistics for all possible sequences. To this end, we compared the number of occurrences of all possible short sequences in the protein-coding regions of a given genome to this same number in the coding regions of randomized versions of the genome that still encode the same set of proteins.
Since the mere counts of short sequences depends on genomic biases in basepair composition, codon-usage, and di-codon counts (Andersson and Kurland 1990; Boycheva et al. 2003; Moura et al. 2007), we used a stringent randomization procedure that preserves all of these properties (Fig. 1A). Specifically, we employed a Markov chain Monte Carlo algorithm that generates a randomized genome by iteratively swapping codons that encode the same amino acid and that are each flanked by the exact same codons (e.g., these two underlined lysine codons, GAG-AAG-TCT and GAG-AAA-TCT, could be swapped) (Fig. 1A). In addition to preserving codon and di-codon counts and thus all in-frame 6-mers, our randomization procedure preserves the exact counts of all short sequences of length 1–4 irrespective of their frame, as well as the exact counts of 5-mers in the 0 and +1 frames. Unlike randomizations that preserve only codon counts, we found that this randomization yields an enrichment of sequences of 6-mers and of longer k-mers that is independent of the absolute number of occurrences of these k-mers in the real genome (Supplemental Fig. S1). Thus, codes that are characterized by short sequences and are enriched or depleted from coding regions beyond the above genomic biases will be detected by our approach, since the number of their occurrences will differ significantly between the real and the randomized genomes.
Figure 1.
Overview of our approach and detection of additional information encoded within protein-coding sequences. (A) Illustration of our method for identifying over- and underrepresented short sequences within coding regions. For each short sequence (here 6-mer sequences are shown), we count the number of its appearances in a given genome's coding sequences and compare that to its average number of appearances in the coding sequences of randomized genomes. The randomization swaps codons from different genomic locations only if they are both flanked by identical codons and, thus, preserves amino acid sequence, codon usage, and di-codon counts. An example of one codon swap is shown (left), and these swaps are repeated iteratively for each randomization, for each species. (B) All genomes contain additional information in their coding sequences. Shown is the Jensen-Shannon information divergence, a measure analogous to information content, between the distribution of all 6-mer sequences when counted out-of-frame in the real and randomized genomes (since our randomization preserves di-codon counts, the counts of 6-mers in-frame are equal in the real and random genomes, by construction). The Jensen-Shannon divergence is shown as a box plot for all organisms in various phyla groups. The red line denotes the median, the blue box delimits 25–75 percentiles, and the outermost bars show the minimum and maximum. The number of species from each phyla group is shown in parentheses. (C) Histograms of log-ratios of number of appearances of the out-of-frame 6-mers in E. coli (black) and out-of-frame 6-mers in randomized E. coli genomes (gray). Box plots of log-ratios for specific families of known biological signals (mononucleotide repeats, restriction enzyme target sites, and bacterial transcription and translation initiation sites) are shown in their appropriate place along the histogram. Histograms were normalized to have a maximum of 1 for ease of comparison. (D) Same as C, but for human.
Phyla-specific overlapping information content in protein-coding regions
We quantify the enrichment or depletion of each short sequence as the ratio between the number of times the sequence appears in the coding regions of the real genome and the average number of times it appears in those of the randomized genomes (Supplemental Fig. S1; Supplemental Tables S1, S2). Applying this approach to the protein-coding regions of hundreds of genomes from diverse phyla, including viruses, bacteria, fungi, vertebrates, and plants, we find that across all genomes, the number of occurrences of short sequences differs substantially between the real and randomized coding regions (Fig. 1B–D). Notably, this difference is more pronounced in bacterial genomes compared with eukaryotic genomes (Kolmogorov-Smirnov test, P < 10−25) (Fig. 1B). This result also holds when constraining all species to similar genome sizes and GC content (Supplemental Fig. S2) and is most likely not affected significantly by genome heterogeneity effects, which may be introduced by factors such as regional GC bias along the genome, horizontal gene transfer, or strand biases such as those caused by transcription-mediated repair (Supplemental Fig. S3; Green et al. 2003). Thus, our results suggest that protein-coding regions carry extensive information beyond that expected from their requirements to encode particular amino acid sequences and to conform to codon usage and di-codon composition.
Known biological codes are among the enriched or depleted sequences
The above deviation from random expectation could simply indicate that protein-coding regions evolve by a more complex evolutionary model than that accounted for by our randomization. A much more intriguing possibility is that the additional information that we find in protein-coding sequences reflects selection for, or avoidance of, meaningful biological codes, and that this additional information has evolved in order to facilitate specific functions. To test for this latter possibility, we asked whether several known biological codes are among the short sequences enriched or depleted above random expectation across various genomes.
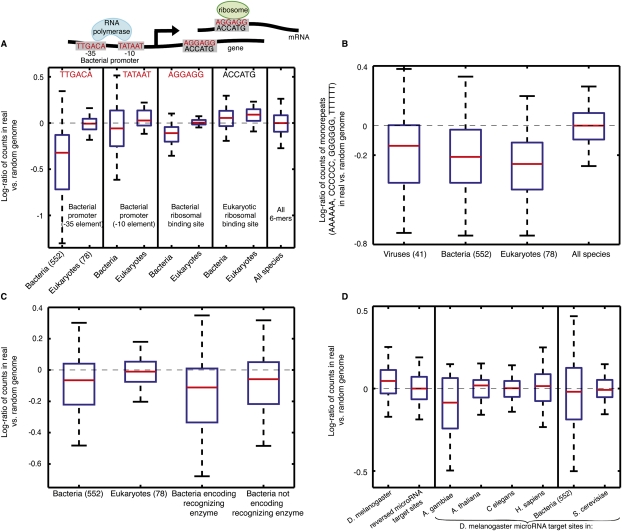
In bacteria, transcription and translation can initiate at any genomic location that contains specific, well-characterized short sequences (Shine and Dalgarno 1975; Haugen et al. 2008). Thus, we may expect such initiation signals to be depleted from coding regions of bacteria in order to protect against aberrant transcription/translation initiation (Hahn et al. 2003). Indeed, we find that the binding sites for sigma factors, a group of ubiquitous prokaryotic transcription initiation factors (Haugen et al. 2008), are highly depleted in bacteria (for the −35 promoter element TTGACA, log-ratio of number of appearances between real and randomized genomes of −0.42 ± 0.02, Kolmogorov-Smirnov of P < 10−152 compared with all 6-mers in all organisms; for the −10 promoter element TATAAT, log-ratio −0.004 ± 0.03, P < 10−20) (Fig. 2A) but not in eukaryotes (log-ratio of 0 ± 0.01 and 0.05 ± 0.03, respectively). We observe a similar bacteria-specific depletion of the Shine-Dalgarno sequence, the bacterial translation initiation site (AGGAGG, log-ratio of −0.13 ± 0.006 in bacteria, P < 10−72, compared with log-ratio of 0.01 ± 0.006 in eukaryotes) (Shine and Dalgarno 1975). Unlike prokaryotic ribosomes, which initiate translation by randomly colliding with the transcript until encountering a position that contains initiation sequences, eukaryotic ribosomes attach to the 5′ UTR of the transcript and linearly scan the transcript, beginning translation at the first initiation signal encountered (Kozak 1991). Thus, eukaryotic initiation signals do not need to be depleted from eukaryotic coding regions since the “correct” initiation signal is invariably encountered by the ribosome before it ever reaches the “wrong” ones. Consistent with this prediction of the eukaryotic ribosome scanning model, the Kozak motif (ACCATG), a strong determinant of translation initiation in eukaryotes (Kozak 1991), is not enriched or depleted in protein-coding regions of either bacteria or eukaryotes (Fig. 2A).
Figure 2.
Coding sequences display phyla-specific enrichment of known biological signals. (A) Box plot of log-ratio of number of appearances between real and randomized genomes, of sequence determinants of transcription (−35 promoter element, TTGACA; −10 promoter element, TATAAT) and translation initiation in bacteria (Shine-Dalgarno motif, AGGAGG) and of translation initiation in eukaryotes (Kozak motif, ACCATG). In each species, each of the above 6-mer sequences is counted out-of-frame in the real genome and in the randomized genome, and the log-ratio of these counts is incorporated into the box plot. The red line denotes the median, the blue box delimits 25–75 percentiles, and the outermost bars show the minimum and maximum. The number of species from each phyla group is shown in parentheses. (B) Same as A, for log-ratios of mononucleotide 6-mers across various phyla. All represents all n-mers in all species. (C) Same as A, for bacterial restriction enzyme sites. The bacteria encoding recognizing enzyme group (third box plot from left) only displays log-ratios of restriction enzyme sites in bacterial genomes that encode the enzymes that recognize those sites, whereas the bacteria not encoding recognizing enzyme group (rightmost box plot) only displays log-ratios of restriction sites in bacterial genomes that do not encode the recognizing enzymes. (D) Same as A, for log-ratios of microRNA target sites from Drosophila melanogaster. The 7-mer seed (reverse complement of nucleotides 2–8 of the microRNA) from each microRNA was taken for the log-ratio computation. The log-ratios are shown in the coding sequences of Drosophila and in several other species, as well as in 552 bacterial genomes. The distribution of the reverse sequences of the microRNA target sites is also shown as a control.
Mononucleotide repeats mutate frequently through single nucleotide insertions or deletions (Ellegren 2004), and we would thus expect these sequences to be depleted from protein-coding regions in order to minimize DNA replication slippage errors that can potentially shift the open reading frame. Indeed, we find a ubiquitous depletion of mononucleotide repeats across all phyla, in line with recent findings (Ackermann and Chao 2006) (log-ratio of number of appearances between real and randomized of −0.11 ± 0.07, P < 10−16; −0.21 ± 0.008, P < 10−225; and −0.27 ± 0.02, P < 10−87 in viruses, bacteria, and eukaryotes, respectively; Fig. 2B).
We next examined the set of cleavage sites for bacterial restriction enzymes. These enzymes cut DNA at specific sequences as a defense mechanism against invading viruses (Roberts et al. 2003; Tock and Dryden 2005), and as such, we expected these sequences to show specific depletion from coding regions of bacteria in order to avoid self-cleavage. Indeed, we find a significant depletion of restriction enzyme sites in bacterial coding regions (log-ratio of −0.14 ± 0.002, P < 10−225), significantly more than in coding regions of eukaryotes (log-ratio of −0.012 ± 0.002). Notably, we find that restriction sites are significantly more depleted in the coding regions of the bacterial genomes that encode their recognizing enzymes compared with their depletion from coding regions of other bacteria (log-ratio of −0.3 ± 0.01 compared with −0.14 ± 0.006, P < 10−17) (Fig. 2C). As restriction enzymes cut nonmethylated sequences, the specific depletion that we find for their sites in bacteria suggests that this avoidance may help to prevent self-cleavage of DNA in cases of leaky bacterial self-methylation of DNA, which is meant to prevent bacterial sequences from being cleaved by their own restriction enzymes (Gelfand and Koonin 1997).
As another type of known biological code, we examined the enrichment of microRNA target sites within protein-coding sequences. MicroRNAs are short RNA sequences that modulate translation by recognizing and binding 6- to 8-bp seeds on the mRNA sequence of their target genes (Bartel and Chen 2004). Although the traditional view is that microRNAs bind within the 3′ UTR of their targets, recent studies demonstrated that microRNA target sites within coding regions are also functional (Duursma et al. 2008; Forman et al. 2008; Rigoutsos 2009). Comparing the number of occurrences of microRNA target site seeds between the real and randomized genomes, we find that microRNA target sites are enriched in the coding regions of the eukaryotic genome that encodes their recognizing microRNA (log-ratio of 0.01 ± 0.004, P < 10−5) but not in eukaryotic genomes that do not encode the recognizing microRNA (log-ratio −0.01 ± 0.01) and not in bacterial genomes (log-ratio −0.026 ± 0.001) (Supplemental Fig. S5), which do not have microRNAs. For example, microRNA target sites in Drosophila melanogaster have a significantly higher log-ratio of occurrences between the real and randomized coding regions of Drosophila, compared with their log-ratios in other eukaryotes in which they do not appear as microRNA target sites (Fig. 2D). As additional evidence that these enrichments likely represent microRNA target sites, we find that in contrast to the significantly high log-ratio of Drosophila microRNA target seeds in the coding regions of Drosophila (0.043 ± 0.01, P < 10−4), the reverse sequences of these microRNA target sites, which are a control not known to be microRNA target sites, are not enriched over random in Drosophila coding regions (−0.01 ± 0.01) (Fig. 2D). Together, these results suggest that eukaryotic genomes may have evolved their protein-coding sequences to harbor microRNA target sites. We note that although significant, these enrichments are relatively small in magnitude compared with the enrichment of signals discussed above.
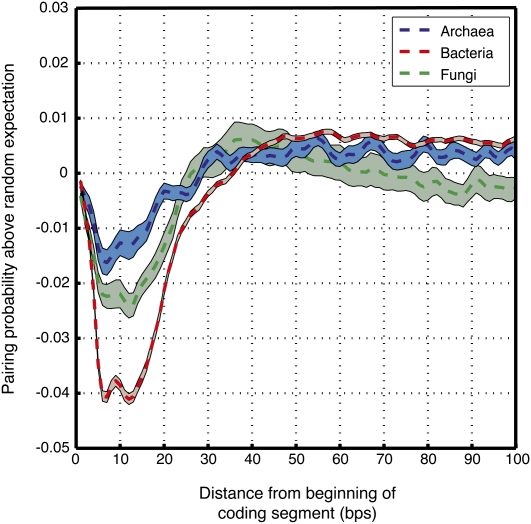
Encoding of RNA secondary structure overlaps coding regions
Overlapping information in coding sequences should not be limited to short contiguous sequences. For example, codon choice can profoundly impact the RNA secondary structure of the resulting mRNA (Katz and Burge 2003), which in turn modulates translation efficiency (Kudla et al. 2009). To explore the extent to which aspects of RNA secondary structure are encoded within protein-coding sequences, we used the Vienna package (Hofacker 2003) to fold the coding segments of the archaeal, bacterial, and sac fungal (ascomycota) genomes (only intronless or intron-poor phyla were analyzed because our data set of coding segments did not keep track of the exon order in eukaryotic genes). We then compared these folds to those of the randomized coding segments of each species. Intriguingly, we find that in all species examined, the probability of being base paired, averaged across all coding segments, is significantly lower in the first 30 bp of coding segments of the real genome compared with the first 30 bp of coding segments in randomized genomes, with bacteria exhibiting the most pronounced deviation from random (Fig. 3). A recent study demonstrated that a low pairing probability in the region spanning positions −4 to +37 around the translation start site is a strong determinant of high translation efficiency in E. coli (Kudla et al. 2009). Thus, our finding of lower than expected pairing probability in this region suggests that protein-coding sequences may have evolved to encode relatively unpaired secondary structures near translation start sites in order to increase the translation efficiency of the corresponding genes.
Figure 3.
Coding regions tend to encode depletion of RNA secondary structure downstream of the start codon. Shown is the difference in the probability of being base-paired between the real and randomized genomes, averaged across the first 100 nt of all coding segments of archaeal (blue), bacterial (red), and fungal (green) genomes. The patches show SE of the difference. Pairing probabilities were predicted by using the Vienna package (Hofacker 2003) to fold the real and randomized genomes. Each curve was smoothed with a 3-bp moving window. Since the first codon in all coding segments has only one flanking codon, it is never swapped by our genome randomization method. Thus, by construction, the first nucleotides of the coding region are more similar between the real and randomized genomes, explaining the lower difference observed in the pairing probability of these nucleotides between the real and randomized genomes.
Phyla-specific enrichment of sequences
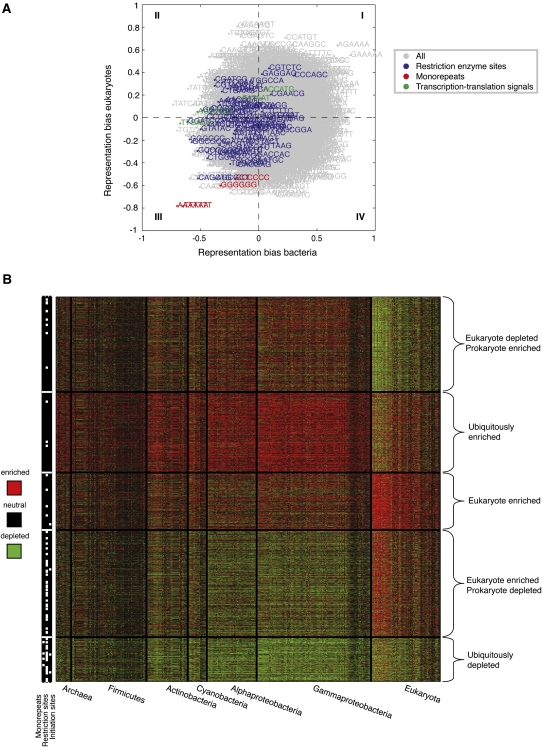
Overlapping codes found within protein-coding sequences are expected to show trends that correlate with the evolutionary history of the species. To obtain a global, unbiased view of the additional information encoded within coding sequences across different groups of species, we first compared the enrichment of short sequences between bacteria and eukaryotes. To this end, we took each short sequence and plotted its overall enrichment in bacteria versus in eukaryotes, where we defined the overall enrichment in each of the two phyla groups as the difference between the fraction of species in the group in which the sequence is enriched in the real versus the randomized genomes (at P < 0.05) and the fraction of species within that phyla in which it is depleted (Fig. 4A; Supplemental Fig. S6). This view reveals several short sequences that have similar behavior in bacteria and eukaryotes, such as the universally depleted mononucleotide repeats. These similarly enriched or depleted sequences contribute to a small but significant correlation between the enrichment of short sequences in bacteria and eukaryotes (R = 0.08, P < 10−6). This correlation is not observed when performing this analysis on randomized versions of representative genomes (R = −0.0001, P < 0.996) (see also Supplemental Fig. S7). A similar small positive correlation can be seen when comparing Z-scores between all organism pairs (Supplemental Fig. S8). The small value of the correlation in Figure 4A suggests that most short sequences exhibit phyla-specific behavior, such as bacterial restriction enzyme sites and bacterial transcription initiation signals, which are depleted in bacteria but not in eukaryotes (Fig. 4A; Supplemental Fig. S6). As an even broader evolutionary view of our catalog of overlapping codes in coding sequences, we clustered the log-ratio enrichment of all short sequences across all species and obtained several clusters, some of which exhibit ubiquitous enrichment or depletion of their member sequences across all species, while others exhibit phyla-specific enrichment or depletion of their member sequences (Fig. 4B).
Figure 4.
A global view of the additional information encoded within protein-coding sequences. (A) Comparison of short sequence enrichments in coding regions between bacteria and eukaryotes. For each short sequence, shown is its overall enrichment in bacteria (x-axis) and eukaryotes (y-axis), where the overall enrichment in each of the two phyla groups is taken to be the difference between the fraction of phyla species in which the sequence is enriched in the real versus randomized genome (at P < 0.05) and the fraction of species within that phyla is which it is depleted. (Red) Sequences that correspond to mononucleotide repeats; (blue) sequences that correspond to bacterial restriction enzyme sites; (green) sequences that correspond to bacterial transcription and translation initiation sites. (B) A clustering representation of the log-ratio coding region enrichment of all 6-mers across all 363 organisms whose coding regions exceed 2 Mbp. Rows, 6-mers; columns, organisms. The data were clustered using k-means clustering (k = 5) of a reduced matrix of representation bias in archaea, bacteria, and eukaryotes. White marks on the left bar indicate three specific sequence families: mononucleotide repeats (left column), restriction enzyme sites (central column), and transcription and translation initiation sites (right column). The organisms are arranged in phyla groups and are shown on the bottom. Red and green denote enrichment and depletion (P < 0.05) above that expected in randomized genomes, respectively.
While our analyses of enrichment or depletion of known biological codes within protein-coding sequences predominantly found depletion of known codes such as transcription and translation initiation sites, mononucleotide repeats and restriction enzyme binding sites, we note that the fraction of short sequences that are overrepresented is similar to the fraction of those that are underrepresented, as apparent by the normal distribution of Z-scores of 6-mers (Supplemental Fig. S11). Some of these overrepresented sequences may be part of yet uncharacterized overlapping codes. To further explore the properties of sequences that are specifically enriched in eukaryotes significantly more than in bacteria (Fig. 4A), we compared, for each 5-mer, the distribution of Z-scores of all of the 6-mer sequences that include it between bacteria and eukaryotes (Supplemental Table S3). We find that sequences containing C(A/G)AGT are the most overrepresented in eukaryotes versus bacteria, followed by C(G/C/A)AGG. It will be interesting to further explore the biological meaning of these sequences and of others that are overrepresented in specific phyla.
Discussion
The way in which genome sequences have evolved to encode information is profoundly different from the way in which information is encoded in human engineered channels. Rather than being a sequentially designed system, genome sequences have evolved under strong, often contradicting constraints and “frozen accidents” (Crick 1968). These constraints include the topology of the genetic code and other existing codes, the structure of the transcription and translation machineries, and constraints set by the mutational process itself. These constraints shape the amino acid content of proteins, the species-specific codon usage, and other short-range correlations between nucleotides. Notably, when comparing real coding sequences to coding sequences of alternative (randomized) genomes that conform to the above constraints, we find that for many short sequences, the number of their appearances in real coding sequences is significantly different than their counts in the coding sequences of alternative genomes. Moreover, we find that several known biological codes are among the short sequences that exhibit significant deviations between real and alternative genomes, suggesting that the additional information that we find in coding sequences may have evolved to facilitate several functions. Although the enrichment or depletion of many of these biological codes was shown, each was shown on a small number of species, which varied across studies. In contrast, our study takes a systematic and comprehensive approach using a common and stringent statistical comparison applied to more than 700 species from diverse phyla to uncover the global use of these codes within protein-coding sequences.
The lower information content that we found in eukaryotes compared with bacteria may be related to the higher-order structure and nuclear compartmentalization of eukaryotic DNA, which may isolate it from interactions with the milieu of proteins and RNA in the cytoplasm, thereby partially alleviating the constraints of avoiding certain signals. Alternatively, the lower information content in eukaryotes may be a result of the increased power of genetic drift related to their larger genomes and smaller effective population sizes (Lynch 2006). Thus, the emergence of overlapping codes, which would, in general, convey a selection advantage at the organism level, could be slower in eukaryotic genomes.
The randomization applied here assumed a homogenous codon and di-codon composition along the genome, an assumption that does not hold in general. Although the Z-score profiles of 6-mers in subsamples of genomes stratified according to either chromosomal content or random selection are well within the range of the profiles for the entire organism (Supplemental Fig. S3), it is possible that some of the codes that we detected may be a result of regional variations in sequence features. These may be introduced by factors such as regional GC bias along the genome, horizontal gene transfer, or strand biases caused by transcription mediated repair (Green et al. 2003). Although the fraction of codes that are common to both the human genome and its chromosomal or random stratifications is ∼70%, significantly higher than the ∼30% codes common to human and other organisms (Supplemental Fig. S4), this figure indicates that many codes may indeed be spuriously detected based on genome heterogeneities. Thus, our analysis serves to generate hypotheses that should eventually be validated by other methods.
Our randomization software, which we have made available in the Supplemental Material, can be used to explore overlapping information in the coding regions of subregions of genomes as well as in additional genomes. Our results may also have important implications for synthetic biology, as expression of synthetic genes in heterologous hosts may be optimized by avoiding or enriching for the overlapping codes within the host (Gustafsson et al. 2004).
While we have shown that our method can readily detect overlapping codes that are represented by short contiguous k-mers, the detection of other codes, which either are of a stochastic nature, such as the recognition site of most transcription factors (Schneider et al. 1986) or are distributed/long codes, such as that of nucleosome positioning (Segal et al. 2006), is more challenging. Although a systematic search for these codes is infeasible due to the prohibitively large sequence space, a more hypothesis-driven approach that specifically examines these codes, such as that which we applied here for studying RNA folding, can be applicable. Overall, our study reveals a wealth of overlapping information encoded within protein-coding sequences and provides an approach and a resource by which overlapping codes may be further explored, with the potential to uncover novel, uncharacterized codes.
Methods
Constructing the database of coding sequences
The database of coding segments (exons in eukaryotes, coding sequences in prokaryotes), organized by species, was constructed using the NCBI Reference Sequence (RefSeq) database as its sole source, accessible at ftp://ftp.ncbi.nih.gov/refseq/release. This database provides a comprehensive, integrated, nonredundant set of coding segments from taxonomically diverse species. We downloaded all of the genomic files in the “complete” version of RefSeq Release 26 (released Nov. 13, 2007). The files were parsed to extract the coding segments and phylogenetic information, and one FASTA file was prepared for each organism, with one coding segment per entry in the FASTA file. Out-of-frame coding segments were brought into frame by trimming either 1 bp or 2 bp, as necessary, from the beginning of the coding segment. After frame-correction, any partial codons (consisting of 1 or 2 nucleotides [nt]) found at the end of coding segments were removed, resulting in all coding segments being fully translatable to amino acid sequences. In cases where the same genomic DNA sequence appeared in multiple coding segments as a result of alternative splicing, the repeated sequence was removed from all but one of the coding segments in which it appeared. Only species with at least 47 × 10 = 163,840 nt in their coding segments, as available from RefSeq, were analyzed, so that every 7-mer had a naïve expected count of at least 10.
Creating the randomized genomes
We created randomized versions of each genome using the following Markov chain Monte Carlo (MCMC) simulation, preserving amino acid sequence and codon and di-codon counts within the genome's protein-coding sequences: (1) Pick two codons at random from the coding sequences of the genome, assuring they code for the same amino acid and have the same two flanking codons (e.g., TCC-AAA-CCA and TCC-AAG-CCA, where the underlined codons are the chosen two codons). (2) Swap the two chosen codons. (3) Repeat steps 1 and 2 until a burn-in of 3nlog(n) swaps have been made (where n is the total number of nucleotides in the genome's coding regions). This ensures that every codon is highly likely to be swapped at least once. (4) Save the current randomized genome, and continue from step 1 if another randomized genome is required. Due to computational constraints, we used 20 randomized genomes for the 6-mer analyses and five randomized genomes for the analyses of 7-mers, since we found that this number of randomizations is sufficient (Supplemental Figs. S9, S10). Note that two adjacent codons may never be swapped with each other because this could change the overall di-codon count as well as disrupt the time-reversibility of the MCMC simulation. In addition, the first and last codons in every coding segment (the “edge” codons) do not have two flanking codons and thus do not participate in any swap. Accordingly, and since coding segments such as exons are considered as disparate units, signals that overlap coding segment edges are not detected.
Another randomization that we performed preserved only codon counts, and not di-codon counts, by swapping synonymous codons irrespective of their flanking codons. This led to 6-mer enrichment scores that were correlated with the number of appearances of the 6-mer (Supplemental Fig. S1B). Observed genomic di-codon biases dictate that abundant codons tend to appear adjacent to abundant codons, possibly to satisfy translation efficiency constraints (Boycheva et al. 2003; Moura et al. 2007). Our more stringent randomization, which preserves di-codons, results in no correlation between 6-mer enrichment scores and 6-mer frequencies, thus controlling for this phenomenon (Supplemental Fig. S1A).
Enrichment scores and P-values
We measured enrichment scores for every 6- and 7-mer sequence in each genome as follows. We counted the number of appearances of each n-mer in the genome's coding sequences, denoted Nreal, as well as the average and standard deviation of its number of appearances in the coding sequences of each of the randomized genomes, denoted Nrand and Srand, respectively. Count values of 0 in either real or randomized genomes were converted to 1 to avoid the zero frequency problem, and the enrichment score was calculated as log2(Nreal/Nrand), where out-of-frame enrichment scores are computed using only out-of-frame n-mer counts. Z-scores ([Nreal − Nrand]/Srand) were computed for each n-mer and converted to P-values for each tail using a normal distribution (Supplemental Fig. S12). For each tail, P-values were defined as significant if they passed a false discovery rate (FDR) threshold of 5%, computed for each genome. Since, by construction, the in-frame 6-mer counts are identical between the real and randomized genomes, only out-of-frame 6-mer counts were considered when analyzing 6-mer results. Note, that by construction, 6-mers are the shortest n-mers for which out-of-frame counts may differ between the real and randomized genomes in both −1 and +1 frames, since 5-mer counts only differ in the −1 frame and 4-mer and 3-mer counts not at all, making the randomization quite stringent. In contrast to 6-mers, analysis of 7-mer counts included counts from all frames. P-values for the comparison of log-ratio distributions between phyla were based on the Kolmogorov-Smirnov test.
Biological sequences
Restriction enzyme target sites were downloaded from the REBASE database (Roberts et al. 2003). Only restriction enzyme target sites of length 6 were considered. As a coarse-graining step and in order to create a match with the RefSeq phyla annotation, all genomes containing each of the following terms were united: Acinetobacter, Yersinia, Bacillus, Escherichia, Pseudomonas, Streptomyces, Deinococcus, Xanthomonas, Vibrio, Staphylococcus, Salmonella, Rhodococcus, Nocardia, Neisseria, Micrococcus, Haemophilus, Citrobacter, and Bifidobacterium. MicroRNA target sites were downloaded from the miRBase database (Griffiths-Jones et al. 2006). Target sites were defined as the reverse complement of positions 2–8 of the microRNA seed.
RNA secondary structure
We ran the RNAfold program of the Vienna package v1.6 (Hofacker 2003) on the coding segments of the archaeal, bacterial, and fungal genomes, as well as on their randomized versions. For each nucleotide in each coding segment, we obtained a “pairness score” representing the probability of that nucleotide being paired with another nucleotide, thus contributing to secondary mRNA structure. For each species, we calculated the mean pairness score for each position on the coding segment relative to segment-start in order to obtain a position-dependent pairness score. We subtracted the randomized genomes' position-dependent pairness score from that of the real genome's to determine the real genome's pairness score above random expectation.
Acknowledgments
We thank Shai Lubliner, Ohad Manor, and Ron Milo for useful discussions. This work was supported by a grant from the European Research Council (ERC) to E.S. E.S. is the incumbent of the Soretta and Henry Shapiro career development chair.
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.105072.110.
References
- Ackermann M, Chao L 2006. DNA sequences shaped by selection for stability. PLoS Genet 2: e22 doi: 10.1371/journal.pgen.0020022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG 1990. Codon preferences in free-living microorganisms. Microbiol Rev 54: 198–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ 2004. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5: 396–400 [DOI] [PubMed] [Google Scholar]
- Boycheva S, Chkodrov G, Ivanov I 2003. Codon pairs in the genome of Escherichia coli. Bioinformatics 19: 987–998 [DOI] [PubMed] [Google Scholar]
- Burge CB, Karlin S 1998. Finding the genes in genomic DNA. Curr Opin Struct Biol 8: 346–354 [DOI] [PubMed] [Google Scholar]
- Burge C, Campbell AM, Karlin S 1992. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci 89: 1358–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH 1968. The origin of the genetic code. J Mol Biol 38: 367–379 [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R 2008. miR-148 targets human DNMT3b protein coding region. RNA 14: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H 2004. Microsatellites: Simple sequences with complex evolution. Nat Rev Genet 5: 435–445 [DOI] [PubMed] [Google Scholar]
- Fickett JW, Tung CS 1992. Assessment of protein coding measures. Nucleic Acids Res 20: 6441–6450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman JJ, Legesse-Miller A, Coller HA 2008. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci 105: 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Koonin EV 1997. Avoidance of palindromic words in bacterial and archaeal genomes: A close connection with restriction enzymes. Nucleic Acids Res 25: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P, Ewing B, Miller W, Thomas PJ, Green ED 2003. Transcription-associated mutational asymmetry in mammalian evolution. Nat Genet 33: 514–517 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J 2004. Codon bias and heterologous protein expression. Trends Biotechnol 22: 346–353 [DOI] [PubMed] [Google Scholar]
- Hahn MW, Stajich JE, Wray GA 2003. The effects of selection against spurious transcription factor binding sites. Mol Biol Evol 20: 901–906 [DOI] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL 2003. Vienna RNA secondary structure server. Nucleic Acids Res 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Alon U 2007. The genetic code is nearly optimal for allowing additional information within protein-coding sequences. Genome Res 17: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L, Burge CB 2003. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res 13: 2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M 1991. Structural features in eukaryotic messenger-RNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870 [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M 2006. The origins of eukaryotic gene structure. Mol Biol Evol 23: 450–468 [DOI] [PubMed] [Google Scholar]
- Moura G, Pinheiro M, Arrais J, Gomes AC, Carreto L, Freitas A, Oliveira JL, Santos MA 2007. Large scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS ONE 2: e847 doi: 10.1371/journal.pone.0000847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark S, Bergstrom S, Edlund T, Grundstrom T, Jaurin B, Lindberg FP, Olsson O 1983. Overlapping genes. Annu Rev Genet 17: 499–525 [DOI] [PubMed] [Google Scholar]
- Rigoutsos I 2009. New tricks for animal microRNAS: Targeting of amino acid coding regions at conserved and nonconserved sites. Cancer Res 69: 3245–3248 [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D 2003. REBASE: Restriction enzymes and methyltransferases. Nucleic Acids Res 31: 418–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Gold L, Ehrenfeucht A 1986. Information content of binding sites on nucleotide sequences. J Mol Biol 188: 415–431 [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J, Dalgarno L 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254: 34–38 [DOI] [PubMed] [Google Scholar]
- Stormo GD 2000. Gene-finding approaches for eukaryotes. Genome Res 10: 394–397 [DOI] [PubMed] [Google Scholar]
- Tock MR, Dryden DT 2005. The biology of restriction and anti-restriction. Curr Opin Microbiol 8: 466–472 [DOI] [PubMed] [Google Scholar]