Abstract
Pancreatobiliary tract strictures result either from malignancies of the biliary tract and pancreas or from nonmalignant etiopathogenesis. The goal of this study was to determine whether KRAS mutations could be identified in residual pancreatobiliary stricture brushings and to compare the performance characteristics of KRAS mutation analysis to cytology and fluorescence in situ hybridization (FISH) for the detection of carcinoma. Residual brushing cytology cell pellets were retrieved from 132 patients with subsequent clinicopathologic follow-up of cholangiocarcinoma (n = 41), pancreatic adenocarcinoma (n = 35), gallbladder cancer (n = 2), and nonmalignant strictures (n = 54). All specimens had a prior cytology and FISH UroVysion results as part of clinical practice. KRAS mutation analysis was performed using the quantitative PCR DxS KRAS Mutation Test Kit. KRAS mutation analysis was successful in 130 of 132 specimens. KRAS mutations and polysomic (ie, positive) FISH results were identified in 24 (69%) and 22 (63%) pancreatic adenocarcinoma specimens, respectively, with a combined sensitivity of 86% (30/35). KRAS mutations and polysomic FISH results were identified in 12 (29%) and 17 (41%) cholangiocarcinoma specimens, with a combined sensitivity of 54% (22/41). KRAS mutations were identified in two patients with primary sclerosing cholangitis, and benign follow-up. Residual cytology specimens can be used to detect KRAS mutations by quantitative PCR. Combined KRAS mutation and FISH analysis appear to increase the cancer detection rate in patients with pancreatobiliary strictures.
Pancreatobiliary tract strictures can result from nonmalignant etiopathogenesis or from pancreatobiliary tract malignancies such as cholangiocarcinoma and pancreatic adenocarcinoma. The diagnosis of malignancy in patients with pancreatobiliary tract strictures can be challenging, especially in patients with primary sclerosing cholangitis (PSC), and relies on clinical examination, biochemical testing (eg, CA 19–9), and endoscopic and imaging procedures such as endoscopic retrograde cholangiopancreatography (ERCP).1 ERCP is a valuable tool for assessing pancreatobiliary tract strictures because it allows for visualization of the biliary tract, therapeutic interventions (eg, placement of biliary stents), and collection of brushing cytology specimens for cytopathologic evaluation. Biliary brushing cytology, although highly specific, has suffered from low to moderate sensitivity (15%−68%).2,3,4,5,6 As a result, ancillary diagnostic techniques including mutation analysis,7,8,9,10,11 DNA ploidy analysis,12,13 methylation analysis,14,15,16,17 and fluorescence in situ hybridization (FISH),2,4,18,19 have been studied to improve tumor detection.
FISH, using the UroVysion probe set, is a molecular cytology/cytogenetic test currently performed as an adjunct to routine cytology. Two types of chromosomal abnormalities are frequently identified with the UroVysion FISH probe set, polysomy and trisomy 7. Polysomy has been defined as a gain of two or more of the four probes in ≥5 cells. Trisomy 7 has been defined as a gain of a single probe targeting the pericentromeric region of chromosome 7 (three signals for the CEP7 probe) with no more than two signals in each of the other three probes in ≥10 cells. A large clinical study recently demonstrated that FISH was more sensitive than cytology for detecting pancreatobiliary tract cancer.2 FISH was able to detect 49/227 (22%) cancers that routine cytology interpreted as nonmalignant without compromising test specificity. However, the overall sensitivity of polysomy FISH (43%) was only moderate. If a FISH-positive diagnosis included patients with a polysomy result or patients with a trisomy 7 result (without polysomy), FISH was able to detect 63% of all cancers. However, the inclusion of trisomy 7 as a criterion for a positive result significantly increases the false positive rate of FISH because only ∼50% of patients with trisomy 7 results have cancer on clinicopathologic follow-up.2 Consequently, although detection rates are improved by FISH testing, additional or substitute molecular markers are needed to further improve the detection of pancreatobiliary tract cancer.
The KRAS (Kirsten rat sarcoma viral oncogene homolog) gene is a member of the RAS family that encodes a G protein involved in signal transduction from cell surface receptors (eg, EGFR) to intracellular targets. Mutations in the KRAS gene (most commonly in codons 12 and 13 of exon 2) impair the GTPase activity of KRAS, which leads to constitutive activation of downstream signaling pathways that control cell proliferation, differentiation, and survival.17,20 Previous studies have shown that between 25%−100% of pancreatic adenocarcinomas harbor a KRAS mutation, depending on the type of specimen (eg, pancreatic tissue, pancreatic juice, etc) analyzed.8,9,10 KRAS alterations have also been identified in a wide range (21%−100%) of cholangiocarcinoma specimens.7 The most frequently observed KRAS mutations in both pancreatic adenocarcinoma and cholangiocarcinoma are mutations in codons 12 and 13, and less frequently, codon 61.7,21 There are no data comparing the performance characteristics of KRAS mutation analysis and aneusomy by FISH on clinical cytology specimens collected during ERCP. The goal of this study was to determine whether KRAS mutations could be identified in residual cytology pancreatobiliary tract specimens and to compare the performance characteristics of KRAS mutation analysis to cytology and FISH results for the detection of carcinoma.
Materials and Methods
Patient Specimens
Residual cytology cell pellets were retrieved from 132 patients (86 males, 46 females) who underwent ERCP evaluation of a pancreatobiliary tract stricture with subsequent clinicopathologic follow-up. The ages of patients included in this study ranged from 15 to 87 years, with a mean and median age of 59 and 60 years, respectively. Specimens were selected from a larger study2 by first subcategorizing patients based on the combination of their FISH and clinicopathology follow-up results. Specimens were included from each of the predetermined categorizations based on whether there was adequate (visible) residual cell pellet available for KRAS testing. This selection of patients was performed to assure that different FISH results were included from patients with nonmalignant (primary sclerosing cholangitis, n = 26; chronic pancreatitis, n = 15; autoimmune pancreatitis, n = 9; acute pancreatitis, n = 1; cholecystitis, n = 1; anastomotic biliary stricture, n = 1; and secondary sclerosing cholangitis secondary to choledochal cysts, n = 1) and malignant pancreatobiliary tract strictures (pancreatic adenocarcinoma, n = 35; cholangiocarcinoma, n = 41; and gallbladder cancer, n = 2). Specimens used for this study were collected for clinical testing between October, 2004 and March, 2009.
All specimens had a prior conventional cytology and FISH (UroVysion probe set, Abbott Molecular Inc., Des Plaines, IL) result as part of routine clinical practice (Figure 1). Slides processed for FISH analysis were assessed by scanning for cytologically atypical cells (eg, nuclear enlargement, irregular nuclear shape) or cells with atypical staining patterns (eg, patchy and/or lighter nuclear DAPI staining), and the number of CEP3, CEP7, CEP17, and 9p21 signals in those cells were determined.4 FISH results were categorized as disomic (negative for malignancy), trisomy (equivocal for malignancy), or polysomy (positive for malignancy; Figure 2). As part of routine clinical testing at our institution,4,12 polysomy is defined as a gain of two or more of the four probes in ≥5 cells. Trisomy 7 has been defined as a gain of a single probe targeting the pericentromeric region of chromosome 7 (three signals for the CEP7 probe) with no more than two signals in each of the other three probes in ≥10 cells. Loss of FISH signal patterns (eg, loss of FISH probe(s) targeting 9p21) were not considered positive as part of routine clinical testing.
Figure 1.
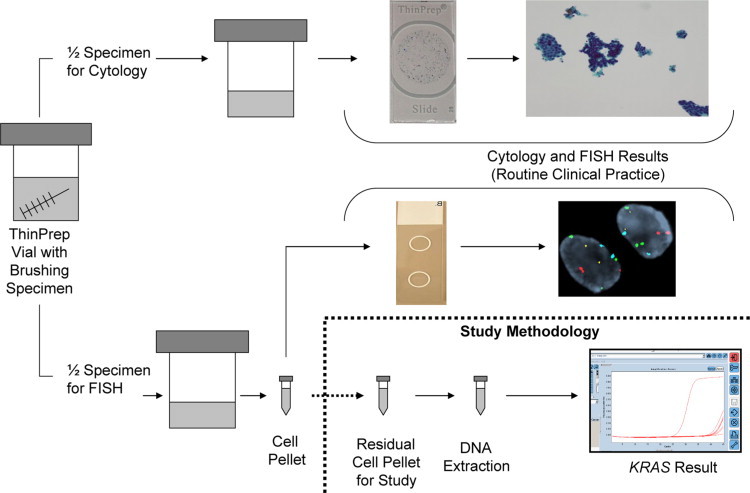
Specimen-processing flowchart, illustrating how pancreatobiliary samples were processed for routine clinical testing and KRAS mutation analysis.
Figure 2.
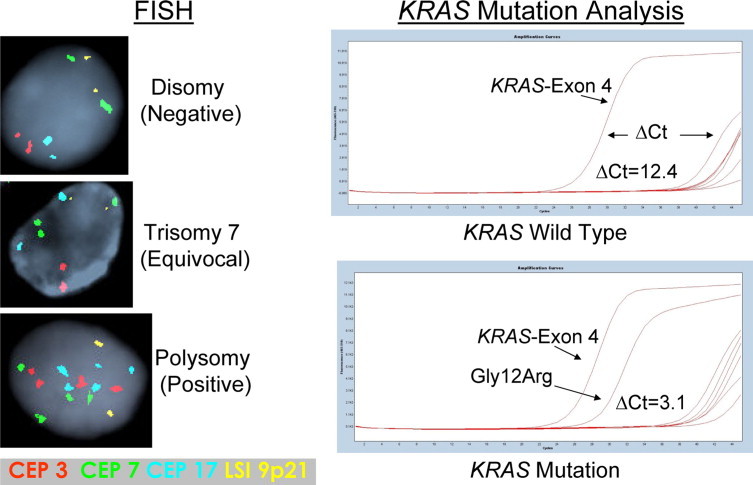
Representative examples of normal and abnormal FISH and KRAS testing results.
DNA Extraction
Residual cell pellets from clinical FISH testing (Figure 1) were retrieved from the −70°C freezer where they were stored in 3:1 methanol/glacial acetic acid. The pellets were centrifuged at 13,200 rpm for 5 minutes, followed by removal of the supernate, vortexing of the pellet, and addition of 1 ml of PBS. These steps were then repeated. This process was repeated a third time but 180 μl of ATL buffer (QIAGEN, Valencia, CA) and 20 μl of Proteinase K (QIAGEN) were added (instead of the 1 ml of PBS used in the prior 2 steps) and incubated at 55°C overnight with gentle agitation. The remainder of the DNA extraction procedure was performed using a QIAamp DNA Mini Kit (QIAGEN) as recommended by the manufacturer.
KRAS Testing
KRAS mutation analysis was performed using the quantitative PCR DxS KRAS Mutation Test Kit (DxS Ltd., Manchester, UK). The DxS KRAS Mutation Test Kit combines two technologies, ARMS and Scorpion to detect seven different mutations in codons 12 and 13 within exon 2 of the KRAS gene by real-time PCR. Allele-specific real-time quantitative PCR (qPCR) was carried out on a LightCycler 480 Real-Time PCR System (Roche Diagnostics, Indianapolis, IN). The DXS kit assesses the number of PCR cycles necessary to detect fluorescent signal above a background signal (exon 4 of the KRAS gene) to determine the cycle threshold (Ct) value. Control Ct values (exon 4 of the KRAS gene) between 20 and 35 were required to assure that there was an appropriate amount of DNA for analysis. The ΔCt value was calculated by subtracting the Ct value from each of the seven assessed mutations by the background or control Ct value for each individual sample. Specimens were considered to have a particular KRAS mutation when the ΔCt values were below the 1% ΔCt value (Figure 2) published by the manufacturer for that mutation. The KRAS exon 2 (codons 12 and 13) mutations and respective ΔCt thresholds included the following: G12A, ≤6.5; G12D, ≤8.0; G12R, ≤8.0; G12C, ≤7.0; G12S, ≤9.0; G12V, ≤6.5; and G13D, ≤9.0. Specimens with all 7 ΔCT values above the aforementioned thresholds were interpreted as wild-type. KRAS testing was performed without knowledge of patients’ previous testing results or clinicopathologic follow-up.
Statistics
Strictures were classified as benign or malignant based on surgical pathology findings when available or >6 months of clinical follow-up demonstrating either no progression (benign) or obvious neoplastic progression (mass lesion, metastasis, and/or death).2 Equivocal FISH results (trisomy 7) were considered negative for statistical analyses. All P values were calculated using a two-tailed Fisher's Exact test with JMP 8.0 statistical software (SAS Institute Inc., Cary, NC). P values ≤ 0.05 were considered statistically significant.
Results
KRAS qPCR testing was successful in 130 of the 132 (99%) specimens. The control Ct values in the 130 specimens ranged from 21 to 32 with a mean and median value of 25. Cell pellets were dark brown and full of noncellular debris in the two specimens where qPCR failed, suggesting that bilirubin, bile salts, or noncellular debris interfered with the PCR reaction. In the 130 specimens where qPCR was successful, there was an identifiable KRAS mutation in 39 (30%) specimens. KRAS G12D (n = 16) and G12V (n = 13) mutations were the most prevalent, followed by G12R (n = 6), G12C (n = 2), G12S (n = 1), and G13D (n = 1) mutations (Figure 3).
Figure 3.
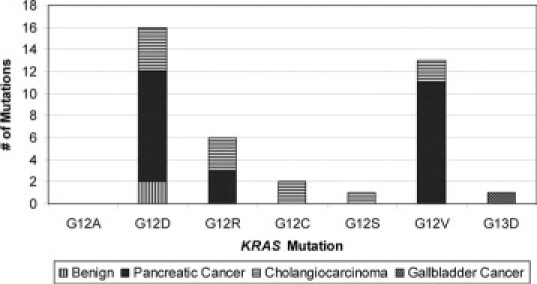
Frequency of specific KRAS mutations detected by qPCR.
The sensitivity of FISH, KRAS, and FISH + KRAS results combined are summarized in Table 1. Overall, FISH + KRAS analyses combined detected significantly (P < 0.001) more cancers (n = 53; 68%) than FISH alone (n = 39; 50%). Among patients with pancreatic adenocarcinoma, FISH + KRAS testing combined detected 86% of tumors which was significantly higher (P = 0.004) than FISH alone (63%). There were mixed results among patients with cholangiocarcinoma. In patients with non-PSC associated (ie, sporadic) cholangiocarcinoma, KRAS + FISH testing combined detected 56% of the cancers which was significantly higher (P < 0.001) than FISH alone which detected 37% of cancers. In PSC-associated cholangiocarcinoma, only four patients had an identifiable KRAS mutation and all four of these patients had a polysomic FISH result. Fifty-eight patients in this study had a disomy (ie, negative) FISH result. Within this group of patients, seven patients had an identifiable KRAS mutation and six of these patients had cancer. Thirty-nine patients had a polysomy FISH result with clinicopathologic evidence of cancer. A KRAS mutation was identified in 23 (59%) of these patients. Thirty-three patients had an equivocal FISH (trisomy 7). Nine of the 33 specimens (27%) had a KRAS mutation, and eight of nine (89%) were from patients with cancer. In contrast, nine of 24 patients (38%) with a trisomy 7 FISH and no KRAS mutation had malignancy.
Table 1.
Sensitivity of FISH and KRAS for Detecting Malignancy
| Cholangiocarcinoma (n = 41) |
|||||
|---|---|---|---|---|---|
| Test | Pancreatic adenocarcinoma (n = 35) | Sporadic | PSC-associated | Combined | All cancers* (n = 78) |
| KRAS | 24/35 (69%) | 8/27 (30%) | 4/14 (29%) | 12/41 (29%) | 37/78 (47%) |
| FISH† | 22/35 (63%) | 10/27 (37%) | 7/14 (50%) | 17/41 (41%) | 39/78 (50%)‡ |
| Combined | 30/35 (86%) | 15/27 (56%) | 7/14 (50%) | 22/41 (54%) | 53/78 (68%)‡ |
Includes two gallbladder cancers.
Only polysomic FISH results were considered positive in this study.
Significant difference (P < 0.001) between FISH and KRAS + FISH results.
The specificity of FISH and KRAS tests are summarized in Table 2. There were two patients who had false positive KRAS results (ie, a mutation was found in a patient without cancer). Interestingly, both of these patients had PSC and ulcerative colitis. There were 16 additional patients with PSC and UC and no identified KRAS mutations. There were no KRAS mutations identified in patients with chronic pancreatitis, autoimmune pancreatitis, acute pancreatitis, cholecystitis, or secondary sclerosing cholangitis.
Table 2.
Specificity of KRAS and FISH Testing
| Test | Fraction of patients without evidence of cancer and negative test result |
|---|---|
| KRAS | 50/52 (96%)* |
| FISH | 52/52 (100%) |
| Combined | 50/52 (96%) |
False-positive KRAS results were identified in two patients with diagnoses of both primary sclerosing cholangitis, and ulcerative colitis.
The cytology interpretation for the 130 patients included negative (n = 85), atypical (n = 11), suspicious (n = 14), and positive (n = 20). Among the 85 patients with a negative cytology result, a KRAS mutation was identified in 15 specimens and 14 (93%) had clinicopathologic evidence of cancer. Eighteen of the 85 (21%) had a positive result by either FISH and/or KRAS analysis and 17 (94%) had clinicopathologic evidence of cancer. Eleven of the 25 patients with equivocal cytology (atypical or suspicious) had a KRAS mutation, and 10 (91%) of these patients had cancer. Nine of the 10 patients had subsequent pathological evidence (resection, n = 7; biopsy, n = 1; and fine needle aspiration, n = 1) of cancer whereas the remaining patient had obvious progression of disease and died from metastatic pancreatic adenocarcinoma. Seventeen of the 25 (68%) equivocal cytology specimens had a FISH and/or KRAS positive result, and 16 (94%) of these patients represented patients with cancer. Twenty patients with malignancy had a positive cytology result, and a KRAS mutation was detected in 13 (65%) of these specimens.
Discussion
In this study we performed qPCR KRAS mutation testing on residual pancreatobiliary tract brushing specimens and found that KRAS mutation analysis, in combination with FISH, could detect significantly more cancers than FISH alone. Previous reports suggest that KRAS mutation analysis can be used to detect cancer in pancreatobiliary tract brushings,10,22 bile fluids,9,23 and fine needle aspirates from pancreatic masses.8,24 Comparisons of KRAS mutation analysis to conventional cytology have generally shown that KRAS mutation analysis increases sensitivity, but often at the expense of test specificity.7,9,10,24,25 FISH testing of brushing specimens has been shown to be a valuable adjunct to routine cytology, and a statistical model that incorporates both of these test results with clinical information (eg, PSC status, age) can be used to predict the risk of malignancy in an individual patient.2 The data from the present study suggests that KRAS mutation analysis combined with FISH testing could provide an additional increase in the diagnostic sensitivity over current testing algorithms. Our results suggest that KRAS testing may be most beneficial in patients with an equivocal FISH result or in patients suspected of having pancreatic adenocarcinoma or sporadic cholangiocarcinoma (Table 1). KRAS mutation analysis appears to be less sensitive for detecting PSC-associated cholangiocarcinoma.
In the present study, we identified KRAS mutations in 37 of the 78 (47%) ERCP brushing specimens from patients with cancer using the TheraScreen DxS quantitative real-time PCR assay. A wide variety of detection methods with varying analytical sensitivities (ie, limits of detection) have been used to assess tissue specimens for KRAS mutations. The most common methods that have been used include Sanger sequencing, pyrosequencing, qPCR, post-PCR fluorescent melting-curve analysis, single stranded conformation polymorphism analysis, and PCR clamping.26,27,28 A more sensitive technique called LigAmp has also been reported.25 It has been suggested that technologies such as LigAmp can detect a KRAS mutation in one of 10,000 cells, whereas the analytic sensitivities of qPCR and Sanger sequencing are approximately 1% and 20%, respectively.25,26,27
The analytical sensitivity required of an assay may vary according to what the test is being used for. To date, KRAS mutation analysis has primarily been used clinically to assess paraffin embedded colorectal and lung tumors for determining whether patients are candidates for anti-EGFR therapies such as Cetuximab or Tarceva.26,27 It is generally easy for a pathologist to select areas within the paraffin-embedded specimen for microdissection that have a tumor percentage that exceeds the limit of detection of the assay being used in the lab.26,27 In this study, we used KRAS analysis to try to detect tumor cells in ERCP brush cytology specimens. The tumor cells generally comprise <20% of the cells in these samples and frequently as little as 1% of all of the cells. Consequently, we needed to use an assay that had an analytical sensitivity approaching 1%.
According to the Sanger Catalog of Somatic Mutation in Cancer Database (http://www.sanger.ac.uk/genetics/CGP/cosmic/, last accessed on May 9, 2010), 58% (2784/4857 specimens) of pancreatic cancers and 31% (459/1463) of biliary tract cancers harbor a KRAS mutation. The most frequently observed KRAS mutations in both pancreatic adenocarcinoma and cholangiocarcinoma are mutations in codons 12 and 13 within exon 2, the mutations analyzed in this study. The data from our study are consistent with the Sanger database with KRAS mutations being detected in 69% and 29% of pancreatic and cholangiocarcinoma specimens, respectively.
The patients with cancer in this study who did not have a detectable KRAS mutation (n = 40) included 11 patients with pancreatic adenocarcinoma and 29 patients with cholangiocarcinoma. Possible explanations for false negative KRAS results include: i) low tumor percentage (ie, the fraction of tumor cells was lower than the analytical sensitivity of the assay); ii) the tumor cells within the specimen do not contain KRAS mutations; or iii) sample collection failure. The ability of KRAS testing, as well as FISH and cytology, to accurately detect cancer depends on the gastroenterologist's ability to brush malignant cells and place them into the vial for analysis. It is well known that the collection of biliary tract specimens is difficult and sampling failure occurs.2 To determine whether sampling failure was a reason for some of the false negative KRAS results in this study, we performed KRAS and FISH testing on paraffin embedded cancer from three patients (two patients with cholangiocarcinoma and one patient with pancreatic adenocarcinoma) who had negative biliary brushing FISH, KRAS, and cytology results. All three of these paraffin-embedded tumor specimens demonstrated polysomic cells by FISH and two specimens (one pancreatic and one cholangiocarcinoma) demonstrated a KRAS mutation. This suggests, at least in these three patients, that the malignant cells were either not present in the specimen collected during ERCP or present in quantities that were not detectable by either FISH or KRAS analysis.
Previous studies have shown that KRAS mutations are present in patients with PSC,23 autoimmune pancreatitis,21 and chronic pancreatitis.29 A current hypothesis suggests that increased Ras activity, either from elevated expression of mutant KRAS gene or by high levels of extrinsic Ras activators (eg, coexpression of TGF-α), may be a cause of inflammatory diseases such as chronic pancreatitis.29 High levels of Ras activity that generate the inflammatory response are postulated to accelerate the genetic changes that promote tumorigenesis.29,30 In the present study, there were 26 patients with PSC, 15 patients with chronic pancreatitis, and 9 patients with autoimmune pancreatitis without clinicopathologic evidence of cancer. The KRAS results from these specimens were negative except for two patients, both of whom had PSC and ulcerative colitis. One of these patients underwent a liver transplant for PSC one month after the biliary brushing diagnosis. It is not possible to determine whether this patient would have progressed to cholangiocarcinoma. The second patient is a 49-year-old male with two years of clinicopathologic follow-up without evidence of cancer. More extensive human studies are needed to better understand possible relationships between KRAS mutations, inflammatory diseases such as PSC, and pancreatobiliary tract cancer.
In conclusion, the results of this study suggest that KRAS mutation analysis can detect cancers not identifiable by FISH. KRAS mutations appear to be more common in pancreatic adenocarcinoma than cholangiocarcinomas. The identification of KRAS mutations in patients with PSC without cholangiocarcinoma appears to be infrequent, but does occur. Additional data are needed to determine the significance of these mutations. This study, like others, demonstrates that the addition of KRAS testing may be beneficial for laboratories performing cytologic testing of pancreatobiliary tract specimens. Further prospective studies are needed to define the algorithm that would be most beneficial to clinicians, clinical laboratories, and patients with pancreatobiliary tract strictures.
Footnotes
K.C.H. and the Mayo Clinic receive royalties from the sale of the FISH probe set (UroVysion, Abbott Molecular Inc., Des Plaines, IL) discussed in this article. K.C.H. also receives grant support from Abbott Molecular Inc.
References
- 1.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritcher EG, Kipp BR, Halling KC, Oberg TN, Bryant SC, Tarrell RF, Gores GJ, Levy MJ, Clayton AC, Sebo TJ, Roberts LR. A multivariable model using advanced cytologic methods for the evaluation of indeterminate pancreatobiliary strictures. Gastroenterology. 2009;136:2180–2186. doi: 10.1053/j.gastro.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 3.Govil H, Reddy V, Kluskens L, Treaba D, Massarani-Wafai R, Selvaggi S, Gattuso P. Brush cytology of the biliary tract: retrospective study of 278 cases with histopathologic correlation. Diagn Cytopathol. 2002;26:273–277. doi: 10.1002/dc.10098. [DOI] [PubMed] [Google Scholar]
- 4.Kipp BR, Stadheim LM, Halling SA, Pochron NL, Harmsen S, Nagorney DM, Sebo TJ, Therneau TM, Gores GJ, de Groen PC, Baron TH, Levy MJ, Halling KC, Roberts LR. A comparison of routine cytology and fluorescence in situ hybridization for the detection of malignant bile duct strictures. Am J Gastroenterol. 2004;99:1675–1681. doi: 10.1111/j.1572-0241.2004.30281.x. [DOI] [PubMed] [Google Scholar]
- 5.Logrono R, Kurtycz DF, Molina CP, Trivedi VA, Wong JY, Block KP. Analysis of false-negative diagnoses on endoscopic brush cytology of biliary and pancreatic duct strictures: the experience at 2 university hospitals. Arch Pathol Lab Med. 2000;124:387–392. doi: 10.5858/2000-124-0387-AOFNDO. [DOI] [PubMed] [Google Scholar]
- 6.Weber A, von Weyhern C, Fend F, Schneider J, Neu B, Meining A, Weidenbach H, Schmid RM, Prinz C. Endoscopic transpapillary brush cytology and forceps biopsy in patients with hilar cholangiocarcinoma. World J Gastroenterol. 2008;14:1097–1101. doi: 10.3748/wjg.14.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nehls O, Gregor M, Klump B. Serum and bile markers for cholangiocarcinoma. Semin Liver Dis. 2004;24:139–154. doi: 10.1055/s-2004-828891. [DOI] [PubMed] [Google Scholar]
- 8.Salek C, Benesova L, Zavoral M, Nosek V, Kasperova L, Ryska M, Strnad R, Traboulsi E, Minarik M. Evaluation of clinical relevance of examining K-ras, p16 and p53 mutations along with allelic losses at 9p and 18q in EUS-guided fine needle aspiration samples of patients with chronic pancreatitis and pancreatic cancer. World J Gastroenterol. 2007;13:3714–3720. doi: 10.3748/wjg.v13.i27.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi C, Fukushima N, Abe T, Bian Y, Hua L, Wendelburg BJ, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. Sensitive and quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chronic pancreatitis, potential for early detection. Cancer Biol Ther. 2008;7:353–360. doi: 10.4161/cbt.7.3.5362. [DOI] [PubMed] [Google Scholar]
- 10.Sturm PD, Rauws EA, Hruban RH, Caspers E, Ramsoekh TB, Huibregtse K, Noorduyn LA, Offerhaus GJ. Clinical value of K-ras codon 12 analysis and endobiliary brush cytology for the diagnosis of malignant extrahepatic bile duct stenosis. Clin Cancer Res. 1999;5:629–635. [PubMed] [Google Scholar]
- 11.Willmore-Payne C, Volmar KE, Huening MA, Holden JA, Layfield LJ. Molecular diagnostic testing as an adjunct to morphologic evaluation of pancreatic ductal system brushings: potential augmentation for diagnostic sensitivity. Diagn Cytopathol. 2007;35:218–224. doi: 10.1002/dc.20608. [DOI] [PubMed] [Google Scholar]
- 12.Baron TH, Harewood GC, Rumalla A, Pochron NL, Stadheim LM, Gores GJ, Therneau TM, De Groen PC, Sebo TJ, Salomao DR, Kipp BR. A prospective comparison of digital image analysis and routine cytology for the identification of malignancy in biliary tract strictures. Clin Gastroenterol Hepatol. 2004;2:214–219. doi: 10.1016/s1542-3565(04)00006-0. [DOI] [PubMed] [Google Scholar]
- 13.Krishnamurthy S, Katz RL, Shumate A, Strohlein K, Khanna A, Tucker SL, Raijman I, Lahoti S. DNA image analysis combined with routine cytology improves diagnostic sensitivity of common bile duct brushing. Cancer. 2001;93:229–235. doi: 10.1002/cncr.9034. [DOI] [PubMed] [Google Scholar]
- 14.Sandhu DS, Shire AM, Roberts LR. Epigenetic DNA hypermethylation in cholangiocarcinoma: potential roles in pathogenesis, diagnosis and identification of treatment targets. Liver Int. 2008;28:12–27. doi: 10.1111/j.1478-3231.2007.01624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhm KO, Lee ES, Lee YM, Kim HS, Park YN, Park SH. Aberrant promoter CpG islands methylation of tumor suppressor genes in cholangiocarcinoma. Oncol Res. 2008;17:151–157. doi: 10.3727/096504008785114110. [DOI] [PubMed] [Google Scholar]
- 16.Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005;18:412–420. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 17.Parsi MA, Li A, Li CP, Goggins M. DNA methylation alterations in endoscopic retrograde cholangiopancreatography brush samples of patients with suspected pancreaticobiliary disease. Clin Gastroenterol Hepatol. 2008;6:1270–1278. doi: 10.1016/j.cgh.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barr Fritcher EG, Kipp BR, Slezak JM, Moreno-Luna LE, Gores GJ, Levy MJ, Roberts LR, Halling KC, Sebo TJ. Correlating routine cytology, quantitative nuclear morphometry by digital image analysis, and genetic alterations by fluorescence in situ hybridization to assess the sensitivity of cytology for detecting pancreatobiliary tract malignancy. Am J Clin Pathol. 2007;128:272–279. doi: 10.1309/BC6DY755Q3T5W9EE. [DOI] [PubMed] [Google Scholar]
- 19.Moreno Luna LE, Kipp B, Halling KC, Sebo TJ, Kremers WK, Roberts LR, Barr Fritcher EG, Levy MJ, Gores GJ. Advanced cytologic techniques for the detection of malignant pancreatobiliary strictures. Gastroenterology. 2006;131:1064–1072. doi: 10.1053/j.gastro.2006.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 21.Kamisawa T, Tsuruta K, Okamoto A, Horiguchi S, Hayashi Y, Yun X, Yamaguchi T, Sasaki T. Frequent and significant K-ras mutation in the pancreas, the bile duct, and the gallbladder in autoimmune pancreatitis. Pancreas. 2009;38:890–895. doi: 10.1097/MPA.0b013e3181b65a1c. [DOI] [PubMed] [Google Scholar]
- 22.van Heek NT, Clayton SJ, Sturm PD, Walker J, Gouma DJ, Noorduyn LA, Offerhaus GJ, Fox JC. Comparison of the novel quantitative ARMS assay and an enriched PCR-ASO assay for K-ras mutations with conventional cytology on endobiliary brush cytology from 312 consecutive extrahepatic biliary stenoses. J Clin Pathol. 2005;58:1315–1320. doi: 10.1136/jcp.2004.022707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicka S, Kuhnel F, Flemming P, Hain B, Kezmic N, Rudolph KL, Manns M, Meier PN. K-ras mutations in the bile of patients with primary sclerosing cholangitis. Gut. 2001;48:403–408. doi: 10.1136/gut.48.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maluf-Filho F, Kumar A, Gerhardt R, Kubrusly M, Sakai P, Hondo F, Matuguma SE, Artifon E, Monteiro da Cunha JE, Cesar Machado MC, Ishioka S, Forero E. Kras mutation analysis of fine needle aspirate under EUS guidance facilitates risk stratification of patients with pancreatic mass. J Clin Gastroenterol. 2007;41:906–910. doi: 10.1097/MCG.0b013e31805905e9. [DOI] [PubMed] [Google Scholar]
- 25.Shi C, Chandrasekaran A, Thuluvath PJ, Karikari C, Argani P, Goggins M, Maitra A, Eshleman JR. Ultrasensitive detection of KRAS2 mutations in bile and serum from patients with biliary tract carcinoma using LigAmp technology. J Mol Diagn. 2009;11:583–589. doi: 10.2353/jmoldx.2009.090061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monzon FA, Ogino S, Hammond ME, Halling KC, Bloom KJ, Nikiforova MN. The role of KRAS mutation testing in the management of patients with metastatic colorectal cancer. Arch Pathol Lab Med. 2009;133:1600–1606. doi: 10.5858/133.10.1600. [DOI] [PubMed] [Google Scholar]
- 27.Plesec TP, Hunt JL. KRAS mutation testing in colorectal cancer. Adv Anat Pathol. 2009;16:196–203. doi: 10.1097/PAP.0b013e3181a9d4ed. [DOI] [PubMed] [Google Scholar]
- 28.Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, Ismail T, Li WQ, Collins P, Ravetto P, Leggett B, Salto-Tellez M, Soong R, Fox S, Scott RJ, Dobrovic A, Iacopetta B. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn. 2009;11:543–552. doi: 10.2353/jmoldx.2009.090057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji B, Tsou L, Wang H, Gaiser S, Chang DZ, Daniluk J, Bi Y, Grote T, Longnecker DS, Logsdon CD. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137:1072–1082. doi: 10.1053/j.gastro.2009.05.052. 1082 e1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohr M, Kloppel G, Maisonneuve P, Lowenfels AB, Luttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]


