Abstract
The standard genetic test for Lynch syndrome (LS) frequently reveals an absence of pathogenic mutations in DNA mismatch repair genes known to be associated with LS. It was recently shown that germ line deletions in the last exons of EPCAM are involved in the etiology of LS. The aim of this study was to evaluate the prevalence of EPCAM deletions in a Spanish population and the clinical implications of deletion. Probands from 501 families suspected of having LS were enrolled in the study. Twenty-five cases with MSH2 loss were identified: 10 had mutations of MSH2, five had mutations of MSH6, and 10 did not show MSH2/MSH6 mutations. These 25 cases were analyzed for EPCAM deletions using multiplex ligation-dependent probe amplification, and deletions were mapped using long-range PCR analysis. One subject with no MSH2/MSH6 mutations had a large deletion in the EPCAM locus that extended for 8.7 kb and included exons 8 and 9. The tumor exhibited MSH2 promoter hypermethylation. EPCAM deletion analysis followed by MSH2 methylation testing of the tumor is a fast low-cost procedure that can be used to identify mutations that cause LS. We propose that this procedure be incorporated into clinical genetic analysis strategies and present a decision-support flow diagram for the diagnosis of LS.
Lynch syndrome (LS) is an autosomal dominant inherited cancer syndrome characterized by early-onset cancers of the colorectum and endometrium and tumors of the stomach, pancreas, small intestine, ovary, bladder, and bile duct.1 In the Spanish population, about 2.5% of colorectal cancers are associated with LS.2 The carcinogenetic etiology of this syndrome involves a DNA mismatch repair (MMR) inactivation caused by a germ line mutation of an MMR gene (MLH1, MSH2, MSH6, or PMS2) followed by somatic inactivation of the second allele.1 As a consequence of MMR inactivation, these tumors exhibit microsatellite instability (MSI) and loss of expression of the mutated MMR gene.1 It was recently shown that germ line deletions involving the last exon of the non-MMR gene, EPCAM (OMIM#185535), may silence its neighboring gene, MSH2 (OMIM#609309), which is located 17 kb downstream of EPCAM, via promoter hypermethylation. This epigenetic inactivation seems to be effective only in tissues in which EPCAM is expressed.3,4 The EPCAM gene codes for the epithelial cell adhesion molecule also known as CD326, which is expressed in all normal epithelial cells and in carcinoma tumors.5 Thus, deletions of the last exon of EPCAM constitute a distinct class mutation associated with LS.
Currently, the standard genetic test for LS (point mutation and large-rearrangement analysis of MLH1, MSH2, MSH6, and PMS2) frequently fails to detect a pathogenic mutation. For this reason, we evaluated the association between EPCAM deletions and LS in a Spanish population and its clinical implications.
Materials and Methods
Patients
A total of 501 index subjects from Spanish families suspected of having LS were recruited from the Genetic Counseling in Cancer units of the La Fe and Elche University Hospitals between 2005 and 2009. All subjects fulfilled the Bethesda Guidelines (Amsterdam II criteria cases included).6 The median age at diagnosis was 49 years (range 21–89 years). Clinical and molecular characteristics of this series are listed in Table 1. Patient selection was based on the results of MSI testing, immunohistochemical (IHC) analysis of MMR proteins, and mutation analysis of MLH1, MSH2, and MSH6. Written consent for the diagnostic genetic tests was obtained from each patient.
Table 1.
Clinical and Molecular Characteristics of Index Subjects
| Variables | n | % |
|---|---|---|
| Sex | ||
| Male | 233 | 46.5 |
| Female | 268 | 53.5 |
| Criteria | ||
| Bethesda | 378 | 75.4 |
| Amsterdam II | 123 | 24.6 |
| Tumor type | ||
| CRC | 463 | 92.4 |
| Endometrial | 24 | 4.8 |
| Others | 14 | 2.8 |
| MSI analysis | ||
| Positive | 102 | 20.4 |
| Negative | 371 | 74.0 |
| Not analyzed | 28 | 5.6 |
| IHQ analysis | ||
| Normal expression | 322 | 64.3 |
| MLH1 loss | 44 | 8.8 |
| MSH2 loss | 25 | 5.0 |
| Not analyzed | 110 | 21.9 |
| BRAF V600E mutation and/or MLH1 hypermethylation analysis* | ||
| Positive | 9 | 20.5 |
| Negative | 35 | 79.5 |
Analysis performed to Bethesda cases with loss of MLH1.
Pregenetic Study of Tumor Tissue: Analysis of Expression of MMR Proteins, MSI, the Presence of the BRAF V600E Mutation, and MLH1 Promoter Hypermethylation
In all cases for which tumor tissues were available, we performed MSI analysis and IHC analysis of MMR proteins (n = 483). Tumors from non-Amsterdam II subjects with loss of MLH1 expression were analyzed for the presence of the BRAF V600E mutation and for MLH1 promoter hypermethylation. The presence of the BRAF V600E mutation or MLH1 hypermethylation indicates that the tumor is of sporadic origin and is disregarded in genetic analysis.7 Samples from subjects with loss of MSH2, MSH6, or MLH1 who did not have the BRAF V600E mutation or MLH1 hypermethylation were analyzed for germ line mutations of the corresponding genes. Samples from subjects for whom tumor tissues were not available but who fulfilled the Amsterdam II criteria were also analyzed for germ line mutations (n = 18).
IHC analysis of MLH1, MSH2, MSH6, and PMS2 expression was performed as previously described.2 To assess MSI, monomorphic markers (BAT26, BAT25, NR21, NR24, and NR27) were analyzed as reported by Buhard et al.8 To detect the BRAF V600E mutation, which is located at exon 15, DNA was sequenced as described by Domingo et al.9 Methylation analysis of the MLH1 gene was conducted using methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA; kit ME011, MRC-Holland, Amsterdam, The Netherlands) according to manufacturer's protocol. The MS-MLPA technique for detecting MLH1 hypermethylation has been validated.10
MLH1, MSH2, and MSH6 Germ Line Mutation Analyses
The selection of genes for analysis was based on IHC results. DNA from peripheral blood leukocytes was used for the analysis. Detection of point mutations was performed using PCR and direct sequencing of the whole coding sequence and the intron–exon boundaries of each gene.11,12,13 Large rearrangements (deletions and insertions) were analyzed using MLPA [kits P003 (MLH1–MSH2), P248 (MLH1–MSH2 confirmation), and P008 (PMS2–MSH6); MRC-Holland] according to the manufacturer's recommended procedure.
EPCAM Germ Line Deletion Analysis
Samples from subjects with loss of MSH2 and MSI (n = 25) were analyzed for large deletions of the EPCAM locus using MLPA (kit P072-B1; MRC-Holland) according to the manufacturer's recommended protocol. This kit contains six probes for the region of interest: four are targeted at EPCAM exons 3, 8, and 9, and two are targeted at the intergenic region between the EPCAM and MSH2 loci (Figure 1). Deletions were confirmed and mapped using multiple long-range PCR analysis of genomic DNA (Expand Long-range dNTP Pack; Roche, Mannheim, Germany) and various combinations of primer pairs that targeted the candidate chromosomal region (Figure 2). DNA sequencing was performed to characterize the deletion breakpoints.14
Figure 1.
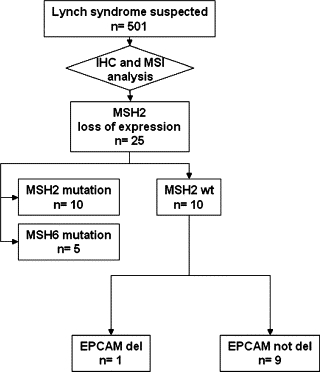
Schematic overview of the MMR and EPCAM genetic analysis. IHC, immunohistochemical; MSI, microsatellite instability.
Figure 2.
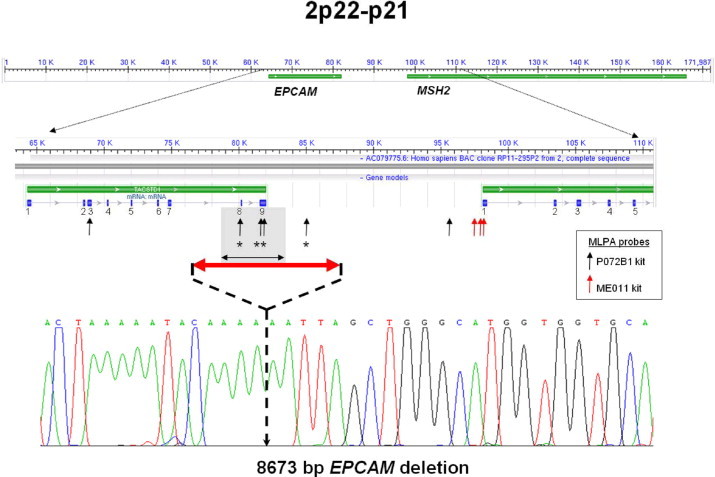
Structural organization of the EPCAM-MSH2 locus. The EPCAM and MSH2 genes are indicated in green, with an expanded region showing positions of relevant exons. Positions of the MLPA probes are indicated by black and red vertical arrows. Arrows with asterisks indicate probes that revealed the EPCAM deletion discovered in this study. The minimal deleted region (5 kb) is depicted as a gray rectangle. The red double arrowhead illustrates the length of the deletion, and a sequencing trace that includes the deletion breakpoint of the 8673-bp deletion is shown.
MSH2 Promoter Hypermethylation Analysis
Subjects with a germ line EPCAM deletion were tested for tumor and peripheral blood leukocyte hypermethylation of the promoter region of the MSH2 gene using MS-MLPA (kit ME011; MRC-Holland). This kit contains three probes for that region. The threshold of methylated versus unmethylated samples was 15% based on a previous study.10
Results
Of the 501 index subjects analyzed, 126 belonged to families that fulfilled the Amsterdam II criteria and 375 subjects fulfilled the Bethesda guidelines. After screening using MSI testing, IHC analysis of MMR proteins, testing for the presence of the BRAF V600 mutation, and MLH1 methylation analysis, samples from 155 subjects were analyzed for point mutations and large rearrangements of MLH1, MSH2, and MSH6 (Figure 1). Mutations of these genes were detected in 51 subjects (32.9%) (details of these mutations will be published elsewhere). Twenty-five subjects had MSH2 loss and MSI. Of these, 10 had a mutation of the MSH2 gene and five had a mutation of the MSH6 gene. The remaining 10 cases did not have alterations of the MSH2 or MSH6 genes. None of the mutated MSH2 or MSH6 genes showed EPCAM deletions, and only one patient with no MSH2–MSH6 mutation harbored a large deletion at this locus (1/10; 10%). This deletion was confirmed by MLPA analysis. We then analyzed samples from the proband's brother, who had colorectal cancer (CRC) that was diagnosed at the age of 36 years, and detected the same EPCAM deletion. This subject also had MSH2 loss and MSI. A subsequent test using the three MS-MLPA probes revealed a clear hypermethylated pattern in the tumors of both subjects. Moreover, no MSH2 methylation was found when peripheral blood leukocytes from these two subjects was analyzed. A mosaic silencing of MSH2 occurs only in tissues that express EPCAM, which is characteristic of these deletions.
Deletion mapping using long-range PCR showed that this deletion extended for 8.7 kb (g.77525_86198del8674 from AC079775.6), including the 5-kb minimal deleted region (EPCAM exons 8 and 9).4
The pedigree of the family is shown in Figure 3. This family fulfilled the Amsterdam I criteria for LS. There were five CRCs in three consecutive generations, four of which were diagnosed before the age of 50. Individuals IV-1 and IV-2 were positive for the EPCAM deletion. Family members are currently undergoing analysis for this mutation to predict their risk of developing LS.
Figure 3.
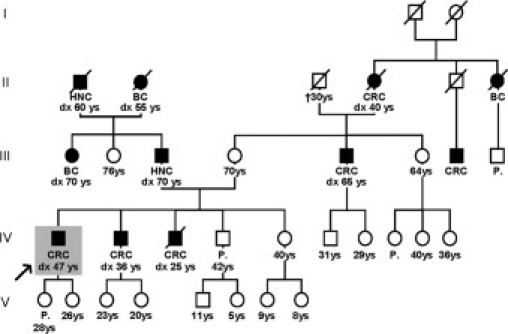
Pedigree of a family with an EPCAM mutation. Family members indicated by solid symbols were diagnosed with CRC at the ages indicated. Index subject IV-1 is indicated by a gray rectangle and an arrow. HNC, Head and neck cancer; BC, Breast cancer; CRC, Colorectal cancer; and P, polyps.
Discussion
The most accepted strategy for LS screening comprises the analysis of samples from patients who fulfill the Bethesda guidelines for DNA MMR status, followed by genetic analysis of MMR genes, if indicated. Markers such as the BRAF V600E mutation and MLH1 promoter hypermethylation may help to distinguish sporadic from familial tumors when MLH1 loss is present in Bethesda-positive cases.7,10 Our laboratory used this strategy to analyze 501 probands recruited over four years from two genetic counseling units of the Comunidad Valenciana (Spain). The efficacy of detection of mutations associated with LS was about 10% for the 501 consultant patients and 33% for the 155 who underwent genetic testing, which is similar to previous results.15 Cost-effective screening methods that are more sensitive and specific than current methods are required.16 Mutations in genes that regulate expression of MMR genes may account for the low rate of detection of mutations associated with LS. Germ line hypermethylation of the MLH1 or MSH2 genes is an atypical alteration.17,18 MSH2 germ line hypermethylation might be caused by the positional effect of large deletions that affect the last exon of the EPCAM gene located 17 kb upstream of MSH2.3 A variety of mechanistically different phenomena classed as negative chromosomal position effects may induce gene silencing through changes in the chromosomal environment rather than by direct targeting of the gene.19 The presence of cis-acting elements would be involved in somatic epigenetic events as reported for MGMT and MLH1 genes.20,21 The EPCAM gene, which lacks a normal polyadenylation signal, may cause mosaic patterns of epigenetic inactivation of its neighboring gene, MSH2, depending on its tissue-specific expression pattern.3 Deletions that remove the transcriptional termination sequences of an upstream gene result in multiple aberrant EPCAM/MSH2 fusion transcripts and consequent inactivation of these two genes. The altered allele may place the MSH2 gene under the control of the EPCAM promoter in cis-.4
We selected subjects with MSH2 loss and MSI who lacked an MSH2 or MSH6 mutation for EPCAM deletion analysis. Heterodimeric proteins containing MSH2 and MSH6 monomers are recognized by the monoclonal antibodies used in the IHC analysis. Mutations in MSH2 or MSH6 may result in IHC loss of both genes.22 Our results showed that 40% (10/25) of subjects with MSH2 loss and MSI lacked a pathogenic mutation. Of these, only one subject had a EPCAM deletion (10%). Likewise, Ligtenberg et al3 detected four mutated cases of 10 unexplained Dutch putative LS families with MSH2 loss and MSI. Niessen et al23 detected three probands with EPCAM deletions of 11 patients from the northern part of the Netherlands who were suspected of having LS. These authors only tested for the presence of the Dutch founder deletion.3 Kovacs et al4 detected four different EPCAM deletions in five of 27 probands selected from clinically well-defined Hungarian LS families. The outcome of our literature review was that only 14 unrelated subjects have been shown to carry one of the six deletions reported to date (Table 2).3,4,21 The alteration reported here is a new deletion and was detected in a family with a very high incidence of early-onset CRC and no extracolonic tumors. The number of potential deletion carriers (ie, unexplained cases of MSH2 loss and MSI) included in our study is small but sufficient to conclude that these types of alterations are present in Spanish LS patients. Taking our results into consideration, the expected proportion of such alterations in cases with MSH2 loss is 10–40%.3 Detection of EPCAM deletions using MLPA followed by MS-MLPA analysis of MSH2 methylation in tumors is a fast low-cost procedure that should be incorporated into clinical LS genetic analysis strategies. We propose a decision-support flow diagram to facilitate genetic analysis, which should include a related cancer patient, if available, to minimize false-positive results. If the two related patients have the same MLPA deletion pattern and the same tumor behavior (eg, MSH2 loss, hypermethylation, and MSI), the alteration can be considered the cause of the LS and therefore genetic counseling should be carried out (Figure 4). Deletion mapping should be performed to characterize and define deletion breakpoints. Although deletion mapping is definitive, it is time-consuming and may delay clinical decisions. For patients in whom MSH2 methylation testing is not possible (eg, inaccessible tumor tissue, low quality or quantity of tumor DNA, etc) deletion mapping is mandatory.
Table 2.
List of Published EPCAM Mutations Related to Lynch Syndrome
| Reference | EPCAM mutation | Frequency* | Ethnicity |
|---|---|---|---|
| Ligtenberg et al3 | c.859-1462_*1999del | 4/10 (40%) | Dutch |
| c.555 + 894_*14194del | 2/? | Chinese | |
| Niessen et al21 | c.859-1462_*1999del | 3/11 (27%) | Dutch |
| Kovacs et al4 | g.77631_92364del14734 | 5/27 (19%) | Hungarian |
| g.77436_86109del8674 | |||
| g.79459_85516del6058 | |||
| g.72468_82822del10355 | |||
| Present study | g.77525_86198del8674 | 1/10 (10%) | Spanish |
Frequency: number of index subjects with EPCAM deletions/number of index subjects analyzed with loss of MSH2 expression and no MSH2/MSH6 mutation.
Figure 4.
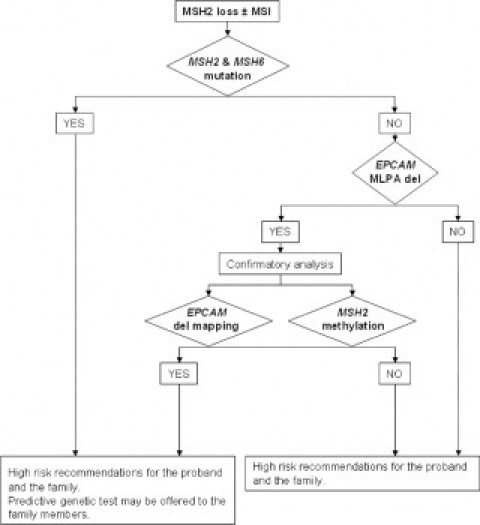
Proposed decision-support flow diagram for incorporating EPCAM deletion testing into the LS genetic diagnosis strategy.
None of the mutated MSH2 or MSH6 genes showed EPCAM deletions, indicating the high specificity of this decision tree for identifying EPCAM deletions. The majority of MSH2 loss subjects who lack pathogenic mutations do not exhibit EPCAM deletions. The underlying causes of these cases are as yet unknown.
In summary, the combination of MLPA analysis for detection of EPCAM germ line deletions and MS-MLPA analysis for MSH2 promoter hypermethylation in tumors can facilitate identification of mutations responsible for LS. These analyses should be incorporated into routine genetic diagnosis protocols for LS.
Acknowledgements
We are indebted to the patients and their families. We thank all members of the Hereditary Cancer Program of the Comunidad Valenciana, Spain.
Footnotes
Supported by the cooperation framework established by the Transversal Cancer Action (ISCIII) and the Biomedical Research Foundation from the Elche University Hospital. C.E. and M.C.A. are recipients of fellowships from the Carolina-BBVA Foundation and Juan Peran-Pikolinos Foundation, respectively.
References
- 1.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 2.Piñol V, Castells A, Andreu M, Castellví-Bel S, Alenda C, Llor X, Xicola RM, Rodríguez-Moranta F, Payá A, Jover R, Bessa X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 3.Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, Tsui WY, Kong CK, Brunner HG, van Kessel AG, Yuen ST, van Krieken JH, Leung SY, Hoogerbrugge N. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 4.Kovacs ME, Papp J, Szentirmay Z, Otto S, Olah E. Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat. 2009;30:197–203. doi: 10.1002/humu.20942. [DOI] [PubMed] [Google Scholar]
- 5.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 6.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasen HF, Möslein G, Alonso A, Bernstein I, Bertario L, Blanco I, Burn J, Capella G, Engel C, Frayling I, Friedl W, Hes FJ, Hodgson S, Mecklin JP, Møller P, Nagengast F, Parc Y, Renkonen-Sinisalo L, Sampson JR, Stormorken A, Wijnen J. Guidelines for the clinical management of Lynch syndrome (hereditary non-polyposis cancer) J Med Genet. 2007;44:353–362. doi: 10.1136/jmg.2007.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF, Duval A, Hamelin R. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol. 2006;24:241–251. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- 9.Domingo E, Laiho P, Ollikainen M, Pinto M, Wang L, French AJ, Westra J, Frebourg T, Espín E, Armengol M, Hamelin R, Yamamoto H, Hofstra RM, Seruca R, Lindblom A, Peltomäki P, Thibodeau SN, Aaltonen LA, Schwartz S., Jr BRAF screening as a low-cost effective strategy for simplifying HNPCC genetic testing. J Med Genet. 2004;41:664–668. doi: 10.1136/jmg.2004.020651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Carbonell L, Alenda C, Paya A, Castillejo A, Barbera VM, Guillen-Ponce C, Rojas E, Acame N, Gutierrez-Aviño FJ, Castells A, Llor X, Andreu M, Soto JL, Jover R. Methylation analysis of MLH1 improves the selection of patients for genetic testing in Lynch syndrome. J Mol Diagn. 2010;12:498–504. doi: 10.2353/jmoldx.2010.090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 12.Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 13.Hampel H, Frankel W, Panescu J, Lockman J, Sotamaa K, Fix D, Comeras I, La Jeunesse J, Nakagawa H, Westman JA, Prior TW, Clendenning M, Penzone P, Lombardi J, Dunn P, Cohn DE, Copeland L, Eaton L, Fowler J, Lewandowski G, Vaccarello L, Bell J, Reid G, de la Chapelle A. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. doi: 10.1158/0008-5472.CAN-06-1114. [DOI] [PubMed] [Google Scholar]
- 14.van der Klift H, Wijnen J, Wagner A, Verkuilen P, Tops C, Otway R, Kohonen-Corish M, Vasen H, Oliani C, Barana D, Moller P, Delozier-Blanchet C, Hutter P, Foulkes W, Lynch H, Burn J, Möslein G, Fodde R. Molecular characterization of the spectrum of genomic deletions in the mismatch repair genes MSH2. MLH1, MSH6, and PMS2 responsible for hereditary nonpolyposis colorectal cancer (HNPCC) Genes Chromosomes Cancer. 2005;44:123–138. doi: 10.1002/gcc.20219. [DOI] [PubMed] [Google Scholar]
- 15.Lagerstedt Robinson K, Liu T, Vandrovcova J, Halvarsson B, Clendenning M, Frebourg T, Papadopoulos N, Kinzler KW, Vogelstein B, Peltomäki P, Kolodner RD, Nilbert M, Lindblom A. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 16.Mvundura M, Grosse SD, Hampel H, Palomaki GE. The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 2010;12:93–104. doi: 10.1097/GIM.0b013e3181cd666c. [DOI] [PubMed] [Google Scholar]
- 17.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 18.Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 19.Barbour VM, Tufarelli C, Sharpe JA, Smith ZE, Ayyub H, Heinlein CA, Sloane-Stanley J, Indrak K, Wood WG, Higgs DR. alpha-thalassemia resulting from a negative chromosomal position effect. Blood. 2000;96:800–807. [PubMed] [Google Scholar]
- 20.Ogino S, Hazra A, Tranah GJ, Kirkner GJ, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Meyerhardt JA, Hunter DJ, Fuchs CS. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28:1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 21.Samowitz WS, Curtin K, Wolff RK, Albertsen H, Sweeney C, Caan BJ, Ulrich CM, Potter JD, Slattery ML. The MLH1-93 G>A promoter polymorphism and genetic and epigenetic alterations in colon cancer. Genes Chromosomes Cancer. 2008;47:835–844. doi: 10.1002/gcc.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halvarsson B, Lindblom A, Rambech E, Lagerstedt K, Nilbert M. Microsatellite instability analysis and/or immunostaining for the diagnosis of hereditary nonpolyposis colorectal cancer? Virchows Arch. 2004;444:135–141. doi: 10.1007/s00428-003-0922-z. [DOI] [PubMed] [Google Scholar]
- 23.Niessen RC, Hofstra RM, Westers H, Ligtenberg MJ, Kooi K, Jager PO, de Groote ML, Dijkhuizen T, Olderode-Berends MJ, Hollema H, Kleibeuker JH, Sijmons RH. Germline hypermethylation of MLH1 and EPCAM deletions are a frequent cause of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:737–744. doi: 10.1002/gcc.20678. [DOI] [PubMed] [Google Scholar]


