Abstract
Microarray technologies provide high-resolution maps of chromosome imbalances and epigenomic aberrations in the cancer cell genome. Such assays are often sensitive to sample DNA integrity, voiding the utility of many archival pathology specimens and necessitating the special handling of prospective clinical specimens. We have identified the remarkable preservation of higher-molecular weight DNA in archival fine-needle aspiration cytopathology specimens from patients greater than 10 years of age. We further demonstrate the outstanding technical performance of 57 fine-needle aspiration cytopathology samples for aberration detection on high-resolution comparative genomic hybridization array, DNA methylation, and single nucleotide polymorphism genotyping platforms. Forty-four of 46 malignant aspirates in this study manifested unequivocal genomic aberrations. Importantly, matched Papanicolaou and Diff-Quik fine-needle aspiration cytopathology samples showed critical differences in DNA preservation and DNA integrity. Overall, this study identifies a largely untapped reserve of human pathology specimens for molecular profiling studies, with ramifications for the prospective collection of clinical biospecimens.
The two most common modalities for collecting patient tissue samples for pathological analysis can be roughly divided into surgical and cytologic. The former includes core needle and surgical biopsy, and these are typically processed as formalin-fixed, paraffin-embedded (FFPE) samples. FFPE surgical pathology specimens currently comprise the foundation for retrospective clinical genetic profiling studies, and also are typically required for prospective patient enrollment in clinical trials. Alternatively, fine-needle aspiration cytology (FNAC) procedures may be used for rapid, cost-effective and accurate diagnosis with reduced patient morbidity.1 Common anatomical sources for diagnostic FNAC specimens include breast, thyroid, lymph nodes, thoracic and visceral organs, and deep soft tissues. FNAC samples may be the only archival materials for certain diagnoses such as small cell lung cancer (SCLC). Body fluids and epithelial scrapes are another source of cytology preparation, including pleural effusion, ascitic fluid, cerebral-spinal fluid, cervical Pap-smears, and bronchial and esophageal brushings. Overall, cytologic preparations represent an estimated 10 to 20% of archival hospital pathology specimens, and may be significantly enriched for particular pathological diagnoses.
We recently reported the excellent performance of archival FFPE samples for large-scale molecular profiling using the GoldenGate methylation assay, a modest-density bisulfite genotyping platform.2 However, the more-informative, higher-resolution genotyping arrays for tissue molecular profiling require DNA of higher molecular weight than is often retrieved from archival FFPE samples, because of requirements for unbiased whole genome amplification, which is compromised in FFPE samples over time. Thus, array-based genome profiling studies may be data-limited when analyzing archived FFPE samples more than a few years old.
Despite their presence in hospital archives alongside FFPE samples, needle aspirate and fluid samples have been largely unutilized for molecular profiling. Nagel et al3 reported the potential value of CGH to support a primary cytologic diagnosis of malignancy in the clinical setting, yet little information is available regarding practical details and prospects for genomic scale molecular profiling of routinely processed archival and prospective cytology specimens. Mattu et al4 reported successful PCR on archival cytology samples up to 5 years old, yet did not assess the DNA yields and integrities from various kinds of cytologic preparations, nor did they explore the feasibility of microarray analysis at a genomic scale. In the current work, we have characterized the suitability of 57 FNAC specimens archived >10 years for high-resolution genotyping, methylation, and/or CGH arrays. Herein we present our method for DNA extraction and observations about the surprising and impressive utility of FNAC samples for molecular profiling; we further discuss the implications of our work for retrospective pathogenomic studies and prospective biospecimen collection.
Materials and Methods
Pathology Specimens
For this study, cases were selected to include FNAC specimens over 10 years of age (range, 11–16 years; n = 57 specimens from 36 distinct patient cases, Table 1), initially collected because of clinical suspicion of SCLC, cancer metastatic to lymph node, and/or primary lymphoma. Aspirates had been performed during a several-year period (1994 to 1997) by numerous physicians including surgeons and interventional radiologists. Aspirate smears and Pap and DQ stains had been prepared by technicians in a single laboratory according to standard procedure, and coverslipped with glass. Following cytologic interpretation, samples had been stored at room temperature under typical pathology archive conditions. On selection for inclusion in this study, patient samples were de-identified, and the cytology slides scanned on an Aperio digital slide scanner (Aperio Technologies, Inc., Vista, CA) to preserve the cytomorphology. Pathology review of slides was performed to confirm the presence of diagnostic cells on smears.
Table 1.
Patient and Pathology Specimen Characteristics
| Case_ID (n=36) | Sample_ID (n=57) | Sample age (yrs*) (avg=13) | Pt. age | Pt. sex | Anatomic site | Type | Stain | FNAC diagnosis | DNA yield (μg) (avg=7.4) | Cell yield (×105) (avg=12) | aCGH label | DLR score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LNFNA_11 | LNFNA_11_DQ1 | 16 | 75 | F | Abdomen | FNA | DQ | Cannot rule out lymphoma‡ | 5.0 | 8 | ULS | 0.04 |
| LNFNA_11 | LNFNA_11_P7 | 16 | 75 | F | Abdomen | FNA | P | Cannot rule out lymphoma‡ | 3.9 | 6 | ULS | 0.08 |
| LNFNA_12 | LNFNA_12_DQ1 | 15 | 58 | M | Low back soft tissue | FNA | DQ | Atypical, suspicious for lymphoma | 0.4 | 1 | — | — |
| LNFNA_12 | LNFNA_12_DQ2 | 13 | 58 | M | Low back soft tissue | FNA | DQ | Atypical, suspicious for lymphoma | 5.9 | 10 | ULS | 0.03 |
| LNFNA_12 | LNFNA_12_P3 | 15 | 58 | M | Low back soft tissue | FNA | DQ | Atypical, suspicious for lymphoma | 1.5 | 2 | — | — |
| LNFNA_16 | LNFNA_16_1_DQ3 | 13 | 73 | F | Thyroid | FNA | DQ | Thyroiditis | 3.1 | 5 | ULS | 0.05 |
| LNFNA_17 | LNFNA_17_DQ2 | 15 | 73 | M | Lung | FNA | DQ | Most likely reactive lymphocytes | 1.2 | 2 | ULS | 0.05 |
| LNFNA_17 | LNFNA_17_DQ4 | 13 | 73 | M | Lung | FNA | DQ | Most likely reactive lymphocytes | 4.7 | 8 | ULS | 0.04 |
| LNFNA_17 | LNFNA_17_P4 | 15 | 73 | M | Lung | FNA | P | Most likely reactive lymphocytes | 13.1 | 21 | ULS | 0.06 |
| LNFNA_19 | LNFNA_19_DQ1 | 15 | 76 | F | Thorax | FLUID† | DQ | Benign | 0.1 | 0.2 | — | — |
| LNFNA_19 | LNFNA_19_P1 | 15 | 76 | F | Thorax | FLUID† | P | Benign | 0.1 | 0.2 | — | — |
| LNFNA_2 | LNFNA_2_DQ | 13 | 72 | M | Low back soft tissue | FNA | DQ | Large cell lymphoma | 8.8 | 14 | — | — |
| LNFNA_21 | LNFNA_21_DQ2 | 13 | 73 | M | Retroperitoneum | FNA | DQ | Most consistent with lymphoma‡ | 5.3 | 9 | ULS | 0.05 |
| LNFNA_21 | LNFNA_21_DQ3 | 15 | 73 | M | Retroperitoneum | FNA | DQ | Most consistent with lymphoma‡ | 4.9 | 8 | ULS | 0.04 |
| LNFNA_21 | LNFNA_21_P4 | 15 | 73 | M | Retroperitoneum | FNA | P | Most consistent with lymphoma‡ | 7.6 | 12 | ULS | 0.20 |
| LNFNA_29 | LNFNA_29_2_DQ3 | 12 | 49 | M | Spleen | FNA | DQ | Diffuse large B-cell lymphoma | 3.6 | 6 | ULS | 0.04 |
| LNFNA_3 | LNFNA_3_1_DQ2 | 13 | 59 | M | Thyroid | FNA | DQ | Diffuse large B-cell lymphoma | 1.7 | 3 | ULS | 0.04 |
| LNFNA_32 | LNFNA_32_DQ1 | 13 | 66 | F | Thorax | FLUID† | DQ | B-cell lymphoma NOS | 2.6 | 4 | — | — |
| LNFNA_32 | LNFNA_32_P2 | 13 | 66 | F | Thorax | FLUID† | P | B-cell lymphoma NOS | 0.2 | 0.3 | — | — |
| LNFNA_33 | LNFNA_33_DQ2 | 11 | 64 | M | Abdomen | FNA | DQ | Follicular lymphoma, low grade | 23.3 | 38 | ULS | 0.04 |
| LNFNA_33 | LNFNA_33_DQ2_rep | 11 | 64 | M | Abdomen | FNA | DQ | Follicular lymphoma, low grade | 23.3 | 38 | ULS | 0.27 |
| LNFNA_36 | LNFNA_36_DQ1 | 11 | 66 | M | Pleural Fluid | FLUID† | DQ | Malignant lymphoproliferation | 8.9 | 14 | — | — |
| LNFNA_37 | LNFNA_37_2_DQ2 | 11 | 81 | M | Cervical lymph nodes | FNA | DQ | Many atypical small lymphocytes‡ | 4.8 | 8 | — | — |
| LNFNA_5 | LNFNA_5_DQ | 14 | 78 | M | Retroperitoneum | FNA | DQ | Follicular lymphoma, low grade | 6.3 | 10 | ULS | 0.04 |
| LNFNA_5 | LNFNA_5_DQ_rep | 14 | 78 | M | Retroperitoneum | FNA | DQ | Follicular lymphoma, low grade | 6.3 | 10 | ULS | 0.43 |
| LNFNA_5 | LNFNA_5_P | 14 | 78 | M | Retroperitoneum | FNA | P | Follicular lymphoma, low grade | 6.3 | 10 | ULS | 0.13 |
| LNFNA_52 | LNFNA_52_DQ2 | 12 | 24 | M | Neck mass | FNA | DQ | High-grade B-cell lymphoma | 20.2 | 33 | ULS | 0.04 |
| LNFNA_52 | LNFNA_52_P3 | 12 | 24 | M | Neck mass | FNA | P | High-grade B-cell lymphoma | 15.0 | 24 | ULS | 0.07 |
| LNFNA_59 | LNFNA_59_DQ1 | 12 | 86 | M | Axillary mass | FNA | DQ | High-grade B-cell lymphoma | 2.7 | 4 | ULS | 0.06 |
| LNFNA_59 | LNFNA_59_P1 | 12 | 86 | M | Axillary mass | FNA | P | High-grade B-cell lymphoma | 2.8 | 5 | ULS | 0.17 |
| LNFNA_65 | LNFNA_65_DQ1 | 16 | 84 | M | Submandibular LN | FNA | DQ | Metastatic neoplasm unknown primary | 7.1 | 12 | ULS | 0.22 |
| LNFNA_65 | LNFNA_65_P3 | 16 | 84 | M | Submandibular LN | FNA | P | Metastatic neoplasm unknown primary | 3.5 | 6 | ULS | 0.36 |
| LNFNA_71 | LNFNA_71_DQ1 | 15 | 50 | F | Thyroid | FNA | DQ | Hashimoto's thyroiditis | 9.4 | 15 | — | — |
| LNFNA_71 | LNFNA_71_P6 | 15 | 50 | F | Thyroid | FNA | P | Hashimoto's thyroiditis | 1.1 | 2 | — | — |
| LNFNA_72 | LNFNA_72_DQ1 | 15 | 74 | F | Inguinal LN | FNA | DQ | Large B-cell lymphoma | 8.2 | 13 | ULS | 0.05 |
| LNFNA_72 | LNFNA_72_P3 | 15 | 74 | F | Inguinal LN | FNA | P | Large B-cell lymphoma | 4.2 | 7 | ULS | 0.08 |
| LNFNA_9_2 | LNFNA_9_2_DQ2 | 16 | 67 | F | Thyroid | FNA | DQ | Anaplastic malignant tumor | 1.5 | 2 | ULS | 0.05 |
| LNFNA_9_2 | LNFNA_9_2_P2 | 16 | 67 | F | Thyroid | FNA | P | Anaplastic malignant tumor | 2.0 | 3 | ULS | 0.15 |
| SCLC_10 | SCLC_10_DQ | 12 | 84 | F | Lung | FNA | DQ | Small cell carcinoma | 21.7 | 35 | ENZ | 0.13 |
| SCLC_10 | SCLC_10_P3 | 12 | 84 | F | Lung | FNA | P | Small cell carcinoma | 9.2 | 15 | ULS | 0.40 |
| SCLC_11 | SCLC_11_DQ1 | 11 | 81 | M | Lung | FNA | DQ | Small cell carcinoma | 5.7 | 9 | ENZ | 0.16 |
| SCLC_18_2 | SCLC_18.2_P_T | 14 | 66 | M | Bronchus | BRUSH | P | Small cell carcinoma | 7.5 | 12 | ULS | 0.12 |
| SCLC_25_2 | SCLC_25_2_DQ1 | 11 | 63 | M | Liver | FNA | DQ | Small cell carcinoma | 4.3 | 7 | ENZ | 0.24 |
| SCLC_28_1 | SCLC_28_1_DQ3 | 11 | 68 | F | Liver | FNA | DQ | Small cell carcinoma | 16 | 26 | ULS | 0.06 |
| SCLC_3_2 | SCLC_3_2_DQ_ENZ | 14 | 65 | M | Mediastinum | FNA | DQ | Small cell carcinoma | 8.1 | 13 | ENZ | 0.24 |
| SCLC_3_2 | SCLC_3_2_DQ_ULS | 14 | 65 | M | Mediastinum | FNA | DQ | Small cell carcinoma | 8.1 | 13 | ULS | 0.07 |
| SCLC_49 | SCLC_49_DQ | 11 | 58 | M | Lung | FNA | DQ | Atypical versus reactive | 6.6 | 11 | ULS | 0.06 |
| SCLC_50 | SCLC_50_DQ2 | 11 | 78 | F | Lung | FNA | DQ | Small cell carcinoma | 27.9 | 45 | ULS | 0.18 |
| SCLC_51 | SCLC_51_DQ1 | 11 | 71 | M | Mediastinum | FNA | DQ | Small cell carcinoma | 3.7 | 6 | ENZ | 0.20 |
| SCLC_52 | SCLC_52_DQ2 | 12 | 82 | F | Mediastinum | FNA | DQ | Small cell carcinoma | 1.7 | 3 | ENZ | 0.21 |
| SCLC_53 | SCLC_53_DQ1 | 12 | 60 | M | Mediastinum | FNA | DQ | Lymphoepithelial-like carcinoma | 1.4 | 2 | ENZ | 0.30 |
| SCLC_55 | SCLC_55_DQ1 | 12 | 68 | F | Mediastinum | FNA | DQ | Small cell carcinoma | 2.7 | 4 | ENZ | 0.27 |
| SCLC_6 | SCLC_6_DQ_1 | 13 | 81 | F | Lung | FNA | DQ | Small cell carcinoma | 17.2 | 28 | ENZ | 0.11 |
| SCLC_6 | SCLC_6_P4 | 13 | 81 | F | Lung | FNA | P | Small cell carcinoma | 7.3 | 12 | ULS | 0.09 |
Notes:
LN, lymph node.
at time of extraction;
filter prep;
subsequent surgical excision diagnosed as lymphoma; — arrayCGH not performed.
DNA Extraction
Slides were de-coverslipped in xylene; in our experience, it can take up to 48 hours for the coverslips to loosen in xylene at room temperature. Xylene-substitute was less efficient at decoverslipping. De-coverslipped slides are soaked an additional 5 minutes in xylene to remove any residual coverslip mounting medium, and then air-dried. Fortunately, we have found that the cellular contents adhere well to the glass slide and not the coverslip during this process. Cellular material is collected by scraping the entire slide with a razor blade; this is facilitated by wetting the slide with a small amount (∼20 μl) of lysis buffer, which helps the material form a clump during scraping. A loose pipette tip is used to push the moist clump from the blade into a 2 ml screwcap tube. Lysis is performed in 100 μl of a 4:1 mixture of ATL and proteinase K (Qiagen, Valencia, CA) at 60°C for 2 to 16 hours. Lysis is usually complete in <2hours, but may be extended overnight for convenience. Lysates were RNase treated and the DNA was subsequently column purified according to the Qiagen DNeasy protocol, with an elution volume of 100 μl. DNA was evaluated for quantity and purity using a Nanodrop ND-1000 UV-Vis Spectrophotometer (Nanodrop Technologies, Wilmington, DE). Two hundred ng DNA was run on a 2% agarose gel alongside 100bp ladder (Invitrogen) and commercial HMW human DNA (Promega) to assess DNA molecular weight range.
Microarray Studies
Archival FNAC DNA extracts were assayed with Agilent 105K or 180K CGH (Agilent, Santa Clara, CA), Illumina Infinium 370K or 610K genotyping (Illumina, San Diego, CA), and/or Illumina Infinium methylation 27K (Illumina) arrays according to the manufacturer's directions. Array image data were analyzed using Nexus (BioDiscovery, Inc., El Segundo, CA), and/or Illumina Bead Studio. The measure of success of profiling for each sample was based on array data sample quality indices (derivative log ratio scores), and genomic structural aberration detection in aspirate smears with a diagnosis of malignancy.
Results
The average DNA yield per case in this study was 7.4 μg (range: 0.1 to 27.9 μg); DQ and Pap-smears from the same FNAC procedure yielded roughly equivalent amounts of DNA (Table 1). Because array CGH assays require 200 to 1000 ng input DNA, while the SNP genotyping arrays require 250 ng, there is ample DNA yield from each sample for numerous profiling assays. Having determined DNA yield for the cases, we reverse calculated cellularity based on a conversion factor of 6pg DNA/cell; this indicated an average recovery of >1 million cells per FNAC case (Table 1).
In contrast with the similar cellularity and total DNA yields from archival DQ versus Pap-stained FNAC samples, a remarkably different molecular weight distribution is observed (Figure 1A). Strikingly, the DQ samples manifest significantly higher molecular weight DNA than the Pap-stained cells from the same needle aspirate. Indeed, the pap-stained cellular DNA is nearly completely degraded to <400bp fragments, while the typical DQ DNA migrates as a smear with over half in the mid-to-high molecular weight range (Figure, 1B and 1C). This observation was validated in 10 matched pairs of DQ and Pap smears, among which every pair manifested superior DQ DNA integrity (Figure 1D).
Figure 1.
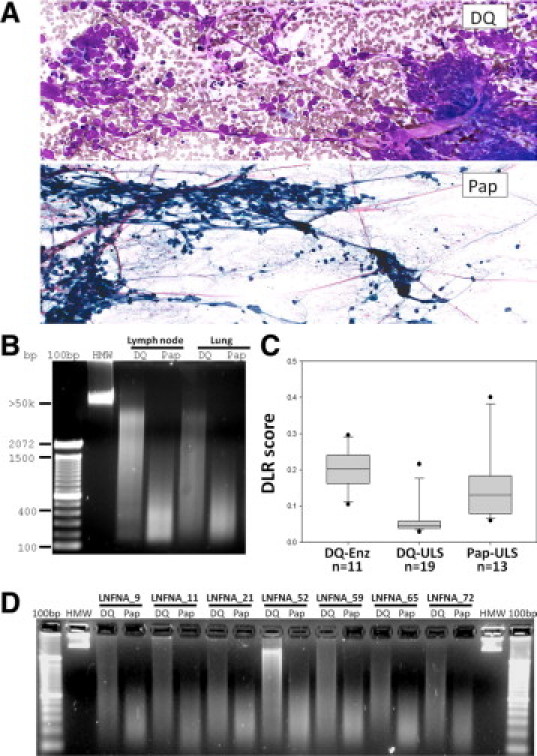
Diff-Quik (DQ) vs. Papanicolaou (Pap) fine-needle aspiration cytopathology (FNAC) preparations. A: Small-cell lung carcinoma smears made from the same thoracic needle aspirate and then processed as either DQ (top panel) or Pap smears (bottom panel). Both stains show cellular clusters of small cells with high nuclear/cytoplasmic ratio and characteristic chromatin stretched accross the slides. B: Striking difference in DNA integrity of iso-aspirate smears prepared as either DQ or Pap. Lane 1: 100-bp DNA ladder (Invitrogen). Lane 2: high molecular weight (HMW) commercial reference DNA. Lanes 3 and 4: DNAs recovered from a lymph node aspirate diagnosed as lymphoma. Lanes 5 and 6: DNAs from a lung aspirate diagnosed as SCLC. C: Boxplot of Agilent array CGH derivative log ratio spreads for DQ versus Pap smear samples; ULS, universal label system (Agilent); Enz: enzymatic DNA labeling. D: Validation set of seven matched pairs of DQ and Pap smears, showing higher-molecular weight DNA in all instances of DQ relative to Pap.
Based on criteria for success, including DNA quantity and quality measures, array performance parameters (ie, derivative log ratio scores, Table 1), and aberration identification, we found that all archival FNAC-DQ extracts were successful for all profiling assays tested (Figures 1–3). Unequivocal genomic structural aberrations were identified in 44 of 46 malignant smears; in one case, LNFNA-5, normal reference DNA from the patient would be needed to confirm that the observed genome structure was mutant and not a copy number variation. Surprisingly, even the degraded archival Pap smear DNAs showed similar structural aberrations as matched DQ samples on Agilent array CGH and Infinium genotyping, although with greater background noise (Figure 3). Universal label system labeling of DNA was superior to enzymatic labeling for array CGH analysis (Table 1, derivative log ratio scores; Figure 1C, boxplot).
Figure 2.
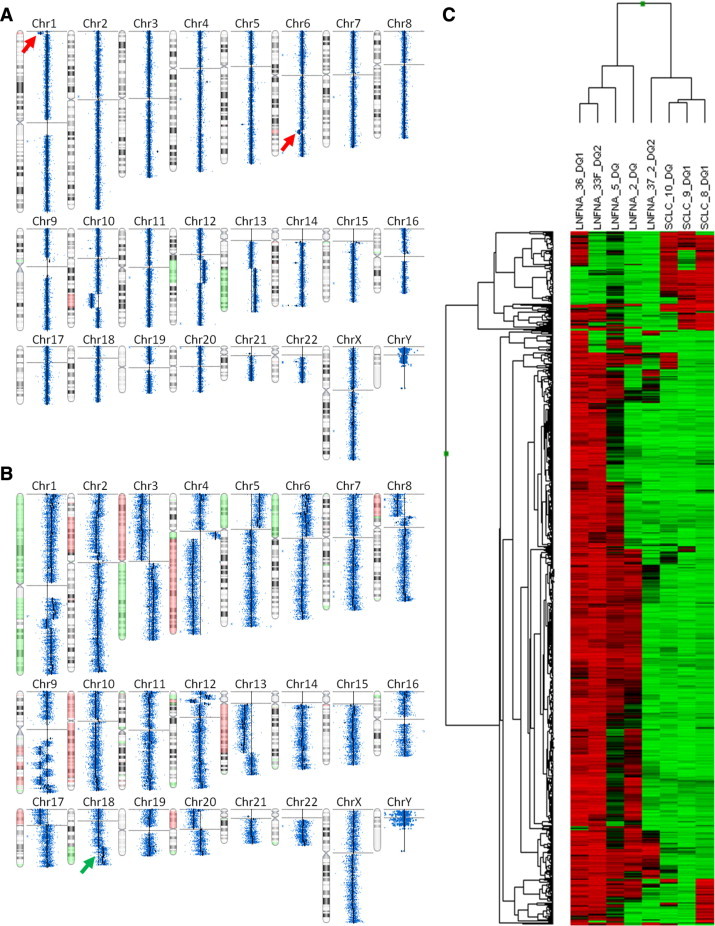
Representative high-resolution microarray plots from archival FNAC-DQ samples. A: Agilent 105K array CGH on sample LNFNA-33DQ2, a case of low-grade follicular lymphoma (FL). The Nexus summary view reveals focal deletions on 1p and 6q (red arrows), recently reported poor prognostic markers in FL.11B: Agilent 105K CGH on SCLC-8-DQ1, a case of SCLC showing BCL-2 amplification (green arrow), reported to predict tumor responsiveness to BCL-2 antagonists.12C: Illumina Infinium 27K methylation assay on eight different FNAC DNA extracts from cases of SCLC and various lymphomas (see Table 1 for sample IDs). Shown is unsupervised cluster after selection for the most highly variable, sex-neutral [(CHR not X) and (methylation β STDEV >0.3); n = 759 of 27,578 total targets] CpG targets among the eight FNAC DNA extracts.
Figure 3.
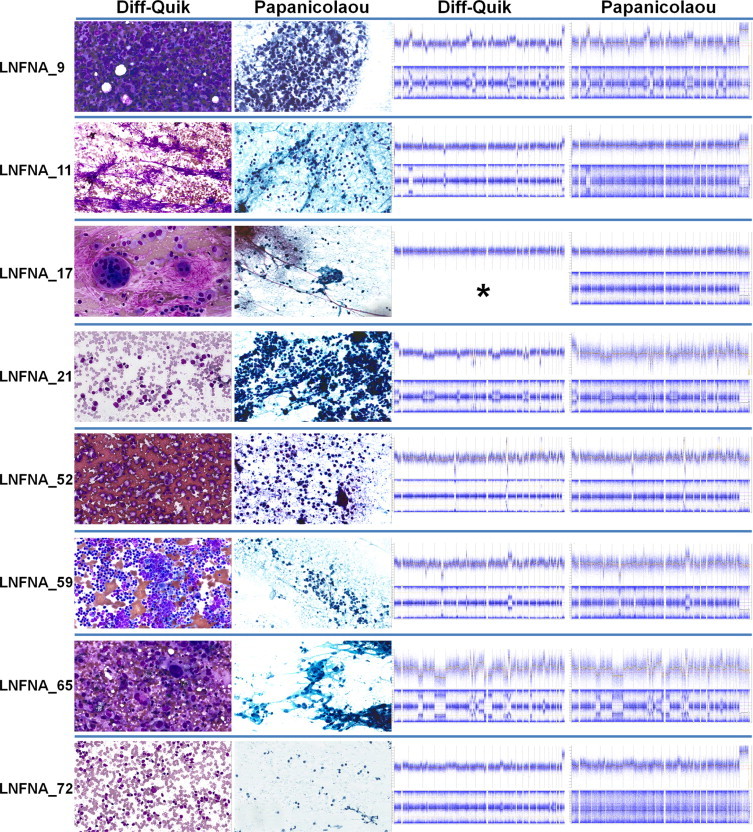
Comparative high-resolution microarray plots from eight matched pairs of archival FNAC DQ and Pap smears. First and second columns from left: representative screenshots of digitally-preserved cytomorphology of DQ and Pap smears. Third and fourth columns from left: top half of row illustrates whole genome view of genome segment copy number derived from array CGH data; bottom half of row shows whole genome view of SNP genotyping array results (B-allele frequency), which highlights regions of allelic imbalance/loss of heterozygosity. Asterisk indicates sample lost to genotype analysis.
Discussion
The observed Pap-specific DNA degradation and DQ DNA preservation is neither widely appreciated nor reported in molecular pathology texts.5,6,7 Mechanistically, the Pap stain process differs from DQ in at least two major ways: one, the Pap smear is fixed in alcohol while the DQ is air dried; two, the Pap cytodyes include hematoxylin, while DQ does not. Whereas alcohol fixation in the Pap process might be expected to better preserve nucleic acid in Pap smears, there is some evidence in the literature for a potentially compromising effect of hematoxylin on DNA amplification8,9,10; however, no studies we could identify report the remarkable contrast in DNA molecular weight in archival samples stained with Pap and/or hematoxylin versus other stains, nor could we find published studies evaluating DNA molecular weight distributions in various types of archival specimens. We hypothesize that hematoxylin may be at least partly responsible for extensive DNA damage in archival Pap smears; if correct, the generalized DNA damaging effect of hematoxylin is largely undescribed at present. Additional studies are warranted to investigate the critical variation among tissue stains in preserving molecular information in these samples.
As a test of the suitability of the archival FNAC DNA extracts for high-resolution genomic profiling, samples were assayed with Agilent CGH (Figures 2 and 3), Illumina Infinium SNP genotyping (Figure 3), and Illumina Infinium methylation arrays (Figure 2C). This is a stringent battery of tests for any DNA extract, as typically only high-purity, fresh or cryopreserved tissue DNA extracts would be suitable for all three of these assays. Based on criteria for success (see Materials and Methods) we found that nearly all archival FNAC extracts were successful for all profiling assays tested (Figures 1–3). Genomic structural aberrations were identified in >95% of malignant smears; the lone exception (LNFNA-5) was a case of low-grade follicular lymphoma in which a deviation from the reference DNA was detected, but normal reference DNA from the patient would be needed to evaluate the somatic mutational status of the variation. Surprisingly, even the degraded archival Pap smear DNAs showed similar structural aberrations as matched DQ samples on Agilent array CGH and Infinium SNP arrays, although with greater background noise. One low-grade follicular lymphoma aspirate clearly manifested (Figure 2A) recently-described poor-prognostic genomic copy number features,11 while some samples of SCLC showed (Figure 2B) 18q amplification encompassing BCL-2, an aberration correlated with responsiveness to certain antitumor drugs.12
Positive identification of gross genomic structural defects in whole slide scrapes of FNAC samples is remarkable, given the potential a priori for low tumor cellular fraction in aspirates. Although cellular heterogeneity in tumor tissues may be expected to confound aberration detection during molecular profiling, our results indicate a sufficient enrichment of tumor cells can be achieved through the FNA technique to permit DNA structural aberration detection without further sample microdissection or fractionation. Samples included in this study were not preselected based on morphological criteria for tumor purity, nor were they microdissected. The sufficient purity of malignant FNA smears for molecular profiling may not be so surprising considering the very focal and targeted lesion sampling of each pass with the needle. The relative tumor purity in FNAC versus tissue extracts, the potential enhancement for biomarker detection, and the utility of a broader spectrum of FNAC samples can now receive greater attention in future studies.
In summary, cytopathology specimens are an excellent potential source of patient materials for clinical molecular profiling, including retrospective genomic analyses and prospective sample collection for individualized therapy or eligibility review for clinical trial enrollment. FNAC samples up to 16 years old yielded copious quantities of genomic DNA suitable for high-resolution genomic and epigenomic profiling. Diff-Quik versus Papanicolaou cytologic staining processes show remarkable differences in DNA preservation possibly implicating hematoxylin as a DNA damaging agent. Over 95% of malignant aspirates in this study manifested clearly discernable genomic aberrations, suggesting that diagnostic FNA samples are often inherently of sufficient purity for genomic scale molecular profiling.
Acknowledgements
We thank Shyam Kalavar, CT, David Hornbeck, Marie Mueller, CTR, and Dr. Eugene Passamani, M.D. for facilitating pathology archive research.
Footnotes
Supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
CME Disclosure: None of the authors disclosed any relevant financial relationships.
References
- 1.DeMay RM, editor. The art & science of cytopathology. ASCP Press; Chicago: 1996. [Google Scholar]
- 2.Killian JK, Bilke S, Davis S, Walker RL, Killian MS, Jaeger EB, Chen Y, Hipp J, Pittaluga S, Raffeld M, Cornelison R, Smith WI, Jr, Bibikova M, Fan JB, Emmert-Buck MR, Jaffe ES, Meltzer PS. Large-scale profiling of archival lymph nodes reveals pervasive remodeling of the follicular lymphoma methylome. Cancer Res. 2009;69:758–764. doi: 10.1158/0008-5472.CAN-08-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagel H, Schulten HJ, Gunawan B, Brinck U, Fuzesi L. The potential value of comparative genomic hybridization analysis in effusion-and fine needle aspiration cytology. Mod Pathol. 2002;15:818–825. doi: 10.1097/01.MP.0000024521.67720.0F. [DOI] [PubMed] [Google Scholar]
- 4.Mattu R, Sorbara L, Filie AC, Little R, Wilson W, Raffeld M, Abati A. Utilization of polymerase chain reaction on archival cytologic material: a comparison with fresh material with special emphasis on cerebrospinal fluids. Mod Pathol. 2004;17:1295–1301. doi: 10.1038/modpathol.3800185. [DOI] [PubMed] [Google Scholar]
- 5.Leonard DGB, Bagg A, Kaul K, Van Deerlin VM, Caliendo AM, editors. Molecular pathology in clinical practice. Oncology. Springer; New York: 2009. [Google Scholar]
- 6.Killeen AA. Principles of molecular pathology. Humana Press; Totowa NJ: 2004. [Google Scholar]
- 7.Coleman WB, Tsongalis GJ, editors. Molecular pathology: the molecular basis of human disease. Academic Press; Burlington, Mass.: 2009. [Google Scholar]
- 8.Burton MP, Schneider BG, Brown R, Escamilla-Ponce N, Gulley ML. Comparison of histologic stains for use in PCR analysis of microdissected, paraffin-embedded tissues. Biotechniques. 1998;24:86–92. doi: 10.2144/98241st01. [DOI] [PubMed] [Google Scholar]
- 9.Kiernan JA. Histological and histochemical methods: theory and practice. Vol x. Butterworth Heinemann; Boston, Oxford: 1999. p. 502. [Google Scholar]
- 10.Murase T, Inagaki H, Eimoto T. Influence of histochemical and immunohistochemical stains on polymerase chain reaction. Mod Pathol. 2000;13:147–151. doi: 10.1038/modpathol.3880028. [DOI] [PubMed] [Google Scholar]
- 11.Cheung KJ, Shah SP, Steidl C, Johnson N, Relander T, Telenius A, Lai B, Murphy KP, Lam W, Al-Tourah AJ, Connors JM, Ng RT, Gascoyne RD, Horsman DE. Genome-wide profiling of follicular lymphoma by array comparative genomic hybridization reveals prognostically significant DNA copy number imbalances. Blood. 2009;113:137–148. doi: 10.1182/blood-2008-02-140616. [DOI] [PubMed] [Google Scholar]
- 12.Olejniczak ET, Van Sant C, Anderson MG, Wang G, Tahir SK, Sauter G, Lesniewski R, Semizarov D. Integrative genomic analysis of small-cell lung carcinoma reveals correlates of sensitivity to bcl-2 antagonists and uncovers novel chromosomal gains. Mol Cancer Res. 2007;5:331–339. doi: 10.1158/1541-7786.MCR-06-0367. [DOI] [PubMed] [Google Scholar]


