Abstract
The definitive identification of malignant pleural mesothelioma (MPM) has significant clinical implications, yet other malignancies often involve the lung pleura, confounding the diagnosis of MPM. In the absence of accurate markers, MPM can be difficult to distinguish from peripheral lung adenocarcinoma and metastatic epithelial cancers. MicroRNA expression is tissue-specific and highly informative for identifying tumor origin. We identified microRNA biomarkers for the differential diagnosis of MPM and developed a standardized microRNA-based assay. Formalin-fixed, paraffin-embedded samples of 33 MPM and 210 carcinomas were used for assay development. Using microarrays, we identified microRNAs differentially expressed between MPM and various carcinomas. Hsa-miR-193–3p was overexpressed in MPM, while hsa-miR-200c and hsa-miR-192 were overexpressed in peripheral lung adenocarcinoma and carcinomas that frequently metastasize to lung pleura. We developed a standardized diagnostic assay based on the expression of these microRNAs. The assay reached a sensitivity of 100% and a specificity of 94% in a blinded validation set of 68 samples from the lung and pleura. This diagnostic assay can provide a useful tool in the differential diagnosis of MPM from other malignancies in the pleura.
Malignant pleural mesothelioma is a relatively rare and aggressive tumor for which several clinical trials using immunomodulating and targeted treatments, whose efficiency relies on an accurate diagnosis, are currently being undertaken.1 It is a solid, locally aggressive tumor of the pleura which leads to a severe clinically symptomatic disease with very poor prognosis.2 Foremost among risk factors for the development of this malignancy is exposure to asbestos,3 often with a 20- to 50-year latency between asbestos exposure and development of the malignancy. Due to the extraordinary fire-resistant properties of asbestos, it was widely used in the United States and Europe, mostly in the shipbuilding and construction industries, between the 1940s and 1979 when the U.S. government limited its use. During that time, an estimated 40% of the entire workforce, or about 27 million individuals, were exposed to asbestos. Exposure to radiation and infection with the SV40 virus have been suggested as additional risk factors; genetic susceptibility and familial clustering have also been observed.4 Alarmingly, the incidence of mesothelioma has clearly grown in recent years in all developed countries of Western Europe and North America, and most probably in developing countries as well.1 Exposure to asbestos is still a major factor that contributes to the continuing growth in the number of cases.
MPM can be divided into three main histological subtypes: epithelioid, which comprises over 60% of MPM tumors; sarcomatoid; and biphasic (mixed).5 The histology of epithelioid MPM is often very similar to that of carcinomas; hence the distinction between epithelioid MPM and carcinomas which involve the lung pleura can be challenging.6 The use of immunohistochemical markers has greatly increased the ability of pathologists to discriminate between pleural malignancies; however there is no single marker which is accurate enough to make the diagnosis. Therefore a panel of immunostains must be used, and the choice of markers, as well as interpretation of the results in equivocal cases, remains subjective.7,8 A reliable and objective assay could help make this distinction with greater confidence. Here we set out to use the tissue-specific expression of microRNA to develop an assay that can identify MPM from lung adenocarcinoma and other carcinomas which may metastasize to the pleura and lung, using a small number of microRNA biomarkers.
MicroRNAs are short (17 to 22 nucleotides) noncoding RNAs that regulate gene expression, and play a major role in oncogenesis.9 Their exceptional tissue specificity has made them potent biomarkers for diagnosing the tissue source of metastatic cancers.10,11,12,13,14 We have previously taken advantage of this property and used microRNAs for the identification of tissue origin of metastatic cancers,12,13 for distinguishing squamous from non-squamous non-small cell lung cancer,15,16 and for distinguishing metastatic from primary tumors of the brain17 and the lung.18 Here we present a study that characterizes microRNA expression in MPM. We identified microRNAs that are differentially expressed between MPM and various carcinoma types and used three of these microRNA biomarkers to develop a diagnostic assay that is able to distinguish between MPM and epithelial cancers involving the lung and pleura with excellent sensitivity and specificity.
Materials and Methods
Patients and Samples
Anonymized formalin-fixed, paraffin-embedded (FFPE) tissue samples from large resection specimens were obtained from Sheba Medical Center, Tel Hashomer, Israel; Rabin Medical Center, Petah Tikva, Israel; Soroka University Medical Center, Beer-Sheva, Israel; ABS Inc., Wilmington, DE; Bnai-Zion Medical center, Haifa, Israel; and Cureline BioPathology, Burlingame, CA. Institutional review approvals were obtained for all samples in accordance with each institute's institutional review board or equivalent guidelines. Tissue from representative blocks was sectioned into 1.5 ml microcentrifuge tubes (three to five 10-μm sections), and H&E-stained slides were obtained from each block, to evaluate percentage of tumor cellular content at sectioning. Validation samples were blinded to the investigators performing the validation assays and analyses.
Histological Diagnosis
For samples used in the biomarker discovery and training phases, tumor tissue diagnosis was based on the medical records. For samples used for validation, the original clinical diagnosis was validated by an independent pathologist. Only samples whose tumor percentage was at least 50% were used for validation. All mesothelioma samples in the validation set except two were examined by immunohistochemistry using at least three different antibodies from a list of potential markers including calretinin, TTF1, and keratin 5/6.
RNA Extraction
Total RNA was extracted as described.12,17 FFPEs were deparaffinized with xylene, washed in ethanol, and digested with proteinase K. RNA was extracted with acid phenol:chloroform followed by ethanol precipitation and DNase digestion.
MicroRNA Microarray
Custom microarrays were prepared as described.12,19 747 DNA oligonucleotide probes representing human microRNAs were spotted in triplicate on coated microarray slides (Nexterion Slide E, Schott, Mainz, Germany). Three to five μg of total RNA were labeled by ligation of an RNA-linker, p-rCrU-Cy/dye (Eurogentec, San Diego, CA; Cy3 or Cy5) to the 3′ end. Slides were incubated with the labeled RNA for 12 to 16 hours at 42°C and then washed twice. Arrays were scanned at a resolution of 10 μm, and images were analyzed using SpotReader software (Niles Scientific, Portola Valley, CA). Microarray spots were combined and signals normalized as described previously.12,19
Microarray Data Analysis and Statistics
Normalized signals were compared between groups of samples (for example MPM versus non-MPM) to find microRNAs which can be used to differentiate between the groups. Only microRNAs which passed the background level criterion of median normalized fluorescence signal greater than 800 in at least one of the two groups were considered. Significance of differences was assessed by a two-sided unpaired t-test. The Benjamini-Hochberg false discovery rate method20 was used to control for multiple hypotheses testing, using a false discovery rate of 0.2. Fold-change was calculated as the ratio of the median values of the normalized fluorescence signals in the two groups. The ability of each microRNA to separate the two groups was characterized by the receiver operating characteristic curve and the area under the receiver operating characteristic curve.
Quantitative Real-Time PCR
MicroRNA amounts were quantified using a quantitative real-time (qRT)-PCR method recently described.15,21 We normalized the CT of each microRNA in each sample by subtracting the CT of U6 small nuclear RNA15 in that sample as follows: normCT[miR-X] = CT[miR-X]–CT[U6]. In Figure 1D, we used an inverted-normalized CT scale such that high values represent high expression (as microarray fluorescence). Inverted-normCT[miR-X] = 15–normCT[miR-X].
Figure 1.
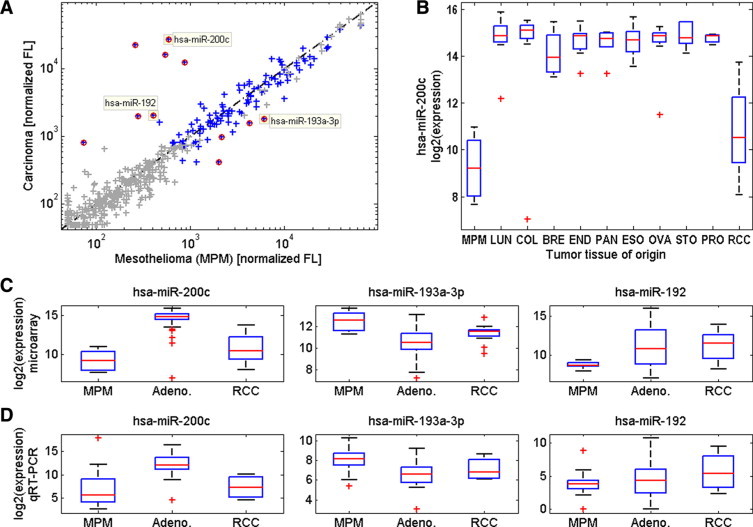
Comparison of microRNA expression levels in MPM and carcinoma samples in the discovery phase. A: Median normalized fluorescence values (microarrays) of microRNAs in seven MPM samples are plotted against the median fluorescence values (microarrays) of these microRNAs in 97 carcinoma samples (Table 1). Gray crosses show control probes and microRNAs whose expression level was at background levels, and red circles mark microRNAs that had statistically significant differences in expression values after false discovery rate correction with at least a twofold change in median expression (see Materials and Methods). Yellow squares highlight the median expression levels of hsa-miR-200c, hsa-miR-192, and hsa-miR-193a–3p. B: Box plot of the expression levels of hsa-miR-200c in tumors of different tissue origin. Tumor origins include MPM (n = 7), RCC (n = 16), and 81 adenocarcinomas from: lung (LUN, n = 15), colon (COL, n = 17), breast (BRE, n = 4), endometrium (END, n = 9), pancreas (PAN, n = 6), esophagus (ESO, n = 11), ovary (OVA, n = 10), stomach (STO, n = 6), and prostate (PRO, n = 3). Units show log2 of the normalized fluorescence signal by microarray. C: Box-plots of the expression levels of hsa-miR-200c, hsa-miR-193a–3p, and hsa-miR-192, in these MPM, adenocarcinoma (Adeno.), and RCC samples. Units show log2 of the normalized fluorescence signal by microarray. D: Box plots of the expression levels of these microRNAs in 22 MPM samples, 39 adenocarcinoma samples, and four RCC samples, measured by qRT-PCR (Table 1). Units show the inverted normalized CT (see Materials and Methods). For all box plots, red crosses indicate outlier values that are defined as being over 1.5 times the distance between the 25th and 75th percentiles above the 75th percentile or below the 25th percentile.
qRT-PCR Assay
For each sample, hsa-miR-200c, hsa-miR-192, hsa-miR-193a-3p, and U6 small nuclear RNA were measured by qRT-PCR in triplicates. The median CT of the triplicates (MedCT-miR-200c, MedCT-miR-192, MedCT-miR-193a-3p, and MedCT-U6) was calculated. Outlier wells were identified (for MedCT < 32, replicates with CT differing by more than 1 cycle from the median; for 32 ≤ MedCT < 37, replicates with CT differing by more than 1.5 cycles from the median and for MedCT ≥ 37, replicates with CT differing by more than 2 cycles from the median). If two or more replicates were undetected, MedCT was set to 50. If any probe had more than one outlier, the assay was repeated. If MedCT-U6 was not between 20 and 32 cycles, or if the MedCT of none of the three assay microRNAs was below 35, the assay was repeated. If the sample failed to meet these criteria a second time, it was classified as an assay failure. For the purpose of calculating the assay scores (as described in the Results section), MedCTs of the assay microRNAs were normalized by subtracting MedCT-U6: NormCT-miR-X = MedCT-miR-X–MedCT-U6.
Results
We developed an assay which differentiates MPM from carcinomas which involve the lung pleura using microRNA expression. The development process consisted of three phases: a discovery phase in which potential markers were identified, a training phase in which a diagnostic protocol that uses the selected markers was established, and a validation phase in which the diagnostic protocol was tested on an independent cohort.
Discovery Phase
To identify microRNAs that can serve as molecular markers for identification of MPM from carcinomas, we used microRNA microarrays and profiled 7 MPM samples from the lung pleura and 97 carcinoma samples of various tissue origins (Table 1). One hundred eleven microRNAs passed the background level criterion and 11 microRNAs showed significant differences in expression between the two groups, with P values smaller than the false discovery rate-corrected threshold and a fold change greater than 2 (see Materials and Methods). Seven of the microRNAs which passed the above criteria were overexpressed in carcinoma samples relative to MPM samples: the hsa-miR-200 family, (which includes hsa-miR-200a/b/c, hsa-miR-141, and hsa-miR-429) and hsa-miR-192/194. The other four microRNAs had higher expression in MPM samples: the hsa-miR-193 family (which includes hsa-miR-193a-3p/5p and hsa-miR-193b) and hsa-miR-152 (Figure 1A and Table 2). The strongest differential expression was identified for the hsa-miR-200 family, and especially hsa-miR-200c and hsa-miR-141; however these microRNAs were less differential between MPM and renal cell carcinoma (RCC) (Figure 1B). We therefore repeated the analysis in two separate steps, comparing microRNA expression between MPM and non-RCC carcinomas, and between MPM and RCC (Table 2). Based on these comparisons, three microRNAs (hsa-miR-200c, hsa-miR-192, and hsa-miR193a-3p) were chosen as candidate biomarkers for the assay (Figure 1C). Hsa-miR-200c expression was high in non-RCC carcinomas and low in MPM, hsa-miR-192 expression was high in RCC and low in MPM and hsa-miR-193a-3p expression was high in MPM compared to both RCC and non-RCC carcinomas.
Table 1.
Tumor Samples Used in the Study
| Discovery |
Training |
Validation | |||
|---|---|---|---|---|---|
| Study phase Cohort | Microarray | qRT-PCR | Full training set | Threshold delineation | Validation set |
| Figure # | Fig. 1ABC | Fig. 1D | Fig. 2AB | Fig. 3ABC | Fig. 4 |
| Samples (new samples) | 104 | 65 (51 new) | 145 (88 new) | 39 (no new) | 63 (all new) |
| Sample type (total N) | |||||
| Mesothelioma (47) | 7 | 22 (15) | 32 (11) | 16 | 14 |
| Epithelioid (29) | 4 | 18 (14) | 21 (3) | 8 | 8 |
| Sarcomatoid (6) | 3 | 3 (0) | 4 (1) | 3 | 2 |
| Biphasic (mixed) (6) | — | 1 (1) | 1 (1) | 1 | 4 |
| Unspecified (6) | — | — | 6 (6) | 4 | — |
| Carcinoma (259) | 97 | 43 (36) | 113 (77) | 23 | 49 |
| Bladder (15) | — | 2 (2) | 14 (13) | 2 | — |
| Breast (26) | 4 | 4 (4) | 9 (9) | 2 | 9* |
| Colon (36) | 17 | 2 (1) | 9 (5) | 2 | 13* |
| Endometrium (12) | 9 | — | — | — | 3* |
| Esophagus (11) | 11 | — | — | — | — |
| Kidney (RCC) (33) | 16 | 4 (2) | 18 (10) | 4 | 5* |
| Kidney (TCC) (1) | — | — | — | — | 1* |
| Liver (HCC) (10) | — | — | 10 (10) | 2 | — |
| Lung (76) | 15 | 25 (25) | 35 (20) | 8 | 16 |
| Ovary (19) | 10 | 4 (1) | 10 (6) | 1 | 2* |
| Pancreas (11) | 6 | 2 (1) | 8 (4) | 2 | — |
| Prostate (3) | 3 | — | — | — | — |
| Stomach (6) | 6 | — | — | — | — |
Table 1 shows the number of tumor samples from different origins or mesotheliomas used in each phase of the study. For the third and fourth columns, the numbers in parentheses indicate the number of new samples that were not used in previous phases. The threshold delineation cohort (fifth column) included only previously used samples, and the validation set (last column) included only new samples. The rows titled Samples, Mesothelioma, and Carcinoma (in bold) indicate the total number of samples used in each of these sets. The samples that were repeated in the threshold delineation cohort were chosen to represent the full range of signals in the full training set (see Results). All other samples that were used in more than one cohort were selected randomly based on the amount of RNA left from the sample. In the first column, the numbers in parentheses indicate the total number of samples of this type.
The validation set included only samples of tumors that were located in the lung and pleura: primary tumors included lung adenocarcinoma and MPM; lung metastases included RCC, TCC, endometrial, breast, and colonic carcinomas; and pleural metastases were from adenocarcinomas of the breast and ovary.
Table 2.
MicroRNAs that Were Differentially Expressed between MPM and Non-RCC Carcinoma or RCC
| Median values |
MPM versus non-RCC carcinoma (Adeno.) |
MPM versus RCC |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| miR name | MPM | Adeno | RCC | P value | Fold-change | AUC | P value | Fold-change | AUC |
| hsa-miR-200c | 590 | 30000 | 1500 | 2.6E-20 | 50.3 | 0.99 | 5.2E-02 | 2.5 | 0.75 |
| hsa-miR-141 | 260 | 25000 | 990 | 2.9E-20 | 96.6 | 0.99 | 6.7E-02 | 3.8 | 0.75 |
| hsa-miR-200b | 550 | 21000 | 5000 | 2.0E-16 | 38.4 | 0.98 | 7.0E-05 | 9.2 | 0.92 |
| hsa-miR-200a | 880 | 15000 | 4400 | 6.7E-12 | 16.8 | 0.98 | 4.6E-04 | 5 | 0.89 |
| hsa-miR-429 | 73 | 980 | 200 | 2.7E-09 | 13.3 | 0.97 | 4.7E-03 | 2.7 | 0.86 |
| hsa-miR-193a-3p | 6100 | 1500 | 3000 | 3.9E-05 | 4.2 | 0.91 | 6.3E-03 | 2.1 | 0.80 |
| hsa-miR-152 | 2200 | 910 | 1600 | 3.5E-03 | 2.4 | 0.89 | 2.9E-01 | 1.4 | 0.64 |
| hsa-miR-193b | 4300 | 1500 | 2500 | 2.1E-03 | 2.9 | 0.85 | 1.2E-01 | 1.7 | 0.72 |
| hsa-miR-193a-5p | 2000 | 380 | 540 | 2.5E-05 | 5.3 | 0.83 | 6.8E-02 | 3.7 | 0.73 |
| hsa-miR-375 | 120 | 960 | 82 | 7.0E-03 | 7.8 | 0.82 | 4.4E-01 | 1.5 | 0.63 |
| hsa-miR-194 | 280 | 1900 | 2400 | 7.9E-03 | 6.8 | 0.81 | 3.3E-04 | 8.6 | 0.94 |
| hsa-miR-221 | 19000 | 12000 | 7800 | 3.9E-03 | 1.6 | 0.79 | 5.2E-03 | 2.5 | 0.88 |
| hsa-miR-192 | 410 | 1800 | 3000 | 1.8E-02 | 4.5 | 0.76 | 1.1E-03 | 7.4 | 0.91 |
| hsa-miR-494 | 3100 | 1600 | 1300 | 2.8E-02 | 2 | 0.76 | 1.2E-02 | 2.4 | 0.8 |
| hsa-miR-143 | 7400 | 13000 | 17000 | 4.8E-02 | 1.8 | 0.74 | 1.6E-03 | 2.3 | 0.92 |
| hsa-miR-130a | 10000 | 3500 | 5200 | 4.7E-02 | 2.9 | 0.73 | 9.2E-02 | 1.9 | 0.75 |
| hsa-miR-30 days | 8800 | 12000 | 20000 | 1.1E-01 | 1.4 | 0.67 | 1.3E-04 | 2.2 | 0.94 |
| hsa-miR-497 | 1300 | 2600 | 5500 | 2.1E-01 | 2 | 0.64 | 9.0E-03 | 4.3 | 0.85 |
| hsa-miR-210 | 5900 | 4000 | 13000 | 2.9E-01 | 1.5 | 0.61 | 3.0E-03 | 2.2 | 0.88 |
| hsa-miR-30a | 7400 | 6100 | 16000 | 4.6E-01 | 1.2 | 0.58 | 5.4E-04 | 2.2 | 0.93 |
| hsa-miR-486-5p | 500 | 590 | 1600 | 4.3E-01 | 1.2 | 0.56 | 4.2E-03 | 3.2 | 0.84 |
| hsa-miR-451 | 1300 | 1200 | 2900 | 8.1E-01 | 1.1 | 0.51 | 2.7E-02 | 2.2 | 0.80 |
| hsa-miR-10b | 350 | 300 | 1200 | 8.3E-01 | 1.2 | 0.5 | 2.3E-02 | 3.4 | 0.85 |
| hsa-miR-126 | 4300 | 5100 | 15000 | 7.8E-01 | 1.2 | 0.49 | 3.7E-03 | 3.5 | 0.87 |
Median values of normalized fluorescence (rounded to two significant digits) in MPM, non-RCC carcinoma (all Adeno. in this set), and RCC samples are shown for microRNAs that were differentially expressed between MPM and either non-RCC carcinoma or RCC, as measured on the microarray platform. A Benjamini-Hochberg false discovery rate of 0.2 was used to identify differentially expressed microRNAs, resulting in P value cutoffs of 0.05 and 0.04 for adenocarcinoma and RCC, respectively. For each of the two comparisons, P values are shown (two-sided unpaired t-test), fold-change of median expression (either up-regulated or down-regulated, as is evident from the median values), and AUC, which is the area under the receiver operating characteristic (ROC) curve, indicative of the classification potential of each microRNA. MicroRNAs are sorted by decreasing values of the AUC of the comparison from MPM versus non-RCC carcinoma.
We verified that the differences in expression of these microRNAs between MPM and carcinomas are platform independent. We used qRT-PCR to measure the expression levels of the three selected microRNAs and of U6 small nuclear RNA which was used for normalization (see Materials and Methods), on a set of 22 MPM samples and 43 carcinoma samples (Table 1). The expression levels of these microRNAs maintained the same pattern (Figure 1D) indicating that the differences in expression are a general property of these tissues that can be robustly measured.
Training Phase
We used qRT-PCR to measure the expression levels of hsa-miR-200c, hsa-miR-193a-3p, hsa-miR-192, and U6 small nuclear RNA, which was used for normalization on a set of 145 samples (Table 1). We first used the full set to define two combinations of microRNAs, which can be used to discriminate MPM from carcinomas. The combination of hsa-miR-200c and hsa-miR-193a-3p expression levels accurately distinguished between MPM and most non-RCC carcinomas, but a large portion of the RCC samples could not be discriminated from the MPM samples using this combination alone. The expression level of hsa-miR-192 helped distinguish MPM from RCC, as well as from hepatocellular carcinoma and adenocarcinomas of the gastrointestinal tract. Figures 2, A and B demonstrate how the above combinations can be used to identify MPM samples in the full training set. The solid lines in the figures indicate cutoffs which were used for a preliminary assessment of the accuracy of a potential assay based on these two combinations. All but one of the 32 MPM samples were located above both of the cutoffs (sensitivity of 97%; 95% confidence interval assuming binomial success distribution: 84%−100%) and 108 of 113 non-MPM samples were located below at least one of the cutoffs (specificity of 96%; confidence interval: 90%−99%).
Figure 2.

The chosen microRNA combinations for differentiation of MPM from confounding cancers on the full training set. A: Expression level (normalized CT, see Materials and Methods) of hsa-miR-192 was measured using qRT-PCR for 32 MPM samples (red stars), 10 hepatocellular carcinoma samples (green circles), 18 RCC samples (blue squares), and 17 adenocarcinomas from the gastrointestinal tract (Adeno-GI; black-gray diamonds). The solid vertical line is a guide to the eye, demonstrating the effectiveness of hsa-miR-192 expression in separating MPM from hepatocellular carcinoma and carcinomas from the GI tract. Two of the MPM samples had much lower expression of hsa-miR-192 (undetected at 40 cycles) and were omitted from the figure for optimal scaling. B: Expression levels of hsa-miR-200c and hsa-miR-193a–3p were measured using qRT-PCR for 32 MPM samples (red stars), 18 RCC samples (blue squares), and 85 adenocarcinoma samples (Adeno; black-yellow diamonds). The solid diagonal line is a guide to the eye, demonstrating the effectiveness of the combined expression levels of hsa-miR-200c and hsa-miR-193a–3p in separating MPM from adenocarcinomas. Five of the adenocarcinoma samples had low expression of hsa-miR-193a–3p (normalized CT between 16.4 and 20.7, with normalized CT of hsa-miR-200c between −0.5 and 2.5) and were omitted from the figure for optimal scaling.
Based on these findings, a diagnostic protocol was established. A subset of 16 MPM samples and 23 carcinoma samples from the full training set (Table 1), which were chosen to represent the range of signals in the cohort, was used to delineate the decision thresholds. Expression levels of hsa-miR-200c, hsa-miR-193a-3p, and hsa-miR-192 were measured in triplicates, and normalized by U6. Two scores were defined for each sample: Score1 was based on the normalized expression of hsa-miR-192 (Score1 = NormCT-miR-192−8.05, see Figure 3A); Score2 was defined as a linear combination of the normalized expression of hsa-miR-193a-3p and hsa-miR-200c (Score2 = NormCT-miR-200c−1.5*NormCT-miR-193a-3p + 6.6, see Figure 3B). We defined a simple classification rule (Figure 3C) to decide whether a given sample is a mesothelioma sample (see Materials and Methods): A sample was identified as mesothelioma if it had Score1 > 0 (Figure 3A) and Score2 > 0 (Figure 3B), otherwise it was identified as non-mesothelioma. Samples whose Score1 and Score2 values were near the cutoff threshold, within margins of 1.5 units from the classification cutoff (indicated by the light-shaded regions and the dotted lines in Figure 3) were designated as “near cutoff” classifications. This margin was designed to take into account possible measurement noise of the assay. The classification rule and possible assay outcomes are demonstrated in Figure 3C. The Score values obtained for MPM and non-MPM samples in this subset of samples is shown in Figure 3D.
Figure 3.
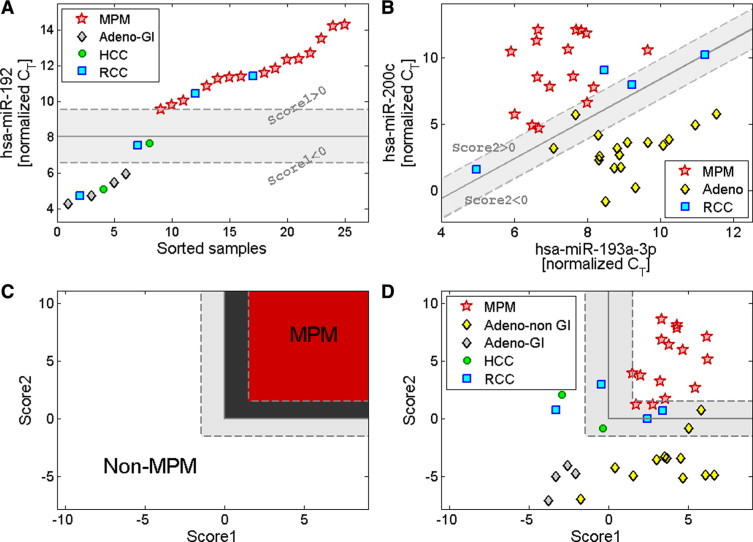
The classification rule for the differential diagnosis of MPM from confounding cancer types. Expression levels (normalized CT, see Materials and Methods) of hsa-miR-192, hsa-miR-193a–3p and hsa-miR-200c were measured using qRT-PCR for 39 samples from the training set selected for threshold delineation (Table 1) including MPM (red stars), hepatocellular carcinoma (green circles), RCC (blue squares), and adenocarcinomas (Adeno; black-yellow diamonds) including those from the gastrointestinal tract (Adeno-GI; black-gray diamonds). A: Threshold delineation for levels of hsa-miR-192. “Score1” was defined as Score1 = (hsa-miR-192 normalized CT) – 8.05. The solid vertical line indicates the cutoff on Score1 at Score1 = 0. The dotted lines and the shaded area mark a region near the cutoff value, with absolute value of Score1 less than 1.5. MPM samples have Score1 greater than 0. One additional MPM sample had much lower expression of hsa-miR-192 (undetected at 40 cycles) and a higher value of Score1, and was omitted from the figure for optimal scaling. B: Threshold delineation for the combined levels of hsa-miR-193a–3p and hsa-miR-200c. “Score2” was defined as Score2 = (hsa-miR-200c normalized CT) – 1.5 * (hsa-miR-193a–3p normalized CT) + 6.6. The solid diagonal line indicates the cutoff on Score2, at Score2 = 0. The dotted lines and the shaded area mark a region near the cutoff value, with absolute value of Score2 less than 1.5. MPM samples have Score2 greater than 0. C: A schematic of the classification rule designed for combining Score1 and Score2 using the training set. Samples are classified as MPM if both Score1 and Score2 are greater than 0 (upper-right region, shaded in red/dark gray), otherwise they are classified as non-MPM. The classification cutoff is marked by an L-shaped solid line. Score values near the cutoff are indicated by dotted lines and dark/light gray shade. D: Data for the samples used for threshold delineation presented on the axes of Score1 and Score2. Two samples, one MPM and one adenocarcinoma, had higher values of Score1, and were omitted from the figure for optimal scaling.
Validation Phase
We tested the assay on an independent validation set of FFPE samples of primary and metastatic tumors excised from the lung or lung pleura. RNA was extracted from 68 FFPE sections and microRNA expression levels were measured according to the assay protocol. Five samples (7%) failed quality assurance criteria indicating insufficient RNA amount (see Materials and Methods) and were excluded. Thus, the assay result was compared to the reference diagnosis for 63 blinded samples of which 14 were MPM and 49 were various carcinomas (Table 1). Assay scores were calculated and each sample was assigned to one of four categories: mesothelioma; mesothelioma (near cutoff); non-mesothelioma (near cutoff); or non-mesothelioma (Figure 3C). The results are summarized in Table 3 and Figure 4: the assay correctly identified 60 of the 63 blinded samples (95%). All of the 14 mesothelioma samples were identified correctly (sensitivity of 100%), and 46 of the 49 carcinomas were identified correctly (specificity of 94%). Fifty-two of the 63 samples (83%) were classified outside of the near cutoff region, and all of these classifications were correct (Table 3). Specifically for lung adenocarcinomas, one of the most important differential diagnoses in the pleura, the assay correctly identified all 16 lung adenocarcinoma samples as non-mesothelioma outside the near cutoff region (Figure 4).
Table 3.
Classification Results of Blinded Validation Set
| Histologic diagnosis | MPM | MPM (near cutoff) | Non-MPM (near cutoff) | Non-MPM | Total |
|---|---|---|---|---|---|
| MPM | 14 | 0 | 0 | 0 | 14 |
| Non-MPM | 0 | 3 | 8 | 38 | 49 |
| Total | 14 | 3 | 8 | 38 | 63 |
Table 3 shows the microRNA-based assay classification results of 63 blinded validation samples. Rows indicate histological diagnoses of samples; columns indicate the microRNA-based classification of the samples into four categories: MPM, MPM (near cutoff), Non-MPM (near cutoff), and Non-MPM (see Results). Numbers in bold indicate samples for which the assay classification and the histological diagnosis were in agreement.
Figure 4.
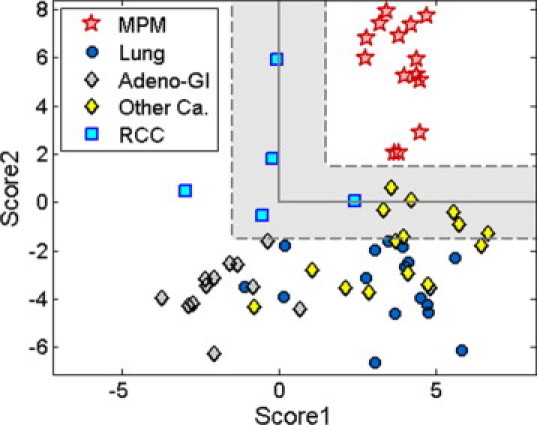
Validation of a microRNA-based qRT-PCR assay for the differential diagnosis of MPM from confounding cancer types. Expression levels (normalized CT, see Materials and Methods) of hsa-miR-192, hsa-miR-193a–3p, and hsa-miR-200c were measured using qRT-PCR for a validation set of 63 samples (Table 1) including MPM (red stars), lung adenocarcinoma (Lung; black-blue circles), adenocarcinomas from the gastrointestinal tract (Adeno-GI; black-gray diamonds), RCC (blue squares), and other carcinomas (Other Ca; black-yellow diamonds). All (14 of 14) MPM samples and 46 of 49 non-MPM samples were classified correctly. Samples with Score values outside of the shaded region (ie, not near the cutoff) were all correctly classified.
Discussion
The pathological assessment of pleural lesions includes a variety of neoplastic conditions that may be difficult to distinguish. One of the most common diagnostic problems is the distinction between epithelioid MPM and carcinomas which invade or metastasize to the lung pleura. Over the past 20 years, immunohistochemistry has become the mainstay of the pathological diagnosis of mesothelioma. However, since there is no single highly sensitive and specific immunostain, the use of immunohistochemical panels is required to accurately discriminate between MPM and other pleural malignancies.7,22
The specificities and sensitivities of various antibodies used to differentiate epithelioid MPM from pulmonary adenocarcinomas and other carcinomas23,24,25 have been evaluated, with inconclusive results.26,27 The differential diagnosis of MPM from carcinomas is therefore based on a combination of positive and negative markers, which are selected from the antibodies available to the pathology department according to the tumors which are suspected in each case based on morphology and patient history.8 There are no predefined panels or criteria on how to interpret problematic cases such as low levels of staining or conflicting results by different antibodies. Thus, the distinction between epithelioid MPM and carcinomas can still be difficult in some cases.28 Due to the low overall incidence of MPM, some pathologists may not be experienced with the histology of this tumor, the selection of the immunohistochemical panel, and the interpretation of the results. Together with the limited sensitivity and specificity of the individual immunohistochemical markers, this can result in inaccuracies in diagnosis.
Two recent papers29,30 studied the microRNA profile of MPM subtypes and normal mesothelium, however they did not compare the microRNA profile of MPM to other carcinomas which invade the lung pleura. Other studies,31,32 describe application of mRNA expression profiling and DNA methylation markers to differentiate MPM from lung adenocarcinoma. However, the large number of markers used in these studies makes them less practical for use as clinical assays. In addition, differentiating MPM from non-lung carcinoma specimens is of clinical value.
Here we present the first study of microRNA expression profiling of MPM aimed at comparing expression levels of microRNAs in MPM samples to samples of carcinomas which invade the lung and pleura. We found that the hsa-miR-200 family is strongly expressed in adenocarcinoma samples from a variety of epithelial tissues, but minimally so in MPM. This finding agrees with previous reports.12,13,33,34,35,36 Hsa-miR-192 was strongly expressed in RCC and gastrointestinal carcinomas, in agreement with previous observations.12,13,37 Differentiation of RCC from MPM was particularly challenging as no microRNA clearly separated the two. By combining the expression patterns of hsa-miR-200c with hsa-miR-193a-3p and hsa-miR-192, RCC could be segregated to the margins of the three-dimensional space occupied by MPM specimen analyses.
We used the microRNA biomarkers we identified to develop a molecular assay to confirm or rule out a diagnosis of MPM. This assay provides a standardized quantitative alternative to the currently applied methods. The small number of microRNAs needed for classification, the high tissue specificity of these microRNAs and their good preservation in archival fixed tissues embedded in paraffin, makes this assay a practical option. The recent demonstration of the preservation of microRNAs and their extraction from body fluids such as serum and plasma21 promises potential application of this assay to pleural effusions and may also signify the future use of microRNAs for early diagnosis of mesothelioma.
The microRNA-based assay that we have developed uses expression levels of only three microRNAs, and is able to accurately diagnose MPM and objectively distinguish it from lung adenocarcinoma and other malignancies involving the lung and pleura with very high sensitivity and specificity. It would be of interest to further characterize the test performance on poorly differentiated tumors and on other types of pathological samples such as small biopsies and pleural effusions. These are particularly challenging for diagnosis with current methods and microRNAs have shown promising results for identifying other lung pathologies in such samples.16
Acknowledgements
We thank Tamara Drozd and Zinaida Rodov (Rabin Medical Center) for their assistance. We thank technicians and researchers at Rosetta Genomics for their assistance and contributions.
Footnotes
Supported by Rosetta Genomics Ltd.
H.B. and D.L. contributed equally to this work.
H.B., D.L., S.R., L.C., H.G., N.B., K.A., E.G., E.M., Y.G., T.B.E., A.C., R.A., Z.B., N.R. & D.C. are or were full-time employees of Rosetta Genomics and/or hold equity in the company, which develops microRNA-based diagnostic products and may stand to gain by publications of these findings. Some of the authors from clinical centers have received research funding from the company as part of this and other collaborative projects. Full disclosures are provided through the online submission forms.
Current address of N.R.: Molecular and Computational Diagnostics Laboratory, Cancer Research UK Cambridge Research Institute, Li Ka Shing Centre, Cambridge UK; and Department of Oncology, University of Cambridge, Cambridge, UK.
References
- 1.Zervos MD, Bizekis C, Pass HI. Malignant mesothelioma 2008. Curr Opin Pulm Med. 2008;14:303–309. doi: 10.1097/MCP.0b013e328302851d. [DOI] [PubMed] [Google Scholar]
- 2.Ismail-Khan R, Robinson LA, Williams CC, Jr, Garrett CR, Bepler G, Simon GR. Malignant pleural mesothelioma: a comprehensive review. Cancer Control. 2006;13:255–263. doi: 10.1177/107327480601300402. [DOI] [PubMed] [Google Scholar]
- 3.Yarborough CM. Chrysotile as a cause of mesothelioma: an assessment based on epidemiology. Crit Rev Toxicol. 2006;36:165–187. doi: 10.1080/10408440500534248. [DOI] [PubMed] [Google Scholar]
- 4.Yang H, Testa JR, Carbone M. Mesothelioma epidemiology, carcinogenesis, and pathogenesis. Curr Treat Options Oncol. 2008;9:147–157. doi: 10.1007/s11864-008-0067-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki Y. Pathology of human malignant mesothelioma–preliminary analysis of 1,517 mesothelioma cases. Ind Health. 2001;39:183–185. doi: 10.2486/indhealth.39.183. [DOI] [PubMed] [Google Scholar]
- 6.Klebe S, Mahar A, Henderson DW, Henderson DW, Roggli VL. Malignant mesothelioma with heterologous elements: clinicopathological correlation of 27 cases and literature review. Mod Pathol. 2008;21:1084–1094. doi: 10.1038/modpathol.2008.125. [DOI] [PubMed] [Google Scholar]
- 7.Hammar SP. Macroscopic, histologic, histochemical, immunohistochemical, and ultrastructural features of mesothelioma. Ultrastruct Pathol. 2006;30:3–17. doi: 10.1080/01913120500313143. [DOI] [PubMed] [Google Scholar]
- 8.Husain AN, Colby TV, Ordonez NG, Krausz T, Borczuk A, Cagle PT, Chirieac LR, Churg A, Galateau-Salle F, Gibbs AR, Gown AM, Hammar SP, Litzky LA, Roggli VL, Travis WD, Wick MR. Guidelines for pathologic diagnosis of malignant mesothelioma: a consensus statement from the International Mesothelioma Interest Group. Arch Pathol Lab Med. 2009;133:1317–1331. doi: 10.5858/133.8.1317. [DOI] [PubMed] [Google Scholar]
- 9.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 10.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2004;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Sherman D, Devor M, Saper BC. A putative flip-flop switch for control of REM sleep. Nature. 2006;441:589–594. doi: 10.1038/nature04767. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. MicroRNAs accurately identify cancer tissue origin. Nature Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 13.Rosenwald S, Gilad S, Benjamin S, Lebanony D, Dromi N, Faerman A, Benjamin H, Tamir R, Ezagouri M, Goren E, Barshack I, Nass D, Tobar A, Feinmesser M, Rosenfeld N, Leizerman I, Ashkenazi K, Spector Y, Chajut A, Aharonov R. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol. 2010;23:814–823. doi: 10.1038/modpathol.2010.57. [DOI] [PubMed] [Google Scholar]
- 14.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, Nonaka D, Li J, Spector Y, Rosenfeld N, Chajut A, Cohen D, Aharonov R, Mansukhani M. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from non-squamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27:2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 16.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 17.Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, Sitbon E, Lithwick Yanai G, Elyakim E, Cholakh H, Gibori H, Spector Y, Bentwich Z, Barshack I, Rosenfeld N. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barshack I, Lithwick-Yanai G, Afek A, Rosenblatt K, Tabibian-Keissar H, Zepeniuk M, Cohen L, Dan H, Zion O, Strenov Y, Polak-Charcon S, Perelman M. MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol Res Pract. 2010;206:578–584. doi: 10.1016/j.prp.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- 21.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beasley MB. Immunohistochemistry of pulmonary and pleural neoplasia. Arch Pathol Lab Med. 2008;132:1062–1072. doi: 10.5858/2008-132-1062-IOPAPN. [DOI] [PubMed] [Google Scholar]
- 23.Carella R, Deleonardi G, D'Errico A, Salerno A, Egarter-Vigl E, Seebacher C, Donazzan G, Grigioni WF. Immunohistochemical panels for differentiating epithelial malignant mesothelioma from lung adenocarcinoma: a study with logistic regression analysis. Am J Surg Pathol. 2001;25:43–50. doi: 10.1097/00000478-200101000-00004. [DOI] [PubMed] [Google Scholar]
- 24.Ordonez NG. The immunohistochemical diagnosis of mesothelioma: a comparative study of epithelioid mesothelioma and lung adenocarcinoma. Am J Surg Pathol. 2003;27:1031–1051. doi: 10.1097/00000478-200308000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Riera JR, Astengo-Osuna C, Longmate JA, Battifora H. The immunohistochemical diagnostic panel for epithelial mesothelioma: a reevaluation after heat-induced epitope retrieval. Am J Surg Pathol. 1997;21:1409–1419. doi: 10.1097/00000478-199712000-00003. [DOI] [PubMed] [Google Scholar]
- 26.Roberts F, Harper CM, Downie I, Burnett RA. Immunohistochemical analysis still has a limited role in the diagnosis of malignant mesothelioma. A study of thirteen antibodies. Am J Clin Pathol. 2001;116:253–262. doi: 10.1309/XL6K-8E62-9FLD-V8Q8. [DOI] [PubMed] [Google Scholar]
- 27.Roberts F, McCall AE, Burnett RA. Malignant mesothelioma: a comparison of biopsy and postmortem material by light microscopy and immunohistochemistry. J Clin Pathol. 2001;54:766–770. doi: 10.1136/jcp.54.10.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ordonez NG. What are the current best immunohistochemical markers for the diagnosis of epithelioid mesothelioma? A review and update. Hum Pathol. 2007;38:1–16. doi: 10.1016/j.humpath.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma—A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]
- 30.Busacca S, Germano S, De Cecco L, Rinaldi M, Comoglio F, Favero F, Murer B, Mutti L, Pierotti M, Gaudino G. MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol. 2010;42:312–319. doi: 10.1165/rcmb.2009-0060OC. [DOI] [PubMed] [Google Scholar]
- 31.Holloway AJ, Diyagama DS, Opeskin K, Creaney J, Robinson BW, Lake RA, Bowtell DD. A molecular diagnostic test for distinguishing lung adenocarcinoma from malignant mesothelioma using cells collected from pleural effusions. Clin Cancer Res. 2006;12:5129–5935. doi: 10.1158/1078-0432.CCR-06-1027. [DOI] [PubMed] [Google Scholar]
- 32.Christensen BC, Marsit CJ, Houseman EA, Godleski JJ, Longacker JL, Zheng S, Yeh RF, Wrensch MR, Wiemels JL, Karagas MR, Bueno R, Sugarbaker DJ, Nelson HH, Wiencke JK, Kelsey KT. Differentiation of lung adenocarcinoma, pleural mesothelioma, and nonmalignant pulmonary tissues using DNA methylation profiles. Cancer Res. 2009;69:6315–6321. doi: 10.1158/0008-5472.CAN-09-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 34.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 35.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]


