Abstract
Natural killer (NK) cells are innate lymphocytes important for early host defense against infectious pathogens and surveillance against malignant transformation. Resting murine NK cells regulate the translation of effector molecule mRNAs (e.g., granzyme B, GzmB) through unclear molecular mechanisms. MicroRNAs (miRNAs) are small noncoding RNAs that post-transcriptionally regulate the translation of their mRNA targets, and are therefore candidates for mediating this control process. While the expression and importance of miRNAs in T and B lymphocytes have been established, little is known about miRNAs in NK cells. Here, we used two next-generation sequencing (NGS) platforms to define the miRNA transcriptomes of resting and cytokine-activated primary murine NK cells, with confirmation by quantitative real-time PCR (qRT-PCR) and microarrays. We delineate a bioinformatics analysis pipeline that identified 302 known and 21 novel mature miRNAs from sequences obtained from NK cell small RNA libraries. These miRNAs are expressed over a broad range and exhibit isomiR complexity, and a subset is differentially expressed following cytokine activation. Using these miRNA NGS data, miR-223 was identified as a mature miRNA present in resting NK cells with decreased expression following cytokine activation. Furthermore, we demonstrate that miR-223 specifically targets the 3′ untranslated region of murine GzmB in vitro, indicating that this miRNA may contribute to control of GzmB translation in resting NK cells. Thus, the sequenced NK cell miRNA transcriptome provides a valuable framework for further elucidation of miRNA expression and function in NK cell biology.
Natural killer (NK) cells are innate immune lymphocytes that are important for early host defense against pathogens and surveillance against malignant transformation (Yokoyama et al. 2004; Caligiuri 2008; Di Santo 2008b; Lanier 2008). NK cells are distinct from adaptive T and B lymphocytes, including unique NK developmental pathways, receptor-based target recognition, trafficking, and functional responses. One distinguishing feature of mature resting murine NK cells is their expression of mRNAs that encode effector molecules, including granzyme B (GzmB), perforin (Prf1), and interferon-gamma (Ifng), which exhibit a post-transcriptional mechanism of regulation (Stetson et al. 2003; Fehniger et al. 2007). Upon activation, these effector molecule mRNAs are rapidly translated, resulting in an armed NK cell ready for a rapid functional response upon triggering by a target cell. The mechanisms responsible for regulating the translation of effector mRNAs, including the critical cytotoxic effector molecule GzmB, in resting NK cells are unclear.
MicroRNAs (miRNAs) comprise a large family of small noncoding RNAs that post-transcriptionally regulate mRNAs and thereby influence fundamental cellular processes and gene expression programs (Bartel 2004). miRNAs are initially transcribed as long primary transcripts (pri-miRNA) that are processed into precursor hairpin intermediates (pre-miRNA), and then to 19–27-nt mature miRNAs through a complex and highly regulated biogenesis process (Davis and Hata 2009). Mature miRNAs are loaded into the RNA induced silencing complex (RISC), which targets specific mRNA causing translational blockade and degradation (Bartel 2009). Each miRNA sequence is predicted to target hundreds of mRNAs, and individual miRNAs have been implicated in regulating multiple, disparate biological processes. As the number of validated miRNA:mRNA target pairs expands, it appears that individual miRNA activity depends on the precise cellular context of expression (Bartel 2009). Since NK cell effector mRNAs have predicted miRNA binding sites in their 3′ untranslated regions (UTRs), we hypothesize that miRNAs may be targeting mRNAs in resting NK cells, thereby dampening protein translation. Currently, the expression of miRNAs and their importance for regulating NK cell function or development are unknown.
Recently, miRNAs have been shown to play important roles in the regulation of the immune system (Baltimore et al. 2008; Xiao and Rajewsky 2009). Studies of mice with T-cell- or B-cell-specific deficiencies in miRNA biogenesis have provided nonspecific evidence of miRNA importance in normal T-cell and B-cell development and function (Muljo et al. 2005; Koralov et al. 2008; Liston et al. 2008; Zhou et al. 2008, 2009). In addition, the expression profiles of miRNAs in adaptive T-cells and B-cells have been reported, allowing for studies of individual or clusters of miRNAs in adaptive immunity (Monticelli et al. 2005; Wu et al. 2007; Basso et al. 2009; Malumbres et al. 2009; Tan et al. 2009; Zhang et al. 2009). Individual miRNAs or clusters have been shown to be important for B-lymphocyte development, class switching, and antibody production (He et al. 2005; Rodriguez et al. 2007; Thai et al. 2007; Xiao et al. 2007, 2008; Ventura et al. 2008), as well as tuning T-cell activation (Li et al. 2007). Thus, miRNAs appear requisite for adaptive lymphocyte development and/or normal function.
Defining the expression profiles of small mature miRNA sequences is an important first step in formulating hypotheses about miRNA function within an individual tissue or cell type. Numerous studies have used hybridization-based techniques such as Northern blots, microarrays, or quantitative real-time PCR (qRT-PCR) to define the relative expression levels of mature miRNA sequences between tissue types or biological conditions. While these approaches have identified important differences in miRNA expression between different cell types, they are limited to detect only known miRNA sequences. Moreover, these techniques currently cannot interrogate the complexity in expressed mature miRNA sequences generated by variable cropping, dicing, or RNA editing events. Small RNA cloning followed by traditional cDNA sequencing has also been applied to profile known and novel miRNA expression, as exemplified in the miRNA “atlas” (Landgraf et al. 2007). While this approach has defined many miRNAs across species and tissues, PCR cloning followed by traditional sequencing remains very labor- and cost-intensive with a limited dynamic range to detect and define relative miRNA expression. Recently, next-generation sequencing (NGS) technologies, including the Illumina Genome Analyzer (GA), Applied Biosystems SOLiD System, and 454 Life Sciences (Roche) FLX instruments, have emerged that sequence DNA in a massively parallel fashion, allowing for the simultaneous generation of millions of sequence reads (Mardis 2008a,b). The Illumina GA has been used in studies to sequence cDNA libraries created from small RNAs from human ES cell lines (Morin et al. 2008b) and transformed mouse pre-leukemia and leukemic cells (Kuchenbauer et al. 2008). Additional studies have sequenced small RNA libraries from plants, Arabidopsis, chickens, and human ovarian cancer using the 454 FLX instrument (Fahlgren et al. 2007; Burnside et al. 2008; Dolgosheina et al. 2008; Morin et al. 2008a; Wyman et al. 2009). An expanded number of sequencing studies on disparate tissues and cell types will be required to fully define miRNA expression and sequence complexity, and identify the full complement of expressed miRNA genes.
Here, we performed Illumina GA and ABI SOLiD sequencing of small RNAs isolated from resting and cytokine-activated NK cells as an initial step in defining their role in NK cell biology. To this end, we developed a bioinformatics approach that allows determination of known miRNA identity and expression using alignments to known miRBase precursor hairpin sequences with the short read mapping package SHRiMP (Rumble et al. 2009). This analysis pipeline also provides a complete depiction of NK cell miRNA sequence variation and isomiRs. Twenty-one novel miRNA genes were identified by aligning short reads to the genome followed by detection of candidate novel precursor hairpins based on miRNA-like predicted secondary structure plus sequence read alignment relationships at a genome locus. miRNA detection in NK cells was also confirmed using microarrays and qRT-PCR. In addition, these data revealed that miR-223, one candidate miRNA predicted to bind to the GzmB 3′ UTR, was down-regulated with IL15-activation. We demonstrate in vitro that miR-223 specifically targets the murine GzmB 3′ UTR, supporting its role in regulating GzmB translation during murine NK cell activation. This study therefore provides a comprehensive quantitative catalog of mature miRNA expression by murine NK cells using multiple platforms and a critical framework for future studies of their impact on NK cell biology. Furthermore, the detailed bioinformatics analysis pipeline for NGS of small RNAs can be applied to a broad range of other rare normal or malignant cell populations to better define miRNAs in health and disease.
Results
Sequencing of small RNAs from primary murine NK cells using the Illumina GA
Splenic NK cells were purified from C57BL/6 (B6) mice by flow cytometric sorting (≥98% NK1.1+CD3−), and lysed immediately for total RNA isolation (resting) or after 24 h of stimulation with recombinant murine IL15 (activated). Next, small 19–26-nt RNAs were isolated and used to generate resting and activated cDNA libraries (see Methods). Each cDNA library was sequenced on three individual lanes of the Illumina GA flow cell (six lanes total), generating a total of 4,643,446 and 4,089,580 sequence reads from the resting and activated NK cells, respectively. These small RNA sequences were analyzed with two primary goals, to define the known miRNA expression in resting and activated NK cells, and to identify and enumerate novel miRNAs expressed in NK cells.
Known microRNAs identified in NK cells using the Illumina GA
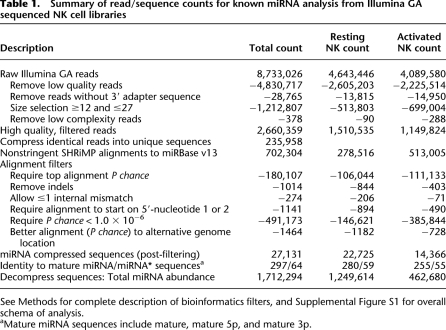
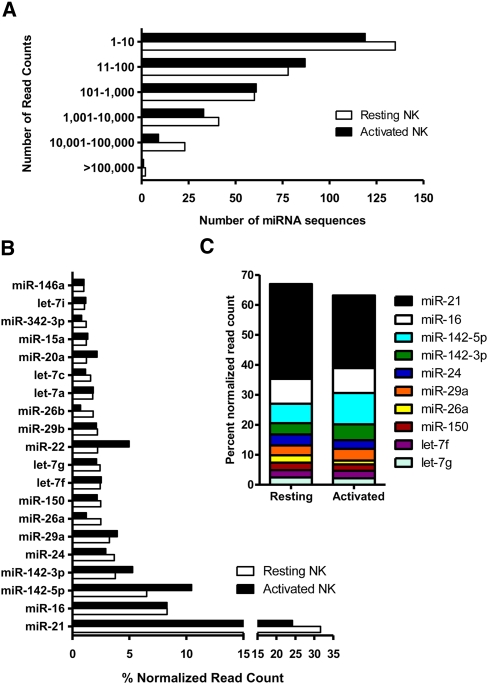
We established a bioinformatics analysis protocol for identifying and enumerating known (miRBase v13) miRNAs based on alignments to mature miRNA and minor miRNA* encoding coordinates in miRNA precursor hairpin sequences using SHRiMP (Supplemental Fig. S1). After initial removal of low-quality reads and those without an identified 3′ adaptor sequence, total reads were “compressed” to unique sequences, retaining their individual read count information (Table 1). These unique sequences were aligned to miRBase v13 miRNA precursor hairpins to detect expression of mature miRNA/miRNA* sequences, and a number of filters were used to ensure accuracy in miRNA identity and read counts as indicated in Table 1. The alignment and read lengths resulting from our approach were consistent with known mature miRNA/miRNA* sequence lengths (Supplemental Fig. S2). Unique sequences representing miRNAs were then “decompressed” to reflect the expression of a given miRNA sequence based on the count of reads corresponding to a given miRNA. Comparing the high-quality reads that were the input for alignment to the subset corresponding to miRNAs, a large proportion of resting (1,249,614 reads, 83%) and activated (462,680 reads, 40%) reads were known miRNA sequences. For miRNA read count abundance data, sequenced reads that mapped with equal probability to different miRNA hairpin precursors (e.g., mmu-mir-16-1 and mmu-mir-16-2) were randomly assigned to only one reference sequence to accurately report total read counts without redundancy. Similar to prior reports of Illumina GA sequencing in human ES cell lines or in a murine leukemia model (Kuchenbauer et al. 2008; Morin et al. 2008b), we observed a large dynamic range of read counts ranging from 1 to 395,295 in primary murine NK cells (Fig. 1A). For all comparisons, the number of reads for a given miRNA was normalized by division of the total number of miRNA reads in that library, yielding the percent normalized read count for each miRNA. miR-21 was the most abundant miRNA detected in resting and activated NK cells, and expression of the top 20 miRNAs is shown in Figure 1B. The top 10 miRNAs by read count corresponded to ∼65% of all miRNA sequence reads in NK cells (Fig. 1C). Differentially expressed miRNAs were defined by fold change in normalized read counts between the resting and activated NK cell sequences (Table 2). A complete list of miRNAs sequenced in each library, total and normalized read counts, and fold differences between resting and activated NK cells is provided in Supplemental Table S1. In addition, a novel miRNA* sequence was identified for 56 known miRNA genes without an annotated miRNA* in miRBase v13 (Supplemental Table S2). Thus, Illumina GA sequencing of small RNAs may be used to identify and enumerate mature miRNAs in this rare primary immune cell type.
Table 1.
Summary of read/sequence counts for known miRNA analysis from Illumina GA sequenced NK cell libraries
See Methods for complete description of bioinformatics filters, and Supplemental Figure S1 for overall schema of analysis.
aMature miRNA sequences include mature, mature 5p, and mature 3p.
Figure 1.
Known miRNAs sequenced in resting and activated NK cells using the Illumina GA. (A) Distribution of miRNA sequences in resting and activated NK cells based on the total read counts. Mature miRNA sequences are categorized by the number of read counts corresponding to each individual miRNA. (B) Top 20 known miRNAs expressed in resting and activated NK cells. The percent contribution of each miRNA sequence to the total pool of known miRNA sequences was calculated by dividing individual miRNA read counts by the total number of known miRNA sequence reads in that cDNA library. (C) Top 10 known miRNAs constitute ∼65% of total miRNA reads in NK cells.
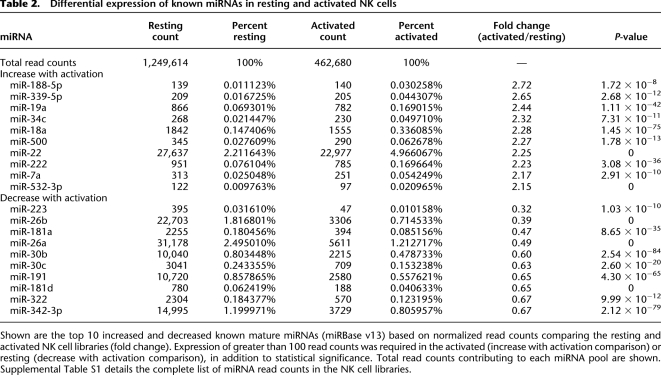
Table 2.
Differential expression of known miRNAs in resting and activated NK cells
Shown are the top 10 increased and decreased known mature miRNAs (miRBase v13) based on normalized read counts comparing the resting and activated NK cell libraries (fold change). Expression of greater than 100 read counts was required in the activated (increase with activation comparison) or resting (decrease with activation comparison), in addition to statistical significance. Total read counts contributing to each miRNA pool are shown. Supplemental Table S1 details the complete list of miRNA read counts in the NK cell libraries.
Complexity of Illumina GA sequenced miRNAs: NK cell isomiRs
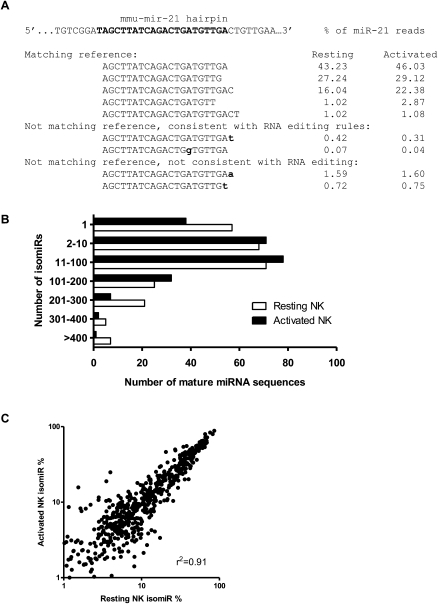
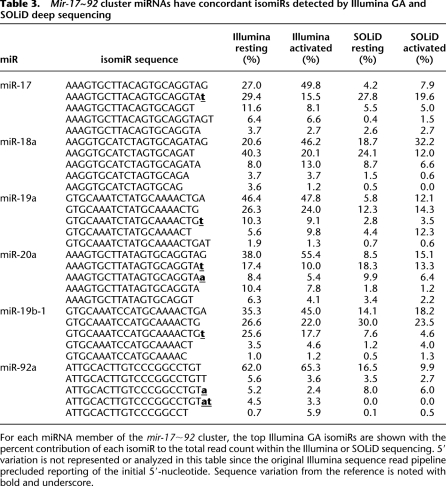
Variability in RNA sequences that comprise an individual miRNA has been designated as “isomiRs” (Morin et al. 2008b). We comprehensively evaluated internal and 3′ variation in miRNA sequences from the Illumina GA, illustrated in Figure 2 with miR-21. After alignment to miRBase v13 precursor hairpins, our analysis identified 3′ variants that matched or did not match the reference miRNA gene. Those sequence alterations that did not match the precursor hairpin sequence were further categorized depending on whether they were consistent with known miRNA editing processes (Fig. 2A). For isomiR analysis, reads that mapped with identical probability to different versions of the miRNA hairpin (e.g., mmu-mir-16-1 and mmu-mir-16-2) were redundantly mapped to both hairpin precursors, since the origin of that sequence could be either reference sequence. IsomiRs were very common (Fig. 2B), with 78% (resting) and 83% (activated) of miRNAs having more than one isomiR sequence identified. A complete depiction of sequenced isomiRs from each individual miRNA is provided as Supplemental material, generated by isomiRview (see Methods). The majority of isomiRs matched the reference genome, and variants likely result from variability in cropping or dicing by the miRNA biogenesis machinery (Davis and Hata 2009). Changes in mature miRNA sequences from the reference were observed that are consistent with known RNA editing of miRNAs (Bass 2002; Rissland and Norbury 2008). We also noted isomiRs with 3′ additions of U(T) or A that also have been reported (Li et al. 2005; Rissland and Norbury 2008; Zhu et al. 2008). Additional changes were noted that do not match the reference miRNA gene and do not follow established RNA editing processes, consisting of nucleotide substitutions or nontemplated 3′ nucleotide additions. These may represent sequencing errors, but could potentially arise from undefined miRNA editing processes (Ebhardt et al. 2009). In many cases, the annotated miRBase mature sequence was not the most abundant 3′ variant isomiR, suggesting that miRNA processing may differ between tissues or cell types, or that the annotated miRBase sequence was derived from a less abundant isomiR. Overall, similar isomiR patterns were identified in resting and activated NK cells, suggesting that global miRNA biogenesis is not dramatically altered with short-term IL15 activation (r2 = 0.91) (Fig. 2C). Notably, sequencing miRNAs using an alternative NGS approach (Applied Biosystems SOLiD system; see below) identified similar isomiRs in resting and activated NK cells (e.g., the mmu-mir-17∼92 cluster of miRNA) (Table 3), suggesting that this isomiR detection is not an artifact from the specific NGS method or platform employed.
Figure 2.
Mature miRNA sequence variation (isomiRs) detected by Illumina GA sequencing of NK cell miRNAs. (A) Example miR-21 isomiRs illustrate the complexity of miRNA sequences identified by Illumina sequencing. Shown are selected isomiR sequences that match or do not match the reference genome and their percent contribution to miR-21 read counts (mature miR-21 sequence annotated in bold). We further divide those reads not matching the reference genome into sequences that may arise by a known RNA editing event and all others. A complete breakdown of isomiR sequence variation for all miRNAs is provided in the Supplemental material. (B) Distribution of miRNA mature sequences by the number of isomiRs identified by Illumina GA sequencing. (C) Comparison of isomiR sequences between resting and activated NK cell libraries. Scatterplot showing the percent contribution of a given isomiR in each library, with each dot representing one isomiR. Analysis was limited to the top five isomiRs and the top 125 miRNAs by expression. In general, isomiRs were found in very similar distributions in resting and activated NK cells (r2 = 0.91).
Table 3.
Mir-17∼92 cluster miRNAs have concordant isomiRs detected by Illumina GA and SOLiD deep sequencing
For each miRNA member of the mir-17∼92 cluster, the top Illumina GA isomiRs are shown with the percent contribution of each isomiR to the total read count within the Illumina or SOLiD sequencing. 5′ variation is not represented or analyzed in this table since the original Illumina sequence read pipeline precluded reporting of the initial 5′-nucleotide. Sequence variation from the reference is noted with bold and underscore.
Novel miRNA genes identified in NK cells using the Illumina GA
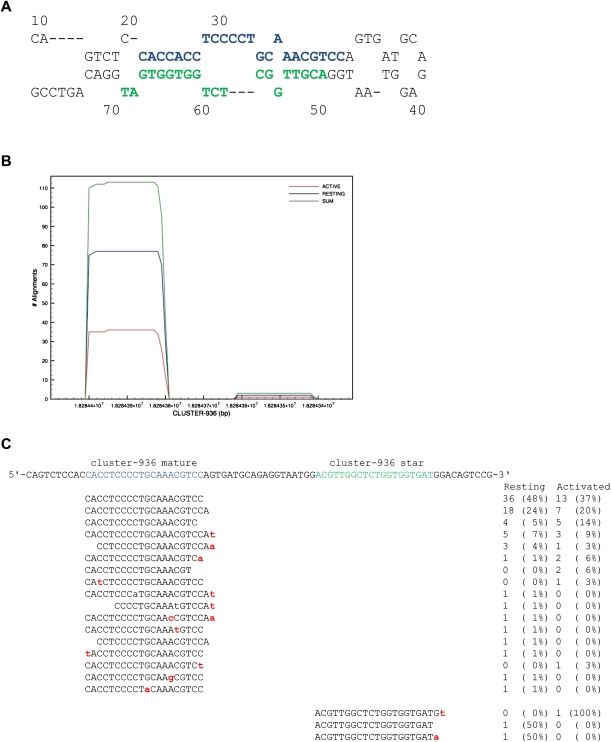
We also used the Illumina GA sequenced small RNAs from NK cells to identify novel miRNA genes (Supplemental Fig. S3). Using SHRiMP, all high-quality small RNA sequences from both NK cell libraries (2,660,359 sequence reads) were aligned to the mouse genome and filtered based on the SHRiMP parameter <P chance> and miRNA characteristics. From these filtered genome alignments, 84,884 alignment clusters were generated, and for the 4886 clusters with greater than 10 reads contributing, cluster sequence coverage maps were built using RefCov (an in-house Perl application; T Wylie, unpubl.). While some of these clusters likely correspond to novel miRNA genes, a large number also could represent mRNA or noncoding RNA degradation products or repetitive sequences. Based on the alignment depth in the coverage map, the top 1483 clusters (read depth > 50) were chosen for additional analysis and annotated for identity with known noncoding RNAs (including miRBase v13 miRNAs) and known protein-coding genes by their genome coordinates (Supplemental Fig. S4). These top 1483 clusters were also analyzed for miRNA precursor characteristics (see Methods), and all clusters that had a predicted negative ΔG of folding were manually reviewed in a custom miRNA validation tool (Supplemental Fig. S5) that simultaneously displays sequence characteristics, predicted folding secondary structure, and small RNA read alignment coverage maps. Known miRNA genes were readily identified using this approach, constituting 210 (45%) clusters that generated spontaneous hairpin folds with miRNA-precursor characteristics. This analysis provided a list of 198 candidate novel miRNA precursor sequences with both a precursor sequence that was predicted to spontaneously form a miRNA-like hairpin, and sequence alignments to the putative miRNA precursor consistent with mature miRNA biogenesis. Putative novel miRNA hairpin sequences were then added to known miRNA precursor hairpin sequences (miRBase v13), and all small RNAs were re-aligned to these novel hairpins using the identical parameters previously used for known miRNAs. Using this combination of read alignments (expression) and precursor characteristics (miRNA biogenesis), 21 novel miRNA genes were identified from NK cells (Table 4; Supplemental Table S3). These sequences varied in abundance from 28 to 993 read counts, and three mature sequences were differentially expressed (fold change ≥1.5 with ≥99 read counts, significant P-value) between resting and activated NK cells (Table 4). An example of a novel miRNA cluster-936 (mmu-mir-1306) is shown in Figure 3 including the precursor hairpin sequence/structure, the read alignment coverage model that includes detection of the miRNA*, and isomiR analysis.
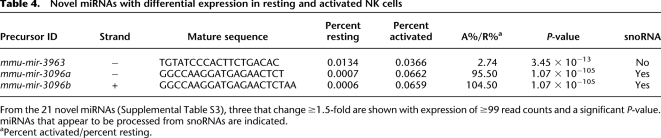
Table 4.
Novel miRNAs with differential expression in resting and activated NK cells
From the 21 novel miRNAs (Supplemental Table S3), three that change ≥1.5-fold are shown with expression of ≥99 read counts and a significant P-value. miRNAs that appear to be processed from snoRNAs are indicated.
aPercent activated/percent resting.
Figure 3.
Novel miRNA genes identified in NK cells. (A) Predicted secondary fold structure of a novel miRNA precursor hairpin cluster-936 (mmu-mir-1306) (16:18284332..18284410) identified in Illumina GA–sequenced NK cell small RNAs (predicted ΔG = −30.7). Mature miRNA is noted in blue, minor star species in green. (B) Read alignment coverage map (RefCov) of novel miRNA gene demonstrating the specific alignment of sequences to the mature miRNA sequence. Shown is the nucleotide position of the precursor (x-axis) and the number of reads aligning to each nucleotide position (y-axis). Coverage models for total (sum = resting and activated), resting, and activated NK libraries are shown. (C) IsomiR analysis of novel miRNA with mature sequence noted in blue, and the read counts and corresponding percentage contribution to the total number of reads aligning to this novel hairpin.
Within the top 1483 genome clusters, we also identified snoRNAs and other noncoding RNA species (Supplemental Fig. S4). Consistent with recent reports that snoRNAs may be processed into functional miRNA (Ender et al. 2008; Saraiya and Wang 2008; Politz et al. 2009; Taft et al. 2009), we identified five snoRNAs with expressed small RNA sequences that aligned to a miRNA-like hairpin precursor structure and were consistent with processing into miRNA (Supplemental Table S3). Since previously reported Illumina GA miRNA sequencing experiments filter out genome locations that correspond to known noncoding RNAs (Kuchenbauer et al. 2008; Morin et al. 2008b), identification of noncoding RNAs that are processed into miRNAs may have been underreported in the past. Thus, sequencing small RNAs in NK cells identified novel murine miRNA genes.
Validation of Illumina GA with ABI SOLiD miRNA sequencing and hybridization-based techniques
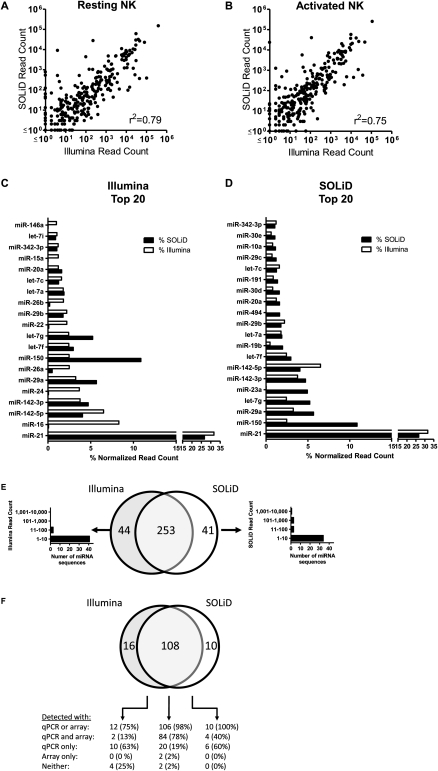
We next compared the miRNA sequencing on Illumina GA results to a similar experiment performed using the ABI SOLiD sequencing instrument. Total RNA from resting and activated NK cells was processed to create libraries for sequencing using the SOLiD small RNA expression kit. SOLiD sequencing of these resting and activated NK cell libraries yielded 20,490,363 and 28,738,852 sequence reads, respectively. SOLiD sequencing reads were aligned to miRBase v13 hairpins in “color-space” using SHRiMP and similar parameters as used for aligning Illumina GA sequences (Supplemental Fig. S1; Supplemental Table S4; Methods). Here, we detected 259/64 and 261/67 miRNAs/miRNA*s in resting and activated NK cells, respectively. A complete list of miRNAs detected by SOLiD sequencing is presented in Supplemental Table S5, while a comprehensive depiction of SOLiD NK isomiRs is available in the Supplemental material. Collectively, there was excellent correlation between the identification and enumeration of known miRNAs on the Illumina GA and SOLiD instruments (Fig. 4A,B). The top 20 miRNAs detected by Illumina (Fig. 4C) and SOLiD (Fig. 4D) are shown. Most cases of differential detection occurred in alignment clusters with relatively low read counts, suggesting that variation in primary cell biological replicates may explain these differences (Fig. 4E); however, a few cases of marked disparity in read count were detected (Fig. 4C,D; Supplemental Table S6).
Figure 4.
Comparison of miRNA detection by Illumina GA and ABI SOLiD sequencing and hybridization-based techniques. Known miRNAs were identified in cDNA libraries created from small RNAs from resting or activated primary mouse NK cells by SHRiMP alignment to miRBase v13 hairpin precursor-miRNAs. For each miRNA detected on either platform, the dot shows the relative expression by normalized read count from Illumina (x-axis) and SOLiD (y-axis) in the resting NK cell (A) or activated NK cell (B) libraries. Overall, the correlation for detection was high (0.75). The top 20 miRNAs detected in both NK cell libraries for Illumina (C) and SOLiD (D) are shown rank ordered based on platform. (E) Venn diagram of overlapping detection in resting and activated NK cell libraries comparing Illumina and SOLiD sequencing. For miRNA detected in only one platform-data set, the normalized read count distribution is shown, highlighting that most discrepant miRNAs are expressed at relatively low levels. (F) Venn diagram showing the breakdown of overlap between Illumina GA sequencing, SOLiD sequencing, AB real-time qRT-PCR, and Agilent microarrays for 136 miRNAs with probes available for both qRT-PCR and microarrays. Detection by qRT-PCR was defined as a ΔCt (compared to MammU6) of <15, and for microarrays a normalized signal intensity of >1.0. Rare cases of major differences in the normalized read counts were detected by Illumina GA and SOLiD (Supplemental Table S4).
We therefore compared the Illumina and SOLiD sequencing results to two separate hybridization-based techniques, limiting the analysis to 134 miRNAs detected by sequencing with primers and/or probes present for qRT-PCR miRNA assays or microarrays (Fig. 4F). Overall, the majority of the miRNAs detected in NK cell libraries via both sequencing platforms were confirmed using the hybridization approaches. Most discordant detection was found in miRNAs with relatively low read counts in NK cells. There were a few notable outliers; for example, miR-16 was detected abundantly by Illumina (8.3% normalized read count), but minimally by SOLiD (0.07 % normalized read count). Since miR-16 was also detected at high levels by both qRT-PCR and microarray analysis, it appeared that in this case SOLiD sequencing failed to detect miR-16. The infrequent cases where marked discordance was noted between Illumina and SOLiD are summarized in Supplemental Table S6. The complete quantitative PCR and microarray profiling results of resting and activated NK cells are reported in Supplemental Tables S7 and S8. Thus, overall SOLiD sequencing confirmed miRNA expression defined by Illumina GA sequencing, and with a few noted exceptions, the sequencing-based results were validated using miRNA qRT-PCR and microarrays.
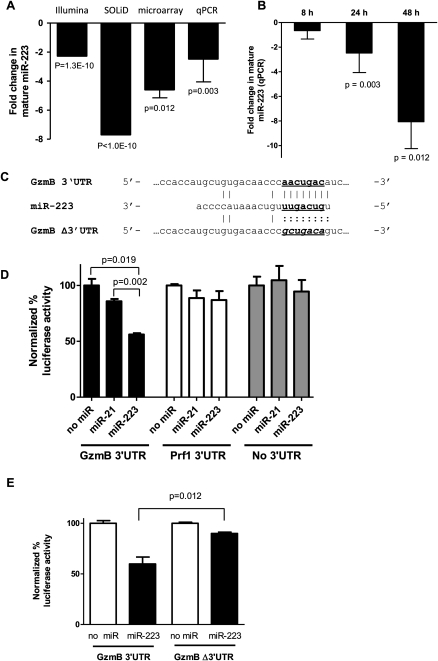
miR-223 is down-regulated with IL15 activation and targets the murine GzmB 3′ UTR
miR-223 was identified as the mature miRNA with the greatest fold decrease after IL15 activation using Illumina GA sequencing (Table 2). All miRNA platforms confirmed mature-miR-223 down-regulation upon NK cell activation (Fig. 5A). The IL15-induced reduction in mature miR-223 is time-dependent (Fig. 5B). GzmB is a cytotoxic effector molecule important for NK cell killing that appears to be regulated at the post-transcriptional level in murine NK cells (Fehniger et al. 2007). miR-223 has a predicted (TargetScanMouse 5.1) target site in the murine GzmB 3′ UTR (Fig. 5C). Utilizing luciferase sensor plasmids, overexpression of miR-223 in 293T cells significantly decreased the luciferase signal controlled by the full-length murine GzmB 3′ UTR (Fig. 5D). Sensor plasmid controls that included no 3′ UTR or an unrelated (Prf1) 3′ UTR exhibited no regulation following miR-223 overexpression. Similarly, an unrelated miR overexpression control (miR-21) not predicted to target GzmB demonstrated no alteration in experimental luciferase signal in the GzmB 3′-UTR sensor plasmid. Moreover, when the GzmB 3′ UTR was mutated at the seed sequence of the predicted miR-223 binding site (GzmBΔ 3′ UTR) (Fig. 5C), there was loss of regulation by miR-223 overexpression (Fig. 5E). Thus, mature miR-223 is expressed in resting NK cells, decreases in abundance with IL15 activation, and directly targets the GzmB 3′ UTR suggesting its role in regulating GzmB translation in resting NK cells.
Figure 5.
miR-223 is down-regulated with IL15 activation and directly targets the murine GzmB 3′ UTR. (A) Mature miR-223 decreases in abundance in resting NK cells following 24 h of IL15 activation on multiple platforms, including Illumina, SOLiD, microarray, and qRT-PCR. (B) Time course of mature miR-223 down-regulation in mouse NK cells at indicated time points after rmIL15 activation assayed by ABI qRT-PCR assay (N = 3–7 independent experiments). (C) Schema of miR-223 binding site in murine GzmB 3′-UTR sequencing (seed sequence highlighted with bold/underline). Mutated GzmB Δ3′ UTR eliminates the seed binding sites (italics). (D) miR-223 selectively targets the GzmB 3′ UTR. psiCheck2 sensor plasmids containing the GzmB 3′ UTR, Prf1 3′ UTR (negative control), or no 3′ UTR (negative control) were cotransfected in 293T cells with an MND-GFP-miRNA overexpression vector containing a mini-gene of mir-223, mir-21 (negative control), or no miRNA (negative control). Overexpression of miR-223 selectively decreased the luciferase expression compared to no miRNA or miRNA-21 controls. No down-regulation of luciferase was observed for negative control (Prf1 3′ UTR or no 3′ UTR) with miR-223 overexpression. (E) miR-223 targeting of GzmB 3′ UTR is direct. miR-223 down-regulates luciferase controlled by the endogenous GzmB 3′ UTR, but fails to down-regulate luciferase controlled by the GzmB Δ3′ UTR (see B), which lacks the predicted miR-223 seed sequence.
Discussion
In this study, we delineate a bioinformatics analysis approach that uses next-generation sequencing of small RNA libraries by the Illumina GA or ABI SOLiD instruments to define the expression of 302 known and 21 novel mature miRNAs and their sequence variation in primary murine NK cells. In general, these two different small RNA cDNA library construction techniques and NGS platforms detected similar mature miRNAs, although a few clear discrepancies were noted. miRNA detection was also validated using qRT-PCR “arrays” and microarrays, with overall agreement between hybridization-based detection and NGS. Using these miRNA expression data, we identified mature miR-223 as significantly down-regulated during NK cell stimulation with IL15. Additional experiments identified that miR-223 specifically targets the murine GzmB 3′ UTR in vitro, implicating this miRNA in the regulation of GzmB translation during NK cell activation. Thus, our NGS miRNA bioinformatics pipeline defined miRNA expression in NK cells, which led to identification and biological confirmation of a novel miRNA:mRNA target interaction. This NK cell miRNA transcriptome provides important information about mature NK cell miRNA expression and a framework to formulate additional questions about the role of miRNAs in many aspects of NK cell biology.
Following initial reports of using Illumina GA sequencing read counts to define miRNA expression in cell lines or murine model systems (Kuchenbauer et al. 2008; Morin et al. 2008b), several miRNA analysis approaches have been recently reported to analyze NGS data (Friedlander et al. 2008; Hackenberg et al. 2009; Linsen et al. 2009; Jagadeeswaran et al. 2010; Nobuta et al. 2010). Many of these approaches share common elements that include aligning small reads to a reference sequence/genome, mapping those reads to known miRNA genes, and using both the secondary RNA structure prediction and the location of the read alignment within a putative novel precursor miRNA to aid in identifying novel miRNA genes. Since miRNA biogenesis generates a distinct pattern of mature 5p/3p small RNA expression abundance compared to star and the remainder of the precursor hairpin (Davis and Hata 2009), it follows that identifying such a pattern provides evidence that these genes are expressed and processed using the miRNA biogenesis molecular machinery. Our own approach uses SHRiMP to align NGS reads to known miRNA precursor hairpins with high stringency requirements for mature/star or 5p/3p identification and enumeration, and also provides a description of isomiR sequence variants, capturing both internal and 3′ alterations (isomiRview). Since small sequence alignment tools are continually in development, new sequence aligners may be integrated into the pipeline if they further improve efficiency and flexibility. Notably, similar isomiRs were identified from different NK cell libraries sequenced on the Illumina and SOLiD platforms, suggesting that isomiRs are not artifacts of an individual library creation or sequencing approach. Overall, we also observed similar isomiR patterns in resting and IL15-activated NK cells, indicating their stability of expression despite sequencing different cDNA libraries with varying cell activation states. While we comprehensively report the internal and 3′ sequence variants, the biological significance of isomiRs is currently unclear. Similar to other studies, most isomiRs in NK cells matched the reference miRNA precursor, suggesting alterations in miRNA cropping or dicing as their etiology, but other types of variants were detected at a lower frequency. Of potential importance, alterations of mature miRNA sequence may result in altered target recognition, and changes in precursor/primary miRNA transcripts may affect processing; however, this requires experimental validation. The ability of our bioinformatics pipeline to identify and track sequence variants will be important for future profiling applications where miRNAs may be mutated or altered (miRNA variants), including the comparison of normal and potentially mutated miRNA sequences and/or genes in malignancy.
This study applies two distinct NGS approaches, both analyzed with a similar bioinformatics pipeline, to the question of miRNA expression in resting and IL15-activated murine NK cells. It is likely that each sequencing approach has some biases (Linsen et al. 2009), and a more complete picture of the NK cell miRNA transcriptome resulted from using complementary approaches. Detection of miRNAs via the two NGS platforms had reasonable concordance, taking into consideration different biological replicates and distinct library construction approaches. In general, we noted fewer discrepancies in known miRNA detection and abundance between the Illumina GA and the qRT-PCR and microarray hybridization techniques, compared to SOLiD (miR-16, miR-24, miR-26a, miR-22, miR-26b, miR-15a, miR-146a). However, in a few instances (miR-19b, miR-494), Illumina GA abundance was discrepant with SOLiD and the hybridization techniques used for validation.
Our analysis pipeline utilizing NGS reads has the added advantage over qRT-PCR or microarrays to identify and enumerate novel miRNAs. In parallel to known miRNA expression analysis, sequence reads were aligned to the reference genome to discover potential novel miRNA precursor genes. The novel miRNA precursors identified with this pipeline meet the currently accepted definition of miRNA genes (required for miRBase curation) based on (1) expression of mature/star or 5p/3p sequences (normalized read counts) and (2) miRNA biogenesis (miRNA precursor identification with reads mapping to mature/star or 5p/3p locations) (Griffiths-Jones et al. 2006, 2008). Utilizing this pipeline, we have identified several novel miRNA genes and sequences, including novel miRNA mature sequences and their corresponding genes, novel miRNA genes that contain known miRNA sequences, and novel minor/star sequences for known miRNAs. Most NGS analyses to date have excluded putative miRNA genes if their genomic alignment location overlaps with other small RNAs. Recently, several reports have identified and experimentally validated that miRNAs may be processed from other small RNAs, such as small nucleolar RNAs (snoRNAs) (Ender et al. 2008; Saraiya and Wang 2008; Scott et al. 2009; Taft et al. 2009) or possibly tRNAs (Cole et al. 2009). Indeed, in our analysis where clusters that mapped to other small RNAs were included in the novel analysis pipeline, we identified six snoRNA genes that are expressed and processed into miRNAs. Thus, while a large proportion of miRNA sequences and genes have been identified over the past decade, continued NGS of different cell types/tissues (including normal and malignant) and species will likely be required to complete the catalog of expressed miRNAs and their genes. Furthermore, altering the input RNA size criteria for future library creation will allow our analysis pipeline to evaluate the expression of other small noncoding RNA species in rare primary cells (Choudhuri 2009; Jacquier 2009).
Recently, Illumina-based miRNA sequencing was reported for a wide array of mouse hematopoietic and immune cells, with an emphasis on B-cell and T-cell development and activation (Kuchen et al. 2010). For comparison purposes in this study, small RNAs from additional immune cell types were also sequenced, including a single library from Rag1−/− splenocytes cultured in high dose IL2 for 8 d and annotated as “NK cells.” Importantly, Rag1−/− splenocytes cultured in IL2 for 8 d do not represent primary murine NK cells in mRNA profiles, phenotype, or functional properties, and are therefore unlikely to represent primary murine NK cell miRNA expression patterns. Since similar Illumina sequencing and normalization approaches were used in the Kuchen et al. (2010) study, we compared normalized read counts from “NK cells” (IL2 activated Rag1−/− splenocytes) in that study to our resting and 24-h IL15-activated primary murine NK cell read counts, which demonstrated no correlation (r2 values of 0.07 and 0.13, respectively). This illustrates the importance of the activation state, purity, and sorting strategy of any primary cell type of interest to accurately profile mature miRNAs.
Thus, this study provides the first comprehensive mature miRNA transcriptome analysis in primary NK cells. All miRNA sequencing and expression data are deposited in the appropriate public repository (microarrays in GEO, sequence data in the SRA). The original hypothesis that led to NK cell miRNA sequencing was intended to determine whether miRNAs expressed in resting NK cells contribute to repressing effector molecule (GzmB and Prf1) mRNA translation in resting NK cells (Fehniger et al. 2007). To identify the most likely candidate miRNAs, we evaluated miRNAs expressed in resting NK cells that decreased more than 1.5-fold with activation. miR-223, originally characterized as important in myeloid development (Johnnidis et al. 2008), was detected in NK cells and decreased approximately threefold with IL15 activation. While the mechanism leading to mature miR-223 decrease in IL15-activated NK cells is unclear, this type of rapid expression change has been previously identified following other cytokine/growth factor stimulation. For example, TGFB/BMP4 have been shown to signal via SMAD protein intermediates, which, in turn, bind to DDX5 (p68) and thereby specifically regulate RNASEN/DGCR8-mediated cropping of pri-mir-21 into pre-mir-21 (Davis et al. 2008). Future studies will evaluate whether downstream signaling intermediates from the IL15 receptor alter miR-223 biogenesis, leading to the selective decrease in mature miR-223 species in NK cells. We also used luciferase sensor plasmids with the wild-type and miR-223 binding-site mutated GzmB 3′ UTR to demonstrate that miR-223 directly targets the murine GzmB 3′ UTR. Collectively, these data support a role for miR-223 in regulating GzmB in resting murine NK cells, although this process is most likely multifactorial. Several candidate miRNAs that also may contribute to the regulation of murine GzmB include miR-24, miR-30b, and miR-30c. Additional studies will evaluate other miRNAs as regulators of NK cell effector molecules and will utilize genetic models to validate candidate miRNAs in NK cells in vivo.
This study also revealed that NK cells contain miRNAs that are expressed and function in other lymphocyte subsets. For example, NK cells express miR-181a, which has been implicated in regulating TCR receptor signaling, thereby providing a mechanism to alter TCR responsiveness during T-cell maturation (Li et al. 2007). In addition, a number of miRNAs appear to be highly expressed in both NK cells and adaptive CD8+ T-lymphocytes (Wu et al. 2007), including prominent expression of miR-21, miR-142-5p, miR-142-3p, miR-16, miR-15b, miR-150, and miR-let-7f. This suggests that these shared miRNAs are not NK-cell-specific, but rather regulate common pathways important for the differentiation, maintenance, or function of the cytotoxic lymphocyte “program.” Comparing the relative abundance of commonly expressed miRNAs between NK cells in our study and other lymphocyte subsets from data in the literature is technically challenging due to differences in cellular preparation, profiling methodology, and bioinformatics analysis. Future experiments using NGS to compare the primary NK cell miRNA transcriptomes to other lymphocyte subsets will be needed to generate a specific NK cell miRNA “signature” and to help to identify those miRNAs that are selectively expressed or down-regulated in primary NK cells. Furthermore, studies of NK cell developmental intermediates and distinct phenotypic and functional NK subsets will be useful to identify miRNAs that regulate different aspects of NK cell differentiation and maturation. Thus far, only limited information is available on the molecular events regulating these processes in NK cells (Di Santo 2006, 2008a; Yokoyama and Kim 2006; Caligiuri 2008), and it is likely that miRNAs regulate multiple facets of NK cell differentiation and maturation, analogous to T- and B-lymphocytes (Baltimore et al. 2008). Furthermore, once miRNA expression has been defined, additional studies will be required to define and biologically verify their relevant mRNA targets (Karginov et al. 2007; Licatalosi et al. 2008; Chi et al. 2009), thereby incorporating miRNA regulatory circuits into the larger framework of NK cell biology.
Methods
NK cell sorting, activation, and total RNA isolation
C57Bl/6 (B6) mice and B6 Rag1−/− mice (used as a source of enriched NK cells) were obtained from Jackson Laboratory. All mice were maintained in SPF housing, and all experiments were conducted in accordance with Washington University animal care and use guidelines. Spleens were harvested from 8- to 12-wk-old mice, single cell suspensions were generated by pressing through sterile 70 μM nylon mesh, and RBCs were lysed using ACK lysis buffer. Viable splenocytes were stained with anti-mouse NK1.1 (PK136) and CD3 (145-2C11) monoclonal antibodies and sorted by flow cytometry (iCyte Reflection). Purified NK cells (≥98% NK1.1+CD3−) were enumerated on a hemacytometer with trypan blue and lysed in TRIzol immediately (resting) or after 24 h of stimulation with recombinant murine IL15 (100 ng/mL; R&D Systems; activated) (Fehniger et al. 2007). Total RNA was extracted from NK cell lysates using the TRIzol reagent according to the manufacturer's instructions (Invitrogen). Total RNA quality was determined by Agilent 2100 bioanalyzer (Agilent Technologies) according to the manufacturer's recommendations.
Small RNA library preparation for Illumina GA sequencing
Total RNA from 7.5 × 106 resting NK and 6.5 × 106 IL15-activated NK cells, pooled from three independent flow sorting experiments, yielded 4.4 μg and 3.0 μg of total RNA, respectively. Small RNA library construction from total RNA followed an adaptation developed by Chen et al. (2005). Briefly, total RNAs were spiked with 32P-labeled 19-mer and 24-mer ribo-oligonucleotides at 10,000 cpm, and small RNAs (19 to 24 nt) were excised following 15% polyacrylamide/urea (PAGEU) SequaGel electrophoresis (National Diagnostics). Recovered RNAs were eluted from acrylamide in 400 mM NaCl and ethanol-precipitated. Suspended small RNAs (2000 cpm) were ligated with 50 pmol of the preadenylated Linker-1 (Linker-1 sequence underscored) oligonucleotide, 5′-rAppCTGTAGGCACCATCAAT/3′ddC (IDT) in the presence of 400 U of T4 RNA ligase 2, truncated (New England BioLabs) for 1 h, at 25°C, in 50 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 10 mM dithiothreitol (DTT), 60 ng/μL BSA. Following 15% PAGEU size selection and purification, 50 pmol of the Illumina SBS-3 ribo-oligonucleotide, 5′-ACACUCUUUCCCUACACGACGCUCUUCCGAUC, was ligated using 40 U of T4 RNA ligase (New England BioLabs) to recovered RNA:Linker-1 ligated molecules (1200 cpm) for 1 h at 37°C, in 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM ATP. A final 15% PAGEU size selection recovered SBS-3:RNA:Linker-1 ligated molecules (350 and 500 cpm for resting and activated libraries, respectively).
Priming first-strand cDNA synthesis used 100 pmol of the 3′ primer, 5′-ATTGATGGTGCCTACAG, annealing to the Linker-1 sequence (underlined) after 15 min of incubation at 65°C and on ice for 5 min. The annealed RNA-primer mixture was incubated in 1× buffer (Superscript III; Invitrogen) containing 10 μmol of dNTPs, 5 mM DTT, 20 U of RNASin (Promega), and 200 U of Superscript III (Invitrogen) and incubated for 1 h at 50°C, 15 min at 72°C, and held at 4°C until recovered. First-strand cDNA was PCR-amplified using one-fourth reaction volume of first-strand cDNA template and was incubated with 100 pmol each of the 5′ and 3′ oligos, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTC and 5′-CAAGCAGAAGAGGGCATACGATTGATGGTGCCTACAG, respectively and included 1× PCR buffer (Roche), 2 μmol of dNTPs, 3 mM MgCl2, and 5 U of Taq DNA polymerase (Roche). Thermal cycling was initiated with a 2-min melt at 94°C, followed by five cycles of denaturing, annealing, and extending at 94°C (15 sec), 54°C (30 sec), and 72°C (30 sec). After five cycles, an additional 17 cycles of 94°C (15 sec), 60°C (30 sec), and 72°C (30 sec) was followed by a final 2-min extension at 72°C. The PCR-amplified micro-cDNAs were suspended in a final 300 mM NaCl solution, phenol/CHCl3-extracted, ethanol-precipitated, and suspended in 20 μL of dH2O. To remove the 19-mer and 24-mer amplified tracers, the micro-cDNAs were treated with the endonuclease PmeI, and the full-length micro cDNAs were gel-purified from 2% LE/TAE low melt agarose (Bio-Rad). Illumina sequencing read analysis used pipeline version 0.2.2 (Firecrest /Bustard 1.8.28) in which the first base (T) was trimmed, and cDNA sequencing was performed in the 5′-to-3′ direction.
Small RNA library preparation for ABI SOLiD sequencing
Small RNA libraries were constructed using the SOLiD small RNA expression kit (SREK) based on the ligase-enhanced genome detection (LEGenD) technology. In brief, 156 ng of resting and 174 ng of activated NK cell total RNA were hybridized with adapter B mix (an RNA/DNA oligonucleotide duplex possessing single-stranded, overhanging degenerate bases) for 10 min at 65°C and cooled to 16°C. Following hybridization between the overhanging degenerate bases and the RNA molecules, the RNA oligonucleotides of the duplex were ligated to the RNA 5′- and 3′-ends for 3 h at 16°C. A 2× reverse transcription (RT) mix containing ArrayScript RT was added to the adapter B–ligated total RNA reactions, and incubated for 30 min at 42°C to synthesize first-strand cDNA. Prior to PCR amplification, 10 μL of the first-strand cDNA/RNA hybrid was treated with kit-supplied RNase H for 30 min at 37°C. In the PCR, 0.5 μL of RNase H–treated cDNA was amplified using suggested inputs of AmpliTaq DNA polymerase, 5′-SOLiD PCR primer, 5′-CCACTACGCCTCCGCTTTCCTCTCTATGGGCAGTCGGTGAT, and the 3′-SOLiD PCR primer, 5′- CTGCCCCGGGTTCCTCATTCTCTAAGCCCCTGCTGTACGGCCAA, and 1× PCR buffer 1 containing 200 nM dNTPs. Cycling conditions were as follows: denaturation for 5 min at 95°C, followed PCR cycling of 30 sec at 95°C, annealing for 30 sec at 62°C, and extension for 30 sec at 72°C for 15 cycles, and a final extension for 7 min at 72°C. Aliquots of 5 μL of resting NK and activated NK cDNAs were collected at cycles 12 and 15, resolved on a 6% DNA Retardation Gel (Invitrogen) with 1 μg of TrackIt 10-bp and 25-bp DNA ladders (Invitrogen), and stained with SYBR-Green stain (Invitrogen). PCR-amplified cDNA products resulted in multiple bands, and we then performed four additional PCR reactions for each sample, 12 cycles per sample, and purified cDNA fragments between 104 and 120 bp, corresponding to cloned small RNAs between 15 nt and 31 nt in length. cDNA fragments were purified by excising the desired gel range and soaking in 400 μL or 400 mM NaCl overnight, adding 2.5 volumes of 100% ethanol and 15 μg of GlycoBlue (Ambion) to eluent, incubating for 4 h at −20°C, and precipitating by centrifugation at 12,000g for 30 min at 4°C. Samples were washed with 70% ethanol, air-dried, and suspended in 20 μL of QIAGEN EB buffer (10 mM Tris-HCl at pH 8.0).
In the processing of our miRNA libraries, the ligation event with adapter mix B results in directional sequencing each miRNA in the 3′-to-5′ direction and in adding SOLiD sequences that enable emulsion PCR (emPCR) and annealing to P1 sequence-containing paramagnetic beads. We used 0.4 pg/μL template in the emPCR, based on the SOLiD version 2 protocol that requires 1120 pg of each cDNA. Post emPCR enrichment recovered 141.6 and 152.8 million resting and activated NK library beads, respectively. Using a slide layout containing quadrants, we deposited 30 million beads in quadrants 1 and 2 and sequenced by ligation per the SOLiD protocol for 25 cycles. Post run metrics identified 20.4 and 28.7 million resting and activated NK reads, respectively. At the time of sequencing, the AB miRNA bioinformatics pipeline was not available. Thus, we used SHRiMP as our color-space alignment tool, which allowed direct comparison to Illumina GA using the same aligner (see analysis methods, below).
Known miRNA identity and enumeration
Sequencing read counts and analysis filters are shown in Table 1 and Supplemental Table S4. Raw Illumina sequencing reads reflect the total unfiltered reads obtained from the Illumina GA. Reads initially were filtered for (1) high quality (Illumina “chastity” filter), (2) the presence of the 3′ adaptor sequence (to ensure a small RNA was ligated and sequenced completely), and (3) size of small RNA reads (12–27 nt). Low complexity reads (>50% homopolymer) were removed. These filtering steps resulted in high-quality, filtered reads representing small RNA sequences of 12–27 nt. These Illumina reads were then “compressed” into unique sequences, and the tallied number of reads corresponding to an individual small sequence (read count) was retained as metadata. Unique sequences were then aligned to the annotated hairpin (miRNA precursor) sequences of miRBase v13 with the following SHRiMP parameters/commands: (rmapper-ls: -s 11111111 -n 1 -w 32 -o 10 -r 27 -h 50%) (probcalc: -s 2) and rmapper-ls -s 11111111 -n 1 -w 32 -o 10 -r 27 -h 50% -R <query file> <subject file> alignments.rmapper, probcalc -p 2 67868 <alignments.rmapper> alignments.probcalc.
For SOLiD small RNA analysis, alignments were performed in a similar fashion as the Illumina-based sequences. However, the issue of di-base (color-space) read alignment unique to the SOLiD platform was dealt with using the SHRiMP aligner's native support (rmapper-cs component) for color-space reads. SOLiD SHRiMP parameters/commands used were: (rmapper-cs: -s 11111111 -n 1 -w 32 -o 10 -r 27 -h 50%) (probcalc: -s 2), and rmapper-cs -s 11111111 -n 1 -w 32 -o 10 -r 27 -h 50% -R <query CSFASTA file> <subject FASTA file> alignments.rmapper, probcalc -p 2 48180 <alignments.rmapper> alignments.probcalc.
A large number of reads generated from small RNA libraries for ABI SOLiD sequencing failed to align to miRNA precursors, consistent with other SOLiD 2.0 miRNA-SEQ experiments (Ramsingh et al. 2009).
Alignment was followed by the post-alignment filter steps noted in Table 1:
Remove sequences with >10 “top” hits/query.
Retain only alignments with a <P Chance> filter of 1 × 10−6 or better (to ensure high-probability alignments only were analyzed).
Remove any alignments with indels.
Remove alignments with greater then one internal mismatch.
Remove all alignments with less than the top <P Chance> for a given query.
Remove alignments that did not start on nucleotide position 1 or 2 of the query.
Remove all alignments with identified sequencing errors (SOLiD/color-space only).
The filtered Illumina sequence reads were analyzed for the length of alignments and overall read length (Supplemental Fig. S2), which verified that mature miRNA-sized RNAs were present and aligned to the miRNA precursor reference. The number of unique sequences that aligned with high confidence to known miRNA is shown, and is decompressed to reflect the total number of sequences corresponding to miRNAs. For read count abundance analysis, any sequence read that passed all filters and mapped with equal probability to two different hairpins (e.g., mmu-mir-16-1 and mmu-mir-16-2) were randomly assigned to one hairpin before summation in order to eliminate redundancy.
IsomiR analysis
An in-house isomiR analysis algorithm was developed to represent the sequence complexity present in reads that aligned to known (or novel) mature 5p/3p or minor star sequences (isomiRview). This application performs an aggregated sequence alignment view localized to known hairpin (or novel miRNA precursor hairpin) reference sequence. Input data required for this process include raw prealignment nucleotide reads, alignment report information, and corresponding reference coordinate and offset values for mature/mature* (or 5p/3p) targets (based on miRBase annotation) or putative novel candidates (identified cluster “hotspot” regions). The subsequent isomiR view report presents read alignments in the context of target mature/mature* candidate sequences, displaying variant sequence forms from the most to least abundant in terms of digital expression. The report also provides detailed information about the localized genome region of interest, as well as a breakdown of alignment contribution in terms of activated and resting reads. The complete isomiRview is presented as a Supplemental material data set and does not represent total read counts since sequencing reads that passed all filters and mapped with equal probability to two hairpins (e.g., mmu-mir-16-1 and mmu-mir-16-2) were redundantly mapped to both hairpins.
Novel miRNA discovery
High-quality NK cell reads from both resting and activated libraries were combined for novel miRNA discovery analysis. First, reads were aligned to the reference mouse genome (M37) using SHRiMP. The parameters/commands used for Illumina and SOLiD sequence data were identical to those defined above for known precursor alignments, except that the <P Chance> filter parameter was modified (1.0 × 10−2) to account for the larger genome search space. Next, clusters of sequence alignments were identified by integrating individual sequence read alignment data using RefCov, which provides a topologic representation of alignments at each nucleotide of a cluster and statistics to represent the depth of maximal alignment (zenith depth)—84,884 clusters of alignment were identified (with a cluster defined as a noncontiguous region of overlapping read alignment), 4886 had a read membership greater than 10 that were processed via RefCov for alignment maps, and the top 1483 clusters by zenith depth were analyzed (range of read alignment depth 51–512,250). Candidate clusters were analyzed for miRNA precursor gene structure by analyzing the predicted secondary structure for a miRNA precursor hairpin. In concert, the location of read alignments was mapped to the putative miRNA precursor in an in-house developed microRNA validation tool. This tool simultaneously provides cluster information (cluster genome location, sequence, zenith depth, annotation of genome location) in a table format, MFold-based secondary structure predictions (Markham and Zuker 2008), and RefCov alignment coverage maps (Supplemental Fig. S5). In addition, the tool allows for flexible adjustment of the candidate precursor location to allow for manual changes to the cluster sequence genomic window based on the initial folding and read alignment results. The flow chart summarizing analysis of the 1483 clusters is shown in Supplemental Figure S4.
Criteria used to define a minimal miRNA-like hairpin/sequence alignment combination included (1) a predicted stem–loop hairpin structure with a −ΔG ≤ 15 with the hairpin structure having the lowest free energy folding if alternative structures were predicted; (2) mapping of the mature miRNA alignments to the stem of the predicted structure without extension through the loop or outside of the stem–loop fold; and (3) mature miRNA length of >18 nt and <27 nt, and lack of a “bulge” constituting >50% of the mature sequence within the stem of the hairpin. In addition, an miRNA was deemed highly probable if a 5p/3p mature configuration or mature/star alignment was identified; however, this was not a requirement. Once possible miRNA candidate precursor genes (and their mature miRNA sequences) were identified, the precursor sequences were added to our database of known miRNA precursors (miRBase v13), and all NK cell small RNA sequences were re-aligned to the new search space with SHRiMP and analysis parameters identical to that used to identify known miRNAs above. Finally, the SOLiD sequencing reads were aligned to the known/novel miRNA precursor hairpins using similar criteria for known miRNAs to determine whether the novel miRNAs were also detected with this second platform. The novel miRNA sequences/genes are summarized in Table 4 and Supplemental Table S3.
Microarray hybridization and analysis
NK cell RNA samples (n = 4 resting, n = 4 IL15-activated) were labeled for miRNA detection using Agilent's miRNA Microarray System version 1.5. Total RNAs (100 ng) were first dephosphorylated with Calf Intestine Alkaline Phosphatase (Takara Bio) and then denatured in 100% DMSO. Samples were labeled by ligation of one cyanine 3-pCP molecule to the 3′-end by T4 RNA Ligase (New England BioLabs). Labeled RNAs were purified with Micro Bio-Spin 6 columns (Bio-Rad) and hybridized to Agilent “pre-release” murine miRNA arrays (array design ID 018077), containing probes to all miRBase v10.0. Microarrays were scanned with an Agilent DNA Microarray Scanner. Images were gridded and analyzed using Agilent feature extraction software version 9.5.3.1. Data were normalized to the 75% median intensity of each array, and values are reported as a total gene signal of multiple probes for each miRNA.
Quantitative real-time PCR assays and analysis
Four-hundred-fifty nanograms of total RNA from each sample (n = 4 replicates of resting, and n = 4 replicates of IL15-activated NK cells) was copied into cDNA using Megaplex RT primers for Rodent Pool “A” and Multiscribe reverse transcriptase per the manufacturer's instructions (Applied Biosystems). qRT-PCR was performed with the corresponding TaqMan Rodent MicroRNA “A” Array Card, loaded according to the manufacturer's instructions. Array cards were run on an Applied Biosystems 7900HT System and analyzed with SDS software v.2.3 and RQ Manager 1.2. MammU6 signal was used as the endogenous control for calculating ΔCt (cycle threshold) values from the raw Ct values generated by the software. ΔCt values were averaged across either resting or activated samples for each miRNA. miRNAs with an average ΔCt (miRNA Ct − MammU6 Ct) of >15 for both resting and activated groups were designated as being below the limit of detection by qRT-PCR. For individual miRNA qRT-PCR assays (Applied Biosystems) to assess miR-223 and miR-21 overexpression following transfection, RNU48 was used as the calibrator.
miR-223 overexpression and psiCheck2 sensor plasmids
miR-223 was overexpressed using a modified pMND-GFP vector with the miRNA “mini-gene” cloned into AscI/SphI sites engineered into the 3′ UTR of GFP. An hsa-mir-223 mini-gene containing the pre-mir-223 hairpin sequence and ∼200 bp of flanking pri-mir-223 sequence was amplified (forward: CTTTACCTGCTTATCTTCAGGATCTCT; reverse: CGTACGCGCCCCCATCAGCACTCT, size 527 nt), and subcloned into the pMND-GFP 3′ UTR. The resultant pMND-GFP-miR-223 vector was transfected into 293T cells, and overexpression was confirmed using qRT-PCR (miR-223 miRNA assay, ABI), normalized to sno135. For a negative control, a pMND-GFP- miR-21 mini-gene was amplified (forward: TTTCCTTTAGGAGCATTATGAGC; reverse: TGGCACAAAAGACTCTAAGTGCCACC, size 375 nt), subcloned in a similar fashion into MND-GFP, and overexpression confirmed via qRT-PCR (ABI). Transient transfection of these vectors using DharmaFECT Duo (Dharmacon) results in robust expression of both GFP (50%–85% GFP+ at 48 h) and miR-223 or miR-21 in 293T cells. To provide a sensor plasmid readout of GzmB 3′-UTR control, the murine GzmB 3′ UTR was amplified by PCR (forward: CTACAGAAGCAACATGGATC; reverse: TTTGATTTTACATCATTTTTGTCCA, size 558 nt), TA cloned (Clonetech), and then subcloned in the psiCheck2 luciferase reporter construct (Promega). This reporter contains both a Renilla luciferase under the control of the subcloned 3′ UTR, and a control firefly luciferase for normalization. A similar cloning approach was used to generate psiCheck2 containing the murine Prf1 3′ UTR (forward: CATAATAACAACAATAACAGCCTG; reverse: GGGAGAAGGGATTTAAAGCTG, size 381 nt). The GzmB 3′ UTR with mutation of the predicted miR-223 seed sequence target site was engineered as shown in Figure 5 using the Quick Change site-directed mutagenesis kit (Stratagene). For luciferase assays, the indicated pMND-GFP-based vector and the indicated psiCheck2-3′-UTR vectors were cotransfected (DharmaFect Duo) into 293T cells, and luciferase activity was determined after 48 h using the Dual-Glo substrate system (Promega) on a standard microplate reader (Beckman Coulter LD400 luminometer). Data are presented as the ratio of experimental (Renilla) luciferase to control (firefly) luciferase.
Statistical comparisons
Read counts from different libraries were normalized and compared using a Fisher exact test with Bonferroni correction (Romualdi et al. 2003; Cummins et al. 2006). For other platforms, means from different conditions were compared using the Student's t-test. A significance threshold of P < 0.05 was used. Linear regressions and Spearman's correlation calculations were performed using Prism v5.00 software (GraphPad).
Acknowledgments
This work was supported by the National Institutes of Health grants K08HL093299 (T.A.F.) and R01DK49786 (T.J.L.). T.A.F. is also supported by the Edward Mallinckrodt Jr. Foundation, the Howard Hughes Medical Institute Physician-Scientist Early Career Award, and the American Society of Hematology Scholar Award. We thank Bill Eades and the Siteman Cancer Center High Speed Cell Sorting Core, the Siteman Cancer Center Bioinformatics Core (Center for Biomedical Informatics), Sanjay Jain for use of the Applied Biosystems 7900HT instrument, and Matt Hickenbotham for SOLiD sequence data generation. We also thank Emily Hodges and Gregory Hannon for technical assistance with the Illumina small RNA isolation and cDNA library creation. We thank R. Wilson and M. Cooper for insightful discussions.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession nos. SRR036363, SRR036364, SRR036206, and SRR036210. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE21003. All novel microRNA sequences have been submitted to miRBase (http://www.mirbase.org).]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.107995.110.
Note added in proof
While this study was under review, Kuchen et al. (2010) published mature miRNA sequencing of high-dose IL2-activated Rag1−/− splenocytes using Illumina GA.
References
- Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD 2008. MicroRNAs: New regulators of immune cell development and function. Nat Immunol 9: 839–845 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K, Sumazin P, Morozov P, Schneider C, Maute RL, Kitagawa Y, Mandelbaum J, Haddad J Jr, Chen CZ, Califano A, et al. 2009. Identification of the human mature B cell miRNome. Immunity 30: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, Meyers BC, Green PJ, Markis M, Isaacs G, Huang E, et al. 2008. Deep sequencing of chicken microRNAs. BMC Genomics 9: 185 doi: 10.1186/1471-2164-9-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caligiuri MA 2008. Human natural killer cells. Blood 112: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. 2005. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev 19: 1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri S 2009. Lesser known relatives of miRNA. Biochem Biophys Res Commun 388: 177–180 [DOI] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G 2009. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. 2006. The colorectal microRNAome. Proc Natl Acad Sci 103: 3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hata A 2009. Regulation of microRNA biogenesis: A miRiad of mechanisms. Cell Commun Signal 7: 18 doi: 10.1186/1478-811X-7-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Santo JP 2006. Natural killer cell developmental pathways: A question of balance. Annu Rev Immunol 24: 257–286 [DOI] [PubMed] [Google Scholar]
- Di Santo JP 2008a. Functionally distinct NK-cell subsets: Developmental origins and biological implications. Eur J Immunol 38: 2948–2951 [DOI] [PubMed] [Google Scholar]
- Di Santo JP 2008b. Natural killer cells: Diversity in search of a niche. Nat Immunol 9: 473–475 [DOI] [PubMed] [Google Scholar]
- Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ 2008. Conifers have a unique small RNA silencing signature. RNA 14: 1508–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhardt HA, Tsang HH, Dai DC, Liu Y, Bostan B, Fahlman RP 2009. Meta-analysis of small RNA-sequencing errors reveals ubiquitous post-transcriptional RNA modifications. Nucleic Acids Res 37: 2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G 2008. A human snoRNA with microRNA-like functions. Mol Cell 32: 519–528 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. 2007. High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219 doi: 10.1371/journal.pone.0000219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ 2007. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 26: 798–811 [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg M, Sturm M, Langenberger D, Falcon-Perez JM, Aransay AM 2009. miRanalyzer: A microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res 37: W68–W76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. 2005. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A 2009. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G, Zheng Y, Sumathipala N, Jiang H, Arese E, Soulages JL, Zhang W, Sunkar R 2010. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics 11: 52 doi: 10.1186/1471-2164-11-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451: 1125–1129 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ 2007. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci 104: 19291–19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, et al. 2008. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132: 860–874 [DOI] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. 2010. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity 32: 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbauer F, Morin RD, Argiropoulos B, Petriv OI, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu AL, et al. 2008. In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res 18: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier LL 2008. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol 8: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. 2007. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129: 147–161 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsen SE, de Wit E, Janssens G, Heater S, Chapman L, Parkin RK, Fritz B, Wyman SK, de Bruijn E, Voest EE, et al. 2009. Limitations and possibilities of small RNA digital gene expression profiling. Nat Methods 6: 474–476 [DOI] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY 2008. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med 205: 1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS 2009. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood 113: 3754–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER 2008a. The impact of next-generation sequencing technology on genetics. Trends Genet 24: 133–141 [DOI] [PubMed] [Google Scholar]
- Mardis ER 2008b. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9: 387–402 [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M 2008. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol Biol 453: 3–31 [DOI] [PubMed] [Google Scholar]
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol 6: R71 doi: 10.1186/gb-2005-6-8-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ 2008a. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008b. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K 2005. Aberrant T cell differentiation in the absence of Dicer. J Exp Med 202: 261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K, McCormick K, Nakano M, Meyers BC 2010. Bioinformatics analysis of small RNAs in plants using next generation sequencing technologies. Methods Mol Biol 592: 89–106 [DOI] [PubMed] [Google Scholar]
- Politz JC, Hogan EM, Pederson T 2009. MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsingh G, Koboldt DC, Fehniger TA, Wylie T, Magrini V, Trissal M, Koul S, Chang LW, Nagarajan R, Ley TJ, et al. 2009. Comprehensive evaluation of microRNA genes and gene expression using next generation sequencing in a patient with acute myelogenous leukemia. Blood 114: A271.[Abstract] [Google Scholar]
- Rissland OS, Norbury CJ 2008. The Cid1 poly(U) polymerase. Biochim Biophys Acta 1779: 286–294 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA 2003. IDEG6: A web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics 12: 159–162 [DOI] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M 2009. SHRiMP: Accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386 doi: 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC 2008. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog 4: e1000224 doi: 10.1371/journal.ppat.1000224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ 2009. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol 5: e1000507 doi: 10.1371/journal.pcbi.1000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM 2003. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med 198: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan LP, Wang M, Robertus JL, Schakel RN, Gibcus JH, Diepstra A, Harms G, Peh SC, Reijmers RM, Pals ST, et al. 2009. miRNA profiling of B-cell subsets: Specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest 89: 708–716 [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. 2007. Regulation of the germinal center response by microRNA-155. Science 316: 604–608 [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N 2007. miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE 2: e1020 doi: 10.1371/journal.pone.0001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O'Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M 2009. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE 4: e5311 doi: 10.1371/journal.pone.0005311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K 2009. MicroRNA control in the immune system: Basic principles. Cell 136: 26–36 [DOI] [PubMed] [Google Scholar]
- Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131: 146–159 [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K 2008. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama WM, Kim S 2006. How do natural killer cells find self to achieve tolerance? Immunity 24: 249–257 [DOI] [PubMed] [Google Scholar]
- Yokoyama WM, Kim S, French AR 2004. The dynamic life of natural killer cells. Annu Rev Immunol 22: 405–429 [DOI] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, et al. 2009. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 113: 4586–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA 2008. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med 205: 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS 2009. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci 106: 10266–10271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C 2008. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res 18: 1456–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]