Abstract
The RING-finger protein Ro52/TRIM21 is known to be an autoantigen and is recognized by anti-Ro/SSA antibodies, which are commonly found in patients with Sjögren's syndrome and systemic lupus erythematosus. We recently showed that Ro52 is an E3 ubiquitin ligase and localizes to cytoplasmic bodies that are highly motile along the microtubule network. To expand our knowledge of Ro52, we searched partners co-operating with Ro52. We performed a yeast two-hybrid screening of a human brain cDNA library with Ro52 as bait. This screening identified several genes encoding Ro52-interacting proteins, including the apoptosis-related proteins, Daxx and FLASH. Further yeast two-hybrid assays revealed that Daxx binds to the B30.2 domain of Ro52 and that FLASH binds to coiled-coil domains of Ro52 through its death-effector domain-recruiting domain. These results suggest that Ro52, Daxx, and FLASH form heteromeric protein complexes. Indeed, this was supported by results of immunoprecipitation experiments in which we found that Daxx is co-immunoprecipitated with Ro52 in the presence of overexpressed FLASH. Importantly, our fluorescence microscopy revealed that, although Daxx is predominantly located in the nucleus, overexpression of both Ro52 and FLASH leads to relocation of Daxx into the cytoplasm. Thus, Ro52 seems to co-operate with FLASH to induce cytoplasmic localization of Daxx in cells.
Keywords: Ro52, TRIM21, Ubiquitin ligase, Daxx, FLASH, Apoptosis
Introduction
Ro52 (also known as TRIM21) possesses a RING-finger domain, a B-box domain, and two coiled-coil domains (see Fig. 1A) and belongs to the tripartite motif (TRIM) protein family (Reymond et al. 2001; Wada and Kamitani 2006a). Ro52 is expressed in most tissues and cells (Sibilia 1998). Clinically, this protein is well known as an autoantigen that is recognized by anti-Ro52 autoantibodies found in the sera of patients with several autoimmune diseases, especially Sjögren's syndrome and systemic lupus erythematosus (Sibilia 1998). Recently, Ro52 has been shown to localize to distinct structures called cytoplasmic bodies that are diffusely located in the cytoplasm (Reymond et al. 2001; Rhodes et al. 2002; Wada et al. 2006b; Yamauchi et al. 2008) and transported along the microtubule network (Tanaka et al. 2010).
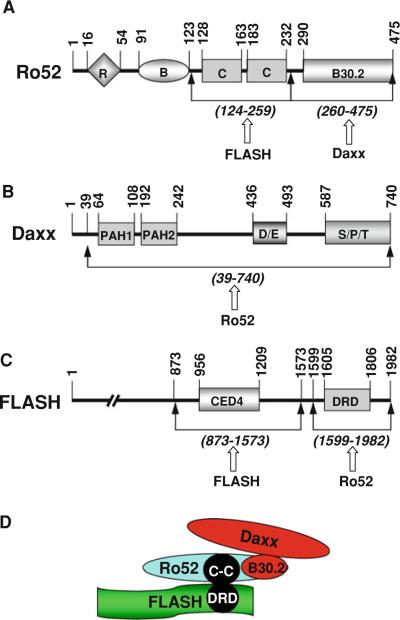
Fig. 1.
Domain structure and possible complex formation of human Ro52, Daxx, and FLASH. A Domain structure of human Ro52. Ro52 possesses a RING finger (R) and a B box (B) at the N-terminal region, two coiled coils (C) at the central region, and a B30.2 domain at the C terminus. B Domain structure of human Daxx. Daxx possesses two paired amphipathic helix (PAH) domains at the N-terminal region, an Asp/Glu-rich (D/E) domain at the central region, and a Ser/Pro/Thr-rich (S/P/T) domain at the C terminus. C Domain structure of human FLASH. FLASH possesses a cell-death-protein 4 (CED4) domain at the central region and a death-effector domain-recruiting domain (DRD) at the C-terminal region. The numbered bars indicate positions of amino acid residue. Mutual interacting regions of each protein are shown by arrows with the name of interacting partner. D Possible protein complex formed by Daxx, Ro52, and FLASH
Previously, we identified Ro52 as an E3 ubiquitin ligase (Wada and Kamitani 2006a). Ro52 has been shown to catalyze ubiquitination of several proteins, including Ro52 itself (Wada and Kamitani 2006a), Usp4 (also known as UnpEL or Unph) (Wada and Kamitani 2006b), IRF-3 (Yoshimi et al. 2009), IRF-8 (Kong et al. 2007), TRIM5α (Yamauchi et al. 2008), and IKKβ (Wada et al. 2009). By ubiquitinating these substrates, Ro52 plays a role in several biological events, including both innate and acquired immunity and NF-κB-dependent inflammatory signaling (Espinosa et al. 2009; Niida et al. 2010; Yoshimi et al. 2009). In addition, Ro52 plays a role in apoptosis because overexpressed Ro52 increases apoptotic cell death in a mouse B cell line (Espinosa et al. 2006). However, the detailed role of Ro52 in apoptosis has been unclear.
FLASH is a huge protein that interacts with the death-effector domain of caspase-8. This protein forms the death-inducing signaling complex (DISC) with the cytoplasmic portion of Fas and is involved in apoptosis induced by TNFα and Fas ligand (FasL) (Choi et al. 2001; Imai et al. 1999). Although FLASH was originally found as a component of a cytoplasmic complex located under the plasma membrane, it has been shown to translocate into the nucleus in response to certain stimuli (Kino and Chrousos 2003; Milovic-Holm et al. 2007).
Daxx is ubiquitously expressed throughout the body with particularly high expression in the thymus and testis (Yang et al. 1997). At the cellular level, Daxx is mainly a nuclear protein that associates with several different subnuclear structures, including the promyelocytic leukemia (PML) nuclear bodies (Salomoni and Khelifi 2006). Originally, however, Daxx was cloned as a Fas-interacting protein that modulates Fas-induced apoptotic cell death (Yang et al. 1997). Thus, Daxx localizes to the cytoplasm as well as the nucleus. In the cytoplasm, Daxx has been reported to interact with various other proteins involved in cell death regulation (Salomoni and Khelifi 2006).
Recently, we performed yeast two-hybrid screening using Ro52 as bait to elucidate the role of Ro52 in biological events. We found that Ro52 interacts with both FLASH and Daxx. In this study, we investigated the role of Ro52 and FLASH in the cytoplasmic relocation of Daxx.
Materials and methods
Cell lines and culture conditions
Human cell lines of embryonic kidney (HEK) 293 and lung fibrosarcoma HT1080 were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics.
Antibodies
Rabbit anti-FLAG antibody was purchased from Sigma Chemical Company (St. Louis, MO). Rabbit anti-HA antibody was purchased from Zymed (South San Francisco, CA). Mouse monoclonal anti-Ro52 (D-12) antibody, rabbit polyclonal anti-Daxx (M-112) antibody, and rabbit polyclonal anti-FLASH (M-300) antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Yeast two-hybrid assay for screening of the human cDNA library
The full-length cDNA of human Ro52 was subcloned into the Gal4 DNA-binding domain vector pGBKT7 (Clontech, Palo Alto, CA) and used as bait to screen a human fetal brain cDNA library in the pACT2 vector (Clontech, Cat. #638804). The yeast two-hybrid screening was performed in the Saccharomyces cerevisiae strain AH109 (Clontech) with a sequential transformation procedure using the lithium acetate method as described previously (Akey et al. 2002; Okura et al. 1996).
Yeast two-hybrid assay for the interaction of Ro52 with Daxx and FLASH
We performed yeast two-hybrid assays to assess the interaction of Ro52 with Daxx and FLASH. First, using a polymerase chain reaction (PCR) with appropriate primers, we prepared cDNAs of full-length and truncated Ro52, Daxx, and FLASH, as described previously (Tanaka et al. 2003; Tanji et al. 2006). The yeast Matchmaker Two-Hybrid System 3 (Clontech) was used to examine the in vivo interaction of these wild-type and truncated proteins. For the assays, the cDNA of Ro52 (full-length or truncated) was subcloned into pGBKT7 (a Gal4 DNA-binding domain vector for Gal4-BD fusion), and the cDNA of Daxx or FLASH (full-length or truncated) was subcloned into pGADT7 (a Gal4-activating domain vector for Gal4-AD fusion). The plasmids of the two fusion constructs were then co-transfected into AH109 yeast cells using the lithium acetate method (Okura et al. 1996). Transformed yeast cells were grown on a His−/Trp−/Leu− synthetic agar plate for 3 days at 30°C. The specific protein–protein interaction was determined by the growth of the yeast cells on the selection plate.
Plasmid construction and transfection
The cDNAs of human Ro52 (Wada and Kamitani 2006a) and Daxx were amplified using PCR with appropriate primers from human cDNA libraries (Invitrogen, Carlsbad, CA). The FLASH cDNA subcloned into a mammalian expression plasmid (pME18S/FLASH-EGFP) was kindly provided by Dr. Tomoshige Kino (National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD). To express proteins tagged with an epitope at the N terminus in human cells, the Daxx, Ro52, and FLASH cDNAs were inserted into the plasmid vectors pcDNA3/HA-N (Kamitani et al. 1997), pcDNA3/FLAG-N (Wada and Kamitani 2006b), and pcDNA3/RH-N (Kamitani et al. 1998), respectively. To express Ro52 fused with enhanced cyan fluorescent protein (ECFP) at the C terminus in human cells, Ro52 cDNA was inserted into pECFP-N1 (Clontech, Palo Alto, CA) and generated pRo52-ECFP. To express Daxx fused with monomeric red fluorescent protein (mRFP) at the N terminus in human cells, we first inserted the mRFP cDNA (a kind gift from Dr. Roger Tsien, University of California San Diego, La Jolla, CA) into pcDNA3 to generate pcDNA3/mRFP-N. Daxx cDNA was then inserted into pcDNA3/mRFP-N to generate pmRFP-Daxx. To express FLASH fused with enhanced green fluorescent protein (EGFP) at the C terminus in human cells, we used pME18S/FLASH-EGFP. These plasmids were transfected into HEK293 or HT1080 cells using FuGENE6 (Roche Applied Science, Indianapolis, IN) or Lipofectamine 2000 (Invitrogen).
Anti-FLAG immunoprecipitation
To immunoprecipitate FLAG-tagged Ro52, HEK293 cells were transfected to express FLAG-Ro52 and other proteins using Lipofectamine 2000 reagent. Twenty hours after transfection, cells were lysed for 30 min at room temperature in 1 ml of lysis buVer (50 mM Tris–HCl [pH 7.5], 100 mM NaCl, 10 mM NaF, 0.05% SDS) containing a protease inhibitor cocktail (Roche Applied Science). After centrifugation at 14,000 rpm for 15 min, the supernatant of the cell lysate was incubated with 40 μl of anti-FLAG M2-agarose beads (Sigma) for 2 h at room temperature. The FLAG M2-agarose beads were washed three times with 750 μl of lysis buffer and incubated for 30 min at 50°C in a sample-treating solution containing 3% SDS and 5% β-mercaptoethanol. After centrifugation, the solubilized proteins in the supernatant were analyzed by western blotting.
Western blotting
Protein samples were treated at 50°C for 1 h in a sample-treating solution containing 3% SDS and 5% β-mercaptoethanol. After SDS-PAGE, western blotting was performed using the protocol provided with the ECL detection system (Amersham Pharmacia Biotech, Piscataway, NJ). As a secondary antibody, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (Santa Cruz Biotechnology) was used.
Immunostaining of endogenous Ro52, Daxx, and FLASH in HT1080 cells
HT1080 cells were cultured on a coverslip in a 3.5-cm dish. After 24 h, the cells were Wxed in a 4% paraformaldehyde solution for 30 min, stained with 0.1 μg/ml of 4′,6-diami-dine-2′-phenylindole dihydrochloride (DAPI; Roche Diagnostics) for 10 min, and permeabilized with 0.1% Triton X-100 for 15 min at room temperature. The cells were first labeled with one of the following primary antibodies overnight at 10°C: mouse monoclonal anti-Ro52 (1:1,000), rabbit polyclonal anti-Daxx (1:1,000), or rabbit polyclonal anti-FLASH (1:1,000). After washing, the cells were labeled with Alexa Fluor 594-conjugated goat anti-mouse IgG or anti-rabbit IgG antibody (Molecular Probes, Eugene, OR) at a dilution of 1:2,000 for 1 h at room temperature. Finally, the cells were analyzed by an Axio Imager M1 fluorescence microscope (Zeiss). The localization of endogenous Ro52, Daxx, and FLASH was shown by the red fluorescence of Alexa Fluor 594.
Results
Interaction of Ro52 with Daxx and FLASH in yeast two-hybrid system
We have previously identiWed Ro52 as an E3 ubiquitin ligase. To investigate the molecular function of Ro52, we searched Ro52-interacting proteins by yeast two-hybrid screening using Ro52 as bait. Because the mRNA of Ro52 was shown to be highly enriched in the microvascular compartment of brain (Shusta et al. 2003), a human fetal brain cDNA library was used for the screening. Approximately 2 × 106 primary library transformants were inoculated onto selection plates. More than 50 colonies grew on the selection plates, 27 of which stained positive when tested for β-galactosidase expression. Subsequent DNA sequencing of the positive clones showed that four clones encoded Daxx (the shortest fragment consisted of amino acid residues 39–740) (Fig. 1B) and one clone encoded FLASH (amino acid residues 1302–1982) (Fig. 1C).
Identification of Ro52-binding site on FLASH
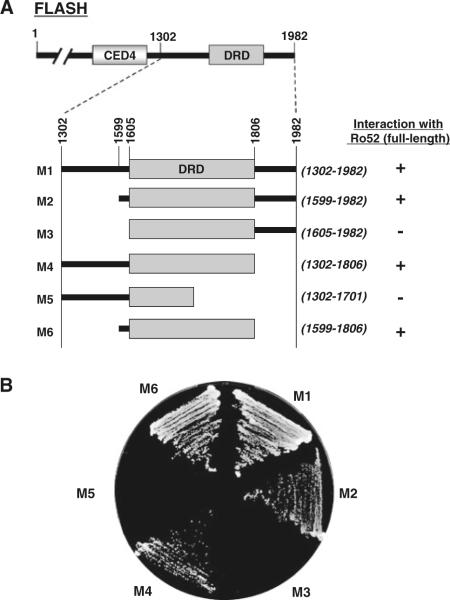
We precisely identiWed the Ro52-binding site on FLASH using deletion mutants of FLASH in a yeast two-hybrid interaction assay. As shown in Fig. 2A, we generated six mutants of FLASH, M1–M6, to examine the interaction with Ro52 (full length). Each mutant has a C-terminal deletion and/or an N-terminal deletion. For example, M1 has an N-terminal deletion from Met-1 to Thr-1301, resulting in the loss of a CED4 domain. M5 has an N-terminal deletion from Met-1 to Thr-1301 and a C-terminal deletion from Glu-1702 to Arg-1982, resulting in the loss of a CED4 domain and a C-terminal half of death-effector domain-recruiting domain (DRD). Using these mutants, we then examined the interaction with Ro52 in yeast cells. In the yeast two-hybrid assay, Ro52 fused to the Gal4 DNA-binding domain was used for the interaction with a panel of FLASH mutants fused to the Gal4-activating domain. As shown in Fig. 2B, Ro52 interacted with FLASH(1302–1982) (M1), FLASH(1599–1982) (M2), FLASH(1302–1806) (M4), and FLASH(1599–1806) (M6), but not with FLASH(1605–1982) (M3) and FLASH(1302–1701) (M5). These results indicate that the Ro52-binding site is located at the C-terminal region of FLASH between amino acid residues 1599 and 1806. Importantly, this region is a DRD domain (see Figs. 1C, D, 2A).
Fig. 2.
Mapping for Ro52-binding site on FLASH using a yeast two-hybrid system. A Summary of interaction between full-length Ro52 and truncated FLASH (M1–M6). B Primary data of interaction between full-length Ro52 and truncated FLASH (M1–M6). The yeast strain AH109 was transformed with pGBKT7/Ro52 and the pGADT7 construct encoding truncated FLASH (M1–M6). Transformed yeast cells were grown on a selection plate to determine the specific protein–protein interaction
IdentiWcation of FLASH-binding site on Ro52
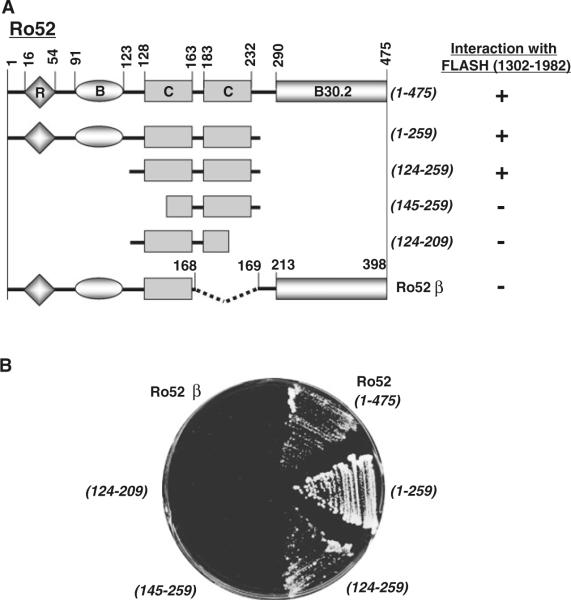
We next identiWed the FLASH-binding site on Ro52 using deletion mutants of Ro52 in a yeast two-hybrid interaction assay. As shown in Fig. 3A, we used wild-type Ro52 and four mutants, Ro52(1–259), Ro52(124–259), Ro52(145–259) and Ro52(124–209). In addition, we used an alternative splicing product Ro52β, which is identical to Ro52 but lacks 77 amino acids (amino acid residues 169–245) inclusive of the second coiled-coil domain with the leucine zipper (Wada et al. 2006a). In the yeast two-hybrid assay, FLASH fused to the Gal4-activating domain was used for the interaction with a panel of Ro52 mutants fused to the Gal4 DNA-binding domain. As shown in Fig. 3B, FLASH interacted with wild-type Ro52, Ro52(1–259), and Ro52(124–259), but not with Ro52(145–259), Ro52(124–209), and Ro52β. These results indicate that the FLASH-binding site is located at the coiled-coil domains of Ro52 between amino acid residues 124 and 259 (see Figs. 1A, D, 3A).
Fig. 3.
Mapping for FLASH-binding site on Ro52 using a yeast two-hybrid system. A Summary of interaction between C-terminal FLASH (1302–1982) and truncated Ro52. B Primary data of interaction between C-terminal FLASH and truncated Ro52. The yeast strain AH109 was transformed with pACT2/FLASH (1302–1982) and the pGBKT7 construct encoding wild-type Ro52 (1–475) and its deletion mutants (1–259, 124–259, 145–259, 124–209, and Ro52 β). Transformed yeast cells were grown on a selection plate to determine the specific protein–protein interaction
IdentiWcation of Daxx-binding site on Ro52
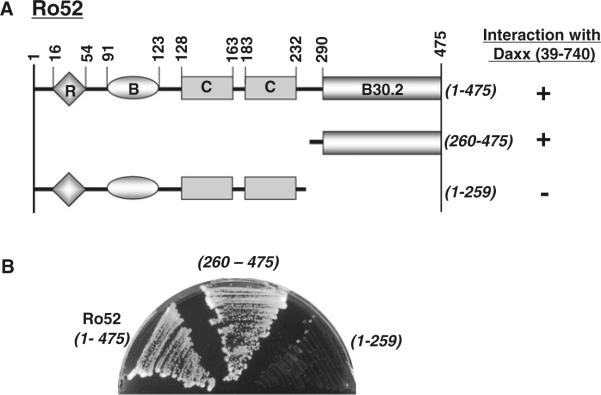
Using the yeast two-hybrid system, we identiWed the Daxx-binding site on Ro52 using deletion mutants of Ro52. As shown in Fig. 4A, we generated two deletion mutants of Ro52. Ro52(260–475) lacks the N-terminal region but possesses a B30.2 domain. Ro52(1–259) lacks the C-terminal B30.2 domain but possesses the N-terminal domains including the RING Wnger, B box, and coiled coils. Using wild-type Ro52 and these mutants, we examined the interaction with Daxx in yeast cells. In the yeast two-hybrid assay, Daxx fused to the Gal4-activating domain was used for the interaction with a panel of Ro52 (wild-type and mutants) fused to the Gal4 DNA-binding domain. As shown in Fig. 4B, Daxx interacted with wild-type Ro52 and Ro52(260–475) but not with Ro52(1–259). These results indicate that the Daxx-binding site is located at the C terminus of Ro52 between amino acid residues 260 and 475. Thus, Daxx binds to the B30.2 domain of Ro52 (see Figs. 1A, D, 4A).
Fig. 4.
Mapping for Daxx-binding site on Ro52 using a yeast two-hybrid system. A Summary of interaction between Daxx (39–740) and truncated Ro52. B Primary data of interaction between Daxx (39–740) and truncated Ro52. The yeast strain AH109 was transformed with pACT2/Daxx (39–740) and the pGBKT7 construct encoding wild-type Ro52 (1–475) and its truncated mutants (260–475 and 1–250). Transformed yeast cells were grown on a selection plate to determine the specific protein–protein interaction
Interaction between Ro52 and Daxx in HEK293 cells
Ro52 interacted with Daxx in yeast cells. Using immunoprecipitation, we examined this interaction in human cells. In brief, FLAG-tagged Ro52 was co-expressed with HA-tagged Daxx and/or RH-tagged FLASH in HEK293 cells by plasmid transfection. FLAG-Ro52 was then immunoprecipitated by anti-FLAG antibody, followed by western blotting using anti-HA antibody to detect the co-immunoprecipitated HA-Daxx. As shown in Fig. 5, HA-Daxx was not co-precipitated with FLAG-Ro52 in the absence of RH-FLASH (lane 6, upper panel). In contrast, HA-Daxx was co-precipitated with FLAG-Ro52 in the presence of RH-FLASH (lane 7, upper panel). This result suggests that FLASH is required for the interaction between Ro52 and Daxx in HEK293 cells.
Fig. 5.
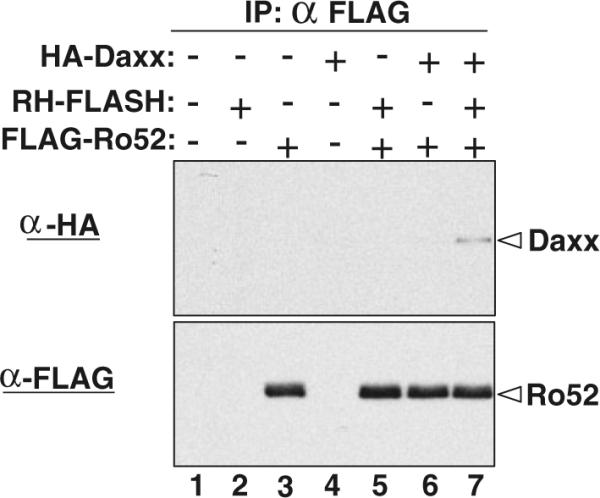
Co-immunoprecipitation of Daxx with Ro52 in HEK293 cells. FLAG-tagged Ro52 was co-expressed with HA-tagged Daxx and RH-tagged FLASH in HEK293 cells as indicated. FLAG-Ro52 in the cell lysates was immunoprecipitated by agarose beads conjugated with mouse anti-FLAG antibody. Co-immunoprecipitated HA-Daxx was detected by western blotting using rabbit anti-HA antibody (upper panel). Immunoprecipitated FLAG-Ro52 was also detected by western blotting using rabbit anti-FLAG antibody (lower panel)
It should be noted that Ro52 has been shown to bind to the Fc portion of IgG (Rhodes and Trowsdale 2007), which is a potential problem in immunoprecipitation experiments. In our experiments, we immunoprecipitated FLAG-Ro52 with anti-FLAG-immobilized beads. Since anti-FLAG M2 antibody is mouse IgG1, the anti-FLAG-immobilized beads also bind to endogenous Ro52 as well as FLAG-Ro52. However, this is not a problem for our immunoprecipitation, because Ro52-interacting proteins can be co-precipitated with both FLAG-Ro52 and endogenous Ro52.
Subcellular localization of Ro52, Daxx, and FLASH in HT1080 cells
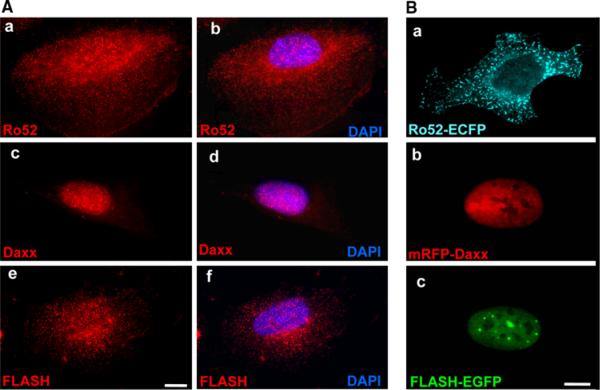
Since immunoprecipitation revealed that Ro52 interacts with Daxx in the presence of FLASH, we hypothesized that these proteins colocalize when co-expressed in human cells. To test this, we Wrst determined subcellular localization of each protein. SpeciWcally, endogenous Ro52, Daxx, and FLASH were immunostained in Xat extended HT1080 cells. As shown in Fig. 6A, endogenous Ro52 localized to cytoplasmic bodies in the cells (Fig. 6A-a, b), which was previously reported by us and several groups (Reymond et al. 2001; Rhodes et al. 2002; Tanaka et al. 2010; Wada et al. 2006a, b; Yamauchi et al. 2008). Endogenous Daxx was detected mainly in the nucleus and weakly in the cytoplasm as small dots (Fig. 6A-c, d), which was consistent with previous reports (Salomoni and Khelifi 2006; Yang et al. 1997). Endogenous FLASH was predominantly located in the cytoplasm, although a small fraction of immunoXuorescence was also detected in the nucleus as reported previously (Kino and Chrousos 2003) (Fig. 6A-e, f).
Fig. 6.
Subcellular localization of Ro52, Daxx, and FLASH in HT1080 cells. A Subcellular localization of endogenous Ro52, Daxx, and FLASH. HT1080 cells were fixed in a 4% paraformaldehyde solution, permeabilized with 0.1% Triton X-100, and immunostained with mouse monoclonal anti-Ro52 antibody (D-12), rabbit polyclonal anti-Daxx antibody (M-112), and rabbit polyclonal anti-FLASH antibody (M-300). After washing, cells were labeled with Alexa Fluor 594-conjugated goat anti-mouse IgG or anti-rabbit IgG antibody. The cells were then analyzed by fluorescence microscopy. The localization of endogenous Ro52, Daxx, and FLASH is shown by the red fluorescence of Alexa Fluor 594. Nuclear counterstaining is shown by the blue fluorescence of DAPI (b, d and f). A scale bar indicates 10 μm. B Subcellular localization of Xuorescent protein-fused Ro52, Daxx, and FLASH. HT1080 cells were transfected to express ECFP-fused Ro52, mRFP-fused Daxx, or EGFP-fused FLASH. Cells were fixed in a 4% paraformaldehyde solution. After washing, the cells were then analyzed by fluorescence microscopy. The localization of Ro52-ECFP is shown by the cyan fluorescence of ECFP. The localization of mRFP-Daxx is shown by the red fluorescence of mRFP. The localization of FLASH-EGFP is shown by the green fluorescence of EGFP. A scale bar indicates 10 μm
Next, we determined subcellular localization of three proteins fused with Xuorescent proteins, such as ECFP, mRFP, and EGFP. We expressed ECFP-fused Ro52, mRFP-fused Daxx, and EGFP-fused FLASH in HT1080 cells, followed by Xuorescence microscopy. As shown in Fig. 6B, Ro52-ECFP was localized to cytoplasmic bodies (Fig. 6B-a). However, the cytoplasmic bodies of Ro52-ECFP were larger than those of endogenous Ro52. The number of cytoplasmic bodies of Ro52-ECFP was less than that of endogenous Ro52 (Fig. 6B-a vs. A-b). Interestingly, the cytoplasmic bodies of Ro52-ECFP showed a rod-like shape, while the shape of the cytoplasmic bodies of endogenous Ro52 was round or irregular as described previously (Tanaka et al. 2010; Wada et al. 2006a, b). mRFP-Daxx was exclusively localized to the nucleus (Fig. 6B-b). FLASH-EGFP was also localized to the nucleus. Specifically, it was detected in the nucleoplasm and nuclear bodies (Milovic-Holm et al. 2007) (Fig. 6B-c). Since endogenous FLASH was predominantly located in the cytoplasm (Fig. 6A-e, f), this result showed the deference between endogenous FLASH and exogenously expressed FLASH-EGFP. The discrepancy of FLASH cytoplasmic versus nuclear localization in the two methods employed may indicate that FLASH shuttles between the cytoplasm and the nucleus as discussed previously (Kino and Chrousos 2003).
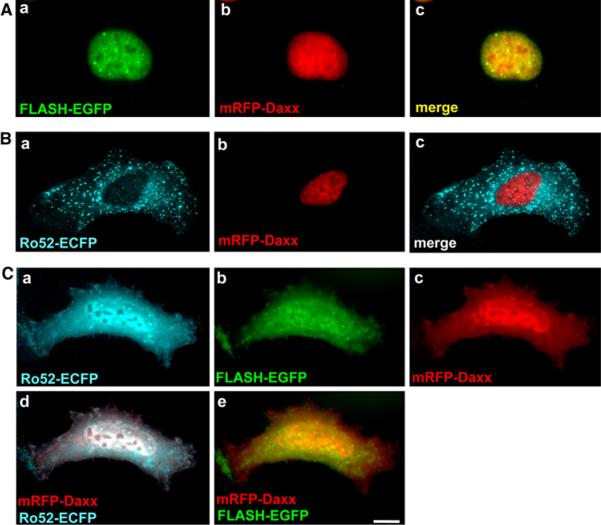
Relocation of mRFP-Daxx in HT1080 cells overexpressing Ro52 and FLASH
To investigate the effect of overexpression of FLASH and Ro52 on subcellular localization of Daxx, mRFP-Daxx was co-expressed with FLASH-EGFP and/or Ro52-ECFP in HT1080 cells. As shown in Fig. 7, mRFP-Daxx localized to the nucleus when co-expressed with FLASH-EGFP (Fig. 7A-b) or Ro52-ECFP (Fig. 7B-b). This nuclear localization was the same as that observed when mRFP-Daxx alone was expressed (see Fig. 6B-b). These results suggest that co-expression with FLASH or Ro52 has no effect on subcellular localization of mRFP-Daxx. However, when mRFP-Daxx was co-expressed with both FLASH-EGFP and Ro52-ECFP in HT1080 cells, mRFP-Daxx strongly localized to the cytoplasm as well as the nucleus (Fig. 7C-c). Importantly, Ro52-positive nuclear bodies almost disappeared and Ro52-ECFP and FLASH-EGFP showed the same localization as that of mRFP-Daxx (Fig. 7C-a to c). The merged images revealed that mRFP-Daxx colocalizes to the cytoplasm and nucleus with Ro52-ECFP and FLASH-EGFP (Fig. 7C-d, e). These results suggest that interactions of three molecules lead to dramatic changes in subcellular location of each molecule.
Fig. 7.
Subcellular localization of mRFP-Daxx in HT1080 cells expressing Ro52-ECFP and/or FLASH-EGFP. HT1080 cells were transfected to express: A FLASH-EGFP and mRFP-Daxx, B Ro52-ECFP and mRFP-Daxx, C Ro52-ECFP, FLASH-EGFP, and mRFPDaxx. The cells were fixed in a 4% paraformaldehyde solution and analyzed by fluorescence microscopy. The localization of Ro52-ECFP is shown by the cyan fluorescence of ECFP. The localization of FLASH-EGFP is shown by the green fluorescence of EGFP. The localization of mRFP-Daxx is shown by the red fluorescence of mRFP. The merged images are shown in A-c, B-c, C-d, and e as indicated. A scale bar indicates 10 μm
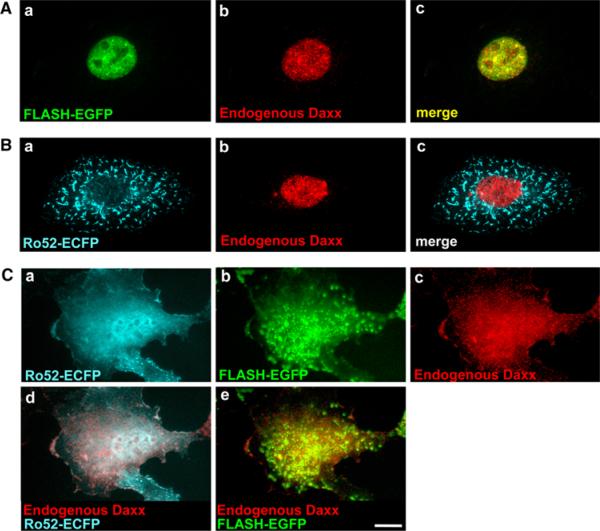
Relocation of endogenous Daxx in HT1080 cells overexpressing Ro52 and FLASH
Next, we immunostained endogenous Daxx to investigate the effect of overexpression of FLASH and Ro52 on subcellular localization of endogenous Daxx. HT1080 cells were transfected for expression of FLASH-EGFP and/or Ro52-ECFP and immunostained with anti-Daxx antibody. As shown in Fig. 8, endogenous Daxx was predominantly localized to the nucleus, when FLASH-EGFP (Fig. 8A-b) or Ro52-ECFP (Fig. 8B-b) was expressed. The nuclear localization was the same as that of endogenous Daxx without transfection (see Fig. 6A-c). These results suggest that expression with FLASH or Ro52 alone has no effect on subcellular localization of endogenous Daxx. However, when both Ro52-ECFP and FLASH-EGFP were expressed in HT1080 cells, endogenous Daxx strongly localized to the cytoplasm as well as the nucleus (Fig. 8C-c). Thus, exogenously co-expressed Ro52 and FLASH caused cytoplasmic relocation of endogenous Daxx (Fig. 8C) as well as mRFP-Daxx (Fig. 7C). Importantly, Ro52-positive nuclear bodies almost disappeared and Ro52-ECFP and FLASH-EGFP showed the same localization as that of endogenous Daxx (Fig. 8C-a, b). The merged images revealed that endogenous Daxx colocalizes to the cytoplasm and nucleus with Ro52-ECFP and FLASH-EGFP (Fig. 8C-d, e). Thus, as described above, the dramatic changes in subcellular location of each molecule appear to be triggered by interactions of three molecules.
Fig. 8.
Subcellular localization of endogenous Daxx in HT1080 cells expressing Ro52-ECFP and/or FLASH-EGFP. HT1080 cells were transfected to express: A FLASH-EGFP, B Ro52-ECFP, C Ro52-ECFP and FLASH-EGFP. The cells were fixed in a 4% paraformaldehyde solution, permeabilized with 0.1% Triton X-100, and immunostained with rabbit polyclonal anti-Daxx antibody (M-112). After washing, cells were labeled with Alexa Fluor 594-conjugated goat anti-rabbit IgG antibody. The cells were then analyzed by fluorescence microscopy. The localization of Ro52-ECFP is shown by the cyan fluorescence of ECFP. The localization of FLASH-EGFP is shown by the green fluorescence of EGFP. The localization of endogenous Daxx is shown by the red fluorescence of Alexa Fluor 594. The merged images are shown in A-c, B-c, C-d, e as indicated. A scale bar indicates 10 μm
Discussion
Ro52 is an E3 ubiquitin ligase that ubiquitinates substrates to modify their function or to change their subcellular location (Kong et al. 2007; Wada and Kamitani 2006a, b; Yamauchi et al. 2008; Yoshimi et al. 2009). For instance, Ro52 interacts with active IKKβ and conjugates a single ubiquitin molecule (monoubiquitin) for translocation of active IKKβ to autophagosomes, resulting in downregulation of NF-κB signaling (Niida et al. 2010; Wada et al. 2009). In this study, using yeast two-hybrid cDNA screening, we found that Ro52 interacts with Daxx and FLASH through its coiled-coil domains and B30.2 domain, respectively. Using immunoprecipitation, we confirmed the interaction between Ro52 and Daxx. Interestingly, Daxx was co-immunoprecipitated with Ro52 in the presence of, but not in the absence of, overexpressed FLASH, suggesting that the interaction between Ro52 and Daxx depends on FLASH. With regard to the interaction between Ro52 and FLASH, in spite of repeated immunoprecipitation experiments, we could not confirm it probably due to very low expression of FLASH (data not shown). Collectively, results of yeast two-hybrid and immunoprecipitation assays suggest that Daxx forms a protein complex with Ro52 in cells, depending on the expression of FLASH.
What is the biological relevance of the interaction of Ro52 with Daxx and FLASH? In general, Ro52 interacts with its substrates and then ubiquitinates them as an E3 ubiquitin ligase (Di Donato et al. 2001; Kong et al. 2007; Wada and Kamitani 2006b; Wada et al. 2009). Because of this enzymatic activity, we initially expected that Ro52 ubiquitinates Daxx and FLASH to regulate apoptosis. In other words, the apoptosis-related proteins, Daxx and FLASH, may be substrates of Ro52. Unexpectedly, however, our ubiquitination assays revealed that Ro52 does not ubiquitinate either of these proteins (data not shown). Ro52 may ubiquitinate unidentified protein(s) in the protein complex containing Daxx and FLASH.
Daxx mainly localizes to the nucleus, but this protein enhances death receptor-induced apoptosis in the cytoplasm. How does the nuclear protein function in the cytoplasm? Previous reports indicate that Daxx relocates from the nucleus to the cytoplasm upon specific stimuli such as crosslinking of Fas (Charette and Landry 2000) and overexpression of the apoptosis signal-regulating kinase 1 (Ask1) (Ko et al. 2001). This cytoplasmic relocation is essential for Daxx to induce apoptotic signaling, because Daxx needs to interact with apoptosis regulators (such as Ask1) in the cytoplasm (Yang et al. 1997). How does the relocation of Daxx occur and how is it regulated? In the studies presented here, we showed that wild-type Ro52 leads to this cytoplasmic relocation of Daxx in the presence of FLASH. Interestingly, however, the inactive mutant Ro52-C16A, which has no ligase activity (Wada and Kamitani 2006a), did not lead to the cytoplasmic relocation of Daxx even in the presence of FLASH (data not shown). These results indicate that although Daxx itself is not ubiquitinated by Ro52, the ligase activity of Ro52 is involved in the cytoplasmic relocation of Daxx. Based on our observations, we hypothesize that Ro52 ubiquitinates unidentified protein(s) in the protein complex containing Daxx and FLASH, leading to the cytoplasmic relocation of Daxx. Further studies are needed to elucidate the role of Ro52-mediated ubiquitination in the cytoplasmic relocation of Daxx and in the receptor-induced apoptosis.
Acknowledgments
We thank Drs. Roger Tsien and Tomoshige Kino for kindly providing mRFP cDNA and FLASH cDNA, respectively. This work was supported in part by National Institutes of Health Grant R01AG024497 (to T.K.).
Abbreviations
- TRIM
Tripartite motif
- HEK
Human embryonic kidney
- HRP
Horseradish peroxidase
- EGFP
Enhanced green fluorescent protein
- ECFP
Enhanced cyan fluorescent protein
- mRFP
Monomeric red fluorescent protein
- PCR
Polymerase chain reaction
- DAPI
4′,6-Diamidine-2′-phenylindole dihydrochloride
- CED4
Cell-death-protein 4
- DRD
Death-effector domain-recruiting domain
References
- Akey DT, Zhu X, Dyer M, Li A, Sorensen A, Blackshaw S, Fukuda-Kamitani T, Daiger SP, Craft CM, Kamitani T, Sohocki MM. The inherited blindness associated protein AIPL1 interacts with the cell cycle regulator protein NUB1. Hum Mol Genet. 2002;11:2723–2733. doi: 10.1093/hmg/11.22.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette SJ, Landry J. The interaction of HSP27 with Daxx identifies a potential regulatory role of HSP27 in Fas-induced apoptosis. Ann N Y Acad Sci. 2000;926:126–131. doi: 10.1111/j.1749-6632.2000.tb05606.x. [DOI] [PubMed] [Google Scholar]
- Choi YH, Kim KB, Kim HH, Hong GS, Kwon YK, Chung CW, Park YM, Shen ZJ, Kim BJ, Lee SY, Jung YK. FLASH coordinates NF-κB activity via TRAF2. J Biol Chem. 2001;276:25073–25077. doi: 10.1074/jbc.M102941200. [DOI] [PubMed] [Google Scholar]
- Di Donato F, Chan EK, Askanase AD, Miranda-Carus M, Buyon JP. Interaction between 52 kDa SSA/Ro and deubiquitinating enzyme UnpEL: a clue to function. Int J Biochem Cell Biol. 2001;33:924–934. doi: 10.1016/s1357-2725(01)00055-3. [DOI] [PubMed] [Google Scholar]
- Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, Horvath L, Wallerskog T, Oukka M, Nyberg F, Kuchroo VK, Wahren-Herlenius M. The Sjögren's syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- Espinosa A, Dardalhon V, Brauner S, Ambrosi A, Higgs R, Quintana FJ, Sjostrand M, Eloranta ML, Ni Gabhann J, Winqvist O, Sundelin B, Jefferies CA, Rozell B, Kuchroo VK, Wahren-Herlenius M. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inXammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J Exp Med. 2009;206:1661–1671. doi: 10.1084/jem.20090585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ETH. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like molecule. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ETH. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273:11349–11353. doi: 10.1074/jbc.273.18.11349. [DOI] [PubMed] [Google Scholar]
- Kino T, Chrousos GP. Tumor necrosis factor alpha receptor- and Fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J Biol Chem. 2003;278:3023–3029. doi: 10.1074/jbc.M209234200. [DOI] [PubMed] [Google Scholar]
- Ko YG, Kang YS, Park H, Seol W, Kim J, Kim T, Park HS, Choi EJ, Kim S. Apoptosis signal-regulating kinase 1 controls the proapoptotic function of death-associated protein (Daxx) in the cytoplasm. J Biol Chem. 2001;276:39103–39106. doi: 10.1074/jbc.M105928200. [DOI] [PubMed] [Google Scholar]
- Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, 3rd, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- Milovic-Holm K, Krieghoff E, Jensen K, Will H, Hofmann TG. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007;26:391–401. doi: 10.1038/sj.emboj.7601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida M, Tanaka M, Kamitani T. Downregulation of active IKKβ by Ro52-mediated autophagy. Mol Immunol. 2010;47 doi: 10.1016/j.molimm.2010.05.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei C-F, Chang HM, Yeh ETH. Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, Sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Rhodes DA, Ihrke G, Reinicke AT, Malcherek G, Towey M, Isenberg DA, Trowsdale J. The 52 000 MW Ro/SS-A autoantigen in Sjögren's syndrome/systemic lupus erythematosus (Ro52) is an interferon-γ inducible tripartite motif protein associated with membrane proximal structures. Immunology. 2002;106:246–256. doi: 10.1046/j.1365-2567.2002.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P, Khelifi AF. Daxx: death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Shusta EV, Li JY, Boado RJ, Pardridge WM. The Ro52/SS-A autoantigen has elevated expression at the brain microvasculature. Neuroreport. 2003;14:1861–1865. doi: 10.1097/00001756-200310060-00021. [DOI] [PubMed] [Google Scholar]
- Sibilia J. Ro(SS-A) and anti-Ro(SS-A): an update. Rev Rhum [Engl Ed] 1998;65:45–57. [PubMed] [Google Scholar]
- Tanaka T, Kawashima H, Yeh ETH, Kamitani T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J Biol Chem. 2003;278:32905–32913. doi: 10.1074/jbc.M212057200. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Tanji K, Niida M, Kamitani T. Dynamic movements of Ro52 cytoplasmic bodies along microtubles. Histochem Cell Biol. 2010;133:273–284. doi: 10.1007/s00418-009-0669-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanji K, Tanaka T, Mori F, Kito K, Takahashi H, Wakabayashi K, Kamitani T. NUB1 suppresses the formation of Lewy body-like inclusions by proteasomal degradation of synphilin-1. Am J Pathol. 2006;169:553–565. doi: 10.2353/ajpath.2006.051067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Kamitani T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem Biophys Res Commun. 2006a;339:415–421. doi: 10.1016/j.bbrc.2005.11.029. [DOI] [PubMed] [Google Scholar]
- Wada K, Kamitani T. UnpEL/Usp4 is ubiquitinated by Ro52 and deubiquitinated by itself. Biochem Biophys Res Commun. 2006b;342:253–258. doi: 10.1016/j.bbrc.2006.01.144. [DOI] [PubMed] [Google Scholar]
- Wada K, Tanji K, Kamitani T. Function and subcellular location of Ro52β. Biochem Biophys Res Commun. 2006a;340:872–878. doi: 10.1016/j.bbrc.2005.12.084. [DOI] [PubMed] [Google Scholar]
- Wada K, Tanji K, Kamitani T. Oncogenic protein UnpEL/Usp4 deubiquitinates Ro52 by its isopeptidase activity. Biochem Biophys Res Commun. 2006b;339:731–736. doi: 10.1016/j.bbrc.2005.11.076. [DOI] [PubMed] [Google Scholar]
- Wada K, Niida M, Tanaka M, Kamitani T. Ro52-mediated monoubiquitination of IKKβ downregulates NF-κB signaling. J Biochem. 2009;146:821–832. doi: 10.1093/jb/mvp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi K, Wada K, Tanji K, Tanaka M, Kamitani T. Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 2008;275:1540–1555. doi: 10.1111/j.1742-4658.2008.06313.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi R, Chang TH, Wang H, Atsumi T, Morse HC, 3rd, Ozato K. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J Immunol. 2009;182:7527–7538. doi: 10.4049/jimmunol.0804121. [DOI] [PMC free article] [PubMed] [Google Scholar]