Abstract
Interpretation of capillary electrophoresis results derived from multiplexed fluorochrome-labeled primer sets can be complicated by small peaks, which may be incorrectly interpreted as clonal T-cell receptor-γ gene rearrangements. In this report, different assay designs were used to illustrate how design may adversely affect specificity. Ten clinical cases, with subclonal peaks containing one of the two infrequently used joining genes, were identified with a tri-color, one-tube assay. The DNA was amplified with the same NED fluorochrome on all three joining primers, first combined (one-color assay) and then amplified separately using a single NED-labeled joining primer. The single primer assay design shows how insignificant peaks could easily be wrongly interpreted as clonal T-cell receptor-γ gene rearrangements. Next, the performance of the one-tube assay was compared with the two-tube BIOMED-2-based TCRG Gene Clonality Assay in a series of 44 cases. Whereas sensitivity was similar between the two methods (92.9% vs. 96.4%; P = 0.55), specificity was significantly less in the BIOMED-2 assay (87.5% vs. 56.3%; P = 0.049) when a 2× ratio was used to define clonality. Specificity was improved to 81.3% by the use of a 5× peak height ratio (P = 0.626). These findings illustrate how extra caution is needed in interpreting a design with multiple, separate distributions, which is more difficult to interpret than a single distribution assay.
Evaluation of clonal T-cell receptor γ gene rearrangements (TCRγGR) using various PCR-based methods is useful in the diagnosis of T-cell malignancies. The TRG@ gene is often used to assess T-cell clonality due to its simplistic structure as compared to the TRA@ and TRB@ genes. Within the TRG@ gene locus on chromosome 7p14, there are 11 variable region genes and five joining region genes involved in the formation of gene rearrangements in tumors. No diversity region genes are present. Various methods have been used in the detection of clonal PCR products including agarose and polyacrylamide gel electrophoresis,1,2 denaturing gradient gel electrophoresis (DGGE),3 heteroduplex analysis4 and single strand conformational polymorphism analysis.5,6 The most common detection method involves fluorochrome-tagged primers and capillary electrophoresis,7,8,9,10 which has good sensitivity and quick turn-around time.
While the method of detection is important for accurate testing, proper PCR assay design is of utmost importance in reducing the number of false positive and false negative results. Multiplex versus monoplex reaction(s), product sizes, and the number of targets amplified (common versus rare rearrangements) are issues that must be thoroughly addressed before detection with capillary electrophoresis. The distribution of primers over the gene segments must be considered, for if not enough segments of the gene are amplified, false negative results may occur. For example, the JγP rearrangement has been unnecessarily excluded in some primer sets due to its rare involvement in T-cell neoplasms and the supposed risk of identifying false positive results.11 The risk however, is based more on assay design, as will be seen later in the results. JγP T-cell receptor gamma primer alignments are available at (http://www.unmc.edu/media/pathology/jp.pdf, last accessed on June 29, 2010). Secondly, the number of primers in a single tube reaction may also affect results. It has been suggested that specific combinations of primers may result in competitive inhibition of the formation of products.11 Finally, the number of size distributions of amplicons may also make interpretation difficult. Multiple protocols have been described for the detection of TCRγGR using capillary electrophoresis. The protocols vary from those that may contain one normal distribution of TCRγGR with a single fluorochrome to protocols with multiple tubes that have multiple polyclonal distributions (eg, BIOMED-2).11 If there are multiple ranges of product sizes, then there will be a reduced number of events to produce polyclonal Gaussian distributions for each individually labeled gene segment. As a result, it may be difficult to accurately evaluate small subclonal peaks in a reduced polyclonal background with the uncommonly used variable or joining gene segments.
Most laboratories use labeled joining region primers in capillary electrophoresis. There are three possible approaches to evaluate TCRγGR using multiple fluorescent labeled joining region primer sets. The first approach consists of a complete multiplex assay using multiple variable and joining region primers within a single tube as previously described.7,12 In this tri-color approach, each primer in a set of joining region primers is labeled with a different fluorochrome tag (Figure 1A). The second complete multiplex approach is similar; however, instead of a different fluorescent label on each primer, the joining primers are labeled with the same fluorochrome tag (one-color, eg, NED) (Figure 1B).7 The third approach involves a partial multiplex reaction using a single fluorochrome-labeled primer for each joining gene segment in separate tubes (Figure 1C). The first goal of this report is to show, in the figures that follow, how assay design can heavily influence the likelihood of generating pseudoclonal peaks that can result in a false positive interpretation of a TCRγGR. The second goal of this work is to compare the performance of two assays with divergent designs; a one-tube assay with one size distribution versus the two-tube TCRG Gene Clonality Assay based on the BIOMED-2 design.11
Figure 1.
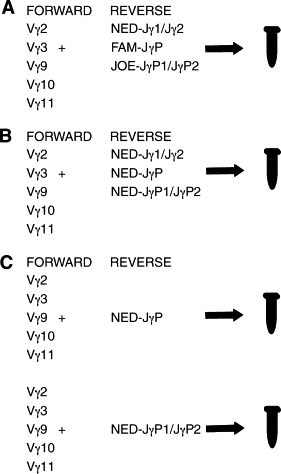
Schematic diagram of multiplex reactions used in Part 1. A: Tri-color assay with a complete multiplex reaction in one tube with three different fluorescently labeled Jγ joining region primers (Jγ1/Jγ2-NED, JγP-FAM, and JγP1/JγP2-JOE) mixed with five unlabeled Vγ variable region primers. B: One-color assay with a similar reaction as in (A); however, all three joining region primers have the same fluorochrome tag (NED). C: Partial multiplex assay is performed in which either JγP1/JγP2-NED or JγP-NED is combined with the five variable region primers in separate tubes.
Materials and Methods
Part I: Illustrating Effects of Design on a TCRγ Assay
Case Selection
Clinical cases were selected, that have not had nor developed a diagnosis of T-cell lymphoma, but which did contain small subclonal peaks, consisting of either a small JγP1/JγP2 (five cases) or JγP (five cases) gene segment, arising within a larger, higher amplitude, polyclonal or oligoclonal background of Jγ1/Jγ2 TCRγGR. DNA was derived from eight fresh tissue cases (peripheral blood, frozen tissue, or bone marrow) and two biopsies from formalin-fixed paraffin-embedded tissue. Three patients had reactive large granular lymphocytosis, three patients had an isolated cytopenia (neutropenia, anemia, or thrombocytopenia) of unknown cause, two patients had atypical lymphoid hyperplasia, one patient had a benign inflammatory process in the skin, and one patient had hairy cell leukemia.
PCR Design
PCR reactions were performed as outlined in Figure 1. First, the cases were amplified in a single multiplex tri-color assay using five unlabeled variable region primers (Vγ2, Vγ3, Vγ9, Vγ10, Vγ11) and three separately labeled joining region primers (Jγ1/Jγ2-NED, JγP1/JγP2-JOE, JγP-FAM) as previously described by Lawnicki et al (Figure 1A).12 These reactions yield products of 190 ± 20 nucleotides in the usual normal distribution. In evaluating for clonal populations with capillary electrophoresis, we used our previously described formula: ratio of peak to background (RPB) must be greater than two times the highest polyclonal peak height, which defines that at least a 2% clonal population is present.7 The height of peaks outside the normal distribution must first be added to the height of the distribution before calculating the ratio.7 Second, the DNA was then amplified with five upstream variable region primers and all three downstream joining primers (Jγ1/Jγ2-NED, JγP1/JγP2-NED and JγP-NED) each labeled with the same fluorochrome (NED) in a single tube (one-color multiplex reaction, Figure 1B). Finally, to illustrate how design flaws can cause interpretation problems, we subsequently analyzed these cases with the individual joining primer in separate tubes (partial multiplex reactions). This third amplification contained all five variable region primers and either a single labeled JγP1/JγP2-NED or JγP-NED primer in separate tubes (Figure 1C). PCR reaction mixes were composed of Perkin Elmer PCR buffer I (Perkin Elmer, Waltham, MA), 100 μmol/L dNTPs, 0.6 μmol/L each Vγ primer, 0.6 μmol/L each Jγ primer (in both complete multiplex or single multiplex assay), and 1.25 U Platinum Taq (Invitrogen, Carlsbad, CA). Amplification was performed on the Omnigene (Omnigene Bioproducts, Woodburn, MA) or MJ Dyad thermocyclers (BioRad, Hercules, CA) using the same protocol for all three approaches. The initial denaturing step (9 minutes at 94°) was followed by 30 cycles with the following time and temperature settings: denaturing, 94°C (75 seconds); annealing, 60°C (75 seconds); extension, 72°C (10 seconds plus 1 second for each cycle). After the reaction, the products were diluted 1:10 with water, and were analyzed using capillary electrophoresis on an ABI 3130xL instrument (Applied Biosystems, Foster City, CA).
Part 2: Comparison of One-Tube Tri-Color Assay with TCRG Gene Clonality Assay
Case Selection
DNA was obtained from 44 clinical cases that had morphological diagnoses and had TRB@ Southern blot results previously performed with a constant region probe. The cases did not have the diagnosis established with either of the PCR methods compared in this manuscript. The series included 28 cases of T-cell malignancy, of which 26 cases had a TRB@ gene rearrangement (Table 1A). One of the two TRB@ negative cases of peripheral T-cell lymphoma had a complex cytogenetic abnormality. The remaining case was regarded as containing lymphoma below the detection limits of Southern blotting as a second biopsy showed definitive peripheral T-cell lymphoma. The series also included eight cases of B-cell malignancy and eight benign specimens, of which six of eight cases had negative TRB@ Southern blot results in each group (Table 1A). B-cell malignancies were included because amplification of small numbers of intermixed T-cells can sometimes result in interpretations of “clonal” TCRγGR that do not represent an aberrant TCRγGR in the B-cell tumor itself. Concurrent IgH and TCRγGR are known to often occur in lymphoblastic lymphomas. The most common source of tissue was from lymph node biopsies (Table 1B).
Table 1A.
Pathologic Diagnoses
| T-cell malignancies | N = 28 |
|---|---|
| Peripheral T-cell lymphoma, not otherwise specified | 18 |
| Peripheral T-cell lymphoma, angioimmunoblastic | 2 |
| Large granular lymphocytic leukemia | 2 |
| Anaplastic large cell lymphoma | 1 |
| Lymphoblastic lymphoma | 1 |
| Subcutaneous panniculitis-like | 1 |
| Prolymphocytic leukemia | 1 |
| Acute lymphoblastic leukemia | 1 |
| Mycosis fungoides | 1 |
| B-cell malignancies | N = 8 |
|---|---|
| Follicular lymphoma | 3 |
| Diffuse large cell lymphoma | 2 |
| Lymphoblastic lymphoma | 1 |
| Chronic lymphocytic leukemia | 1 |
| Hodgkin's lymphoma | 1 |
| Benign disorders | N = 8 |
|---|---|
| Atypical lymphoid hyperplasia | 4 |
| Reactive lymphocytosis | 2 |
| Follicular hyperplasia | 1 |
| Negative marrow | 1 |
| Table 1B. Tissue sources N = 44 | |
|---|---|
| Lymph node | 27 |
| Peripheral blood | 8 |
| Bone marrow | 4 |
| Skin | 2 |
| Other-uterus, pheresis product, and pleural fluid | 3 |
PCR Reactions
Archival DNA (1 μg) was amplified for TCRγGR in a single tube with the tri-color multiplex PCR assay [Jγ1/Jγ2 NED (black), JγP1/JγP2 JOE (green), and JγP FAM (blue) labeled primers and five unlabeled variable region primers].7 A one-color method with the same NED label on the same three Jγ primers was also performed as it is easier to interpret.7 Secondly, the BIOMED-2-based TCRG Gene Clonality Assay was used to amplify 1 μg of DNA per the manufacturer's directions in a 50 μl reaction using the following cycling program: 95°C 7 minutes, 35 cycles of 95°C 45 seconds, 60°C 45 seconds, 72°C 90 seconds; final 72°C 90 seconds. Whereas the exact primer sequences are proprietary information, the assay is based on the BIOMED-2 primers described by van Dongen et al.11 The products, which were expected to range from 80 to 200 nucleotides and 145 to 255 nucleotides in the two tubes, were analyzed by capillary electrophoresis on an ABI 3130XL instrument using 3130 Performance Optimized Polymer-4 (Applied Biosystems, CA). A positive result was defined as a peak height with a ratio greater than two times (2×) the polyclonal background peak height (RPB) for the one-tube methods.7 Two RPB methods of greater than two (2×) or five times (5×) were used for the TCRG Gene Clonality Assay. Oligoclonal results, defined as cases with three or more peaks present, were regarded as negative for the calculations of sensitivity and specificity. The assay results were then compared with the original morphological diagnoses to determine sensitivity and specificity. The presence of pseudoclonal spikes was defined for each method as: One-tube method, peaks between 1× and 2× RPB; TCRG Gene Clonality Assay, peaks between 2× and 5× RPB.
Statistical Analysis
The Chi Square Goodness of Fit Test statistic was calculated to assess the significance of the results.
Results
Part I: Illustrating Effects of Design on a TCRγ Assay
Examples of positive clonal results with Jγ1/Jγ2, JγP1/JγP2 and JγP using the one-tube, tri-color method are identified in Figure 2A. For a negative polyclonal result, peripheral blood lymphocyte DNA from a normal patient sample demonstrates polyclonal distributions in both the tri-color and one-color approaches (Figure 2B). An expected polyclonal background is identified with the separate JγP1/JγP2 assay (Figure 2B). However, the separate JγP assay shows a more prominent peak in the central portion of the curve, that is now easily more than two times the height of the polyclonal background, which may incorrectly suggest a clonal rearrangement (Figure 2B). This JγP peak, which likely represents canonical rearrangements, corresponds to the same small blue peak in the tri-color assay, wherein the tri-color assay clearly shows it does not exceed the overall Jγ1/Jγ2 polyclonal distribution and therefore contains far less than a 2% population necessary for interpreting it as a clonal result. This limited JγP distribution observed is due to the known rarity of JγP rearrangements used by T-cells, thereby resulting in the low number of events in the JγP polyclonal distribution.
Figure 2.
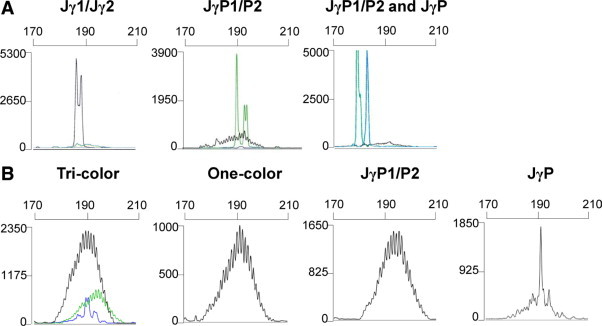
A: Three representative cases of clonal rearrangements from paraffin-embedded T-cell lymphomas, using either Jγ1/Jγ2 (left panel), JγP1/P2 (biallelic, middle panel), or JγP1/P2 plus JγP (biallelic, right panel). Each reaction contained all five forward Vγ variable region primers and all three differentially labeled Jγ joining primers. B: Polyclonal peripheral blood lymphocyte DNA amplified with five forward Vγ variable region primers and labeled Jγ joining primers. Tri-color: a complete multiplex reaction using three separate fluorochrome tags on joining primers (Jγ1/Jγ2-NED-black, JγP-FAM-blue, and JγP1/JγP2-JOE-green) produces three polyclonal populations of one size range. One-color: the same multiplex reaction is performed; however, all three joining primers are labeled with the same NED fluorochrome, demonstrating a combination of all three polyclonal populations into one distribution. JγP1/P2 or JγP: DNA was amplified using five variable primers and a single joining region primer, JγP1/JγP2-NED or JγP-NED, respectively. Note the single peak in the JγP figure, which could be mistaken for a clonal rearrangement if not compared to the larger distribution of rearrangements seen in the tri-color or one-color figure. (x axis scale = bp; y axis = relative fluorescence units).
Illustrative Case Series of Effects of Design
Ten clinical cases are shown in which insignificant small green JγP1/JγP2 (Figure 3A, first column) or blue JγP (Figure 3B, first column) peaks are present in the tri-color assay. While the cases are confirmed as polyclonal by the one-color multiplex reactions (second column), the cases give prominent peaks in separate assays using only the single joining region primer (third column). Using the standard definition for clonality described in the methods (2× RPB), these insignificant peaks would become false positive pseudoclonal peaks with this separate primer approach (Figure 3, A and B; third columns).
Figure 3.
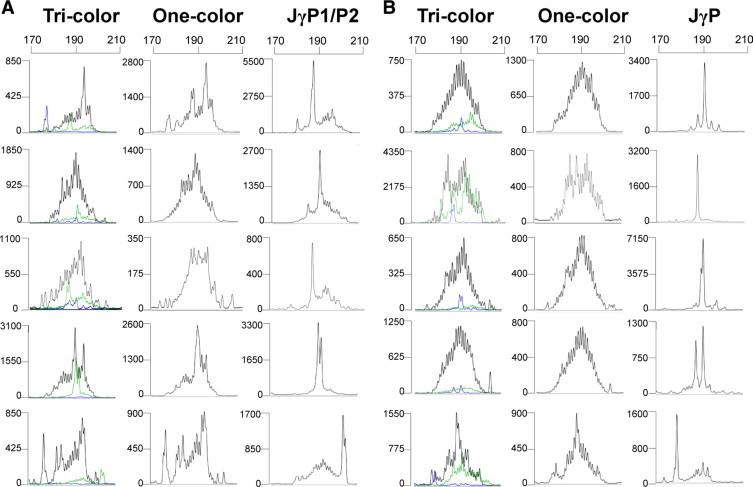
Ten clinical cases amplified by three approaches. Tri-color (left column): the 10 cases are amplified with Jγ1/Jγ2-NED, JγP1/JγP2-JOE, and JγP-FAM labeled primers in a single tube labeled with three separate fluorochrome tags. Note that amplification with JγP1/2 (green) in five cases (A) or JγP (blue) in five cases (B) yields a small population of rearrangements with no peak greater than two times the height of the Jγ1/Jγ2 polyclonal background. One-color (middle column): the three NED-labeled joining primers yield a single polyclonal distribution. Separate JγP1/P2 or JγP (right column): Five variable region primers and a single fluorescent NED-labeled joining primer [JγP1/P2 (A) or JγP (B)]. Each case now shows one or two peaks that are more than two times the JγP1/P2 (A) or JγP (B) polyclonal background. The electropherograms in the right hand column would produce an interpretation of a false-positive clonal TCRγGR as a result of assay design using the same primers. (x axis scale = bp; y axis = relative fluorescence units).
Part 2: Comparison of One-Tube Method with the TCRG Gene Clonality Assay
The sensitivity between both methods was not significantly different (χ2 0.35, P = 0.55). Clonal TCRγGR results were seen in 92.9% (26 of 28) of the cases of T-cell malignancies by the One-tube method whereas 96.4% (27 of 28) were positive by the TCRG Gene Clonality assay, regardless of whether a RPB of 2× or 5× was used for assessing the peaks in the latter assay. A JγP rearrangement, that was seen with the one-tube method in a case of peripheral T-cell lymphoma, was not identified with the TCRG Gene Clonality Assay, as the JγP primer is not included in the design of the assay (Figure 4D). The case status (clonal) was not affected as a biallelic TCRγGR with a common joining gene, which occurs about 35% of the time, was detected by both methods (Figure 4D).
Figure 4.

Representative comparison of cases analyzed with the tri-color, one-color, and the IVS TCRG Gene Clonality Assay (Tubes A and B). A: PBL, 1 μg DNA. Note pseudoclonal spikes in tubes A and B of the TCRG Gene Clonality Assay. B: Diffuse large B-cell lymphoma, Southern blot result was negative for a TRB@ gene rearrangement. Both the one-tube assays and the BIOMED-2 assay show an oligoclonal result with multiple pseudoclonal spikes present. C: Reactive lymphocytosis, Southern blot result negative for a TRB@ gene rearrangement. Whereas the one-tube assays are negative, two peaks greater than 2× RPB are identified in IVS tube A (arrows) that would be interpreted as clonal using the 2× RPB threshold, but would be interpreted as negative if a 5× RPB threshold was used. D: Peripheral T-cell lymphoma, Southern blot result positive for a TRB@ gene rearrangement. Discordant results with a biallelic Jγ1/Jγ2 and a JγP rearrangement identified in the one-tube, tri-color method, the latter not identified in the IVS System. The peak in tube B is one-fifth the intensity of the clonal peak in A and is not JγP.
Specificity was assessed in the cases of B-cell malignancy combined with benign tissues. The specificity of the one-tube method was 87.5% (14 of 16 cases) whereas the TCRG Gene Clonality Assay had a specificity of only 56.3% (9 of 16 cases) when using the RPB of 2× (χ2 3.86, P = 0.049). Specificity was improved to an equivalent 81% (13 of 16 cases) when an RPB of 5× was used in the TCRG Gene Clonality Assay (χ2 0.237, P = 0.626). There was greater specificity in the one-tube method because there were fewer spikes outside the polyclonal background. Seven percent of the cases had spikes (3 of 44 cases) in the one-tube assay, whereas the TCRG Gene Clonality Assay had spikes in 38.6% of cases (17 of 44 cases) (χ2 10.6; P = 0.001). Tube B in the TCRG Gene Clonality Assay had the greatest percentage (88%) of the spikes in 15 of 17 cases.
In the analysis of peripheral blood lymphocytes (PBLs) one can see the difficulty of using a uniform peak height ratio rule for two differently designed assays. While there are no prominent peaks in PBL with the one-tube assay, a prominent peak (spike) could be called positive with a RPB ratio of greater than 2 times the height of the polyclonal background in the TCRG Gene Clonality Assay using the same DNA (Figure 4A). In a case of TCRβ Southern blot negative diffuse large B-cell lymphoma, multiple spikes are seen in both IVS tubes that would be regarded as oligoclonal, thus, there would be no adverse clinical outcome (Figure 4B). However, in a case of reactive lymphocytosis that is negative by Southern blot and negative with the one-tube assays, there are two prominent spikes between 2× and 5× RPB in the TCRG Gene Clonality Assay that could result in a clonal interpretation by a lab using a 2× RBP threshold (Figure 4C).
Discussion
T-cell receptor gamma gene rearrangement analysis is useful as an ancillary test in the diagnosis of T-cell neoplasms. However, interpretation can be complicated by small, but observable, peaks, which may be incorrectly interpreted as clonal rearrangements. This phenomenon is most likely to occur in assays where multiple tubes, size distributions and colors are used.
The number of primers, size distributions and reaction tubes varies between methods used for the evaluation of TCR-γGR. Table 27,8,9,10,11,12,13,15,22,23,24,25,26,27,28,29,30,31,32,33,34,35 shows the increasing complexity one can expect from the way assays are designed, which can result in an increase in the difficulty in interpretation. Detailed information regarding primer locations for each Vγ and Jγ gene segment (V1-8, V9, V10, V11, J1&2, JP1 & JP2, and JP, as well as the legend for the T-cell receptor gamma primer alignments) of the referenced authors’ protocols are available by following the related links at the following website http://www.unmc.edu/pathology/index.cfm?conref=74, last accessed on June 29, 2010. Complete primer mixes in one-tube, partial multiplex, and monoplex (single primer pair) assays have been used, of which the latter have reported high specificities. Benhattar et al2 has used a single primer set, targeting a Vγ4-5 region and Jγ1-Jγ2 region, yielding product sizes ranging 160 to 190 bp by gel electrophoresis, which resulted in a specificity of 100% as compared to Southern analysis.2 However, other groups using multiple primer sets have described the presence of prominent peaks in either normal or benign, reactive specimens. Lou et al,10 in comparing detection by capillary electrophoresis and DGGE, used two partial multiplex reactions, with one tube containing Vγ1-8 and three Jγ primers (Jγ1/2, JγP1/2 and JγP) and the other tube containing the Vγ9, Vγ10, Vγ11 and Vγ12 primers with the same three Jγ primers. Interestingly, two cases of benign control specimens were described in which “dominant” peaks appeared against the polyclonal background (one in each partial multiplexed tube and one in each different size distribution).10 According to the authors, these would have been considered indeterminate peaks, based on their definition of clonality (peak height ratio = peak height/average of two adjacent peak heights).10 Similarly, Lee et al13 reported in a one-color assay with one size distribution that up to 20% of benign specimens may have a “pseudo-spike” defined as small peaks with a ratio of 0.5 to 1.5 compared to the polyclonal background.
Table 2.
Characteristics of Capillary Electrophoresis Protocols
 |
Vega et al9 described a protocol using four separately labeled variable region primers (Vγ1, Vγ2, Vγ3, Vγ4) and four joining region primers (Jγ1, JγP, JγP1, JγP2) in a single multiplex PCR reaction that produces four different size distributions. The authors described the formation of “pseudoclonality” in inflammatory or reactive conditions.9 It is well known that false positive pseudoclonal peaks may occur when small numbers of T-cells are present. This stresses the need for a large polyclonal background with which to compare a potential clonal peak.
Subsequently, Shadrach et al14 showed that assays using separate tubes with fluorescent-labeled variable region primers can lead to false positive results. Small spikes may be interpreted as clonal populations when there are insufficient polyclonal T-cell distributions. They demonstrated how a rare Vγ11 region peak, identified when separate fluorochrome-labeled primers were used, was rendered insignificant if included with other variable region primers containing the same fluorochrome label (one-color approach) in a multiplex reaction. We have shown how similar effects occur when separating labeled joining region primers. Thus, one can expect interpretation difficulties when either rare or infrequent joining or variable labeled gene segments (JγP, Vγ11, JγP1/JγP2, Vγ10) are displayed in separate distributions.
Duplicate analysis on each specimen is one good solution to help reduce false positive results in any assay design. In a seminested, monoplex study of cutaneous T-cell lymphomas and benign lesions, three cases of psoriasis or eczematous dermatitis demonstrated “pseudo-monoclonality” in which a small clonal, dominant population was initially present. However, repeat PCR experiments showed inconsistent differences in the size of the “clonal” population, leading the authors to both define these populations as “pseudo-monoclonal” and recommend repeat analysis.15 Duplicate analyses alone will not overcome all design limitations of a multiple distribution assay.
In the initial report of the BIOMED-2 Concerted Action,11 a two-tube system was used in which one tube contained two variable region primers (Vγ1–8 and Vγ10) and a second tube contained two other variable region primers (Vγ9 and Vγ11). While both tubes contained the same joining region primers (Jγ1/2 & JγP1/2), JγP was not included in the BIOMED-2 primer mix11 to help avoid false positive results, despite the fact that JγP rearrangements are seen in 3% of T-cell neoplasms.12 There are eight size distributions and two fluorochromes that spread out small populations of polyclonal cells in the commercial version of the assay known as the TCRG Gene Clonality Assay. Separating TCRγGR amplicons into multiple distributions decreases the symmetry of the normal distributions, which increases the likelihood of calling an oligoclonal peak as clonal among the few T-cells in the polyclonal background, especially in Tube B. Table 312 shows the low frequency of TCRγGR of the uncommonly used gene segments expected in tube B, where Vγ11/Jγ1/2, Vγ11/JγP1/2 and Vγ9/JγP1/2 include 3%, 1% and 3% of TCRγGR respectively, calculated from our previous report of gene segment use in TCRγGR.12 In contrast, there is only one distribution in Tube A with Vγ10/JγP1/2 TCRγGR that has a small number (2%) of expected rearrangements. Based on these data one would predict a priori that the greatest difficulty in interpretation would occur in tube B. Indeed, in the BIOMED-2 report, it was noted that there is a potential risk of false positive results due to over interpretation of small peaks, most notably with variable region primers, which amplify infrequently rearranged variable regions such as Vγ10, Vγ9 and especially the rarely used Vγ11.11 This may be especially true when T-cells are limited in the DNA sample.
Table 3.
Expected Percentage of TCRγGR in Each Distribution of the TCRG Gene Clonality Assay
| Tube A | Vγ10 J1γ1/2 | Vγ10 JγP1/2 | Vγ1-8 Jγ1/2 | Vγ1-8 JγP1/2 |
| 15% | 2% | 46% | 8% | |
| Tube B | Vγ11 + Jγ1/2 | Vγ11 + JγP1/2 | Vγ9 + Jγ1/2 | Vγ9 + JγP1/2 |
| 3% | 1% | 17% | 3% | |
Percentages are calculated from the distribution of variable and joining gene segments used in TCRγGR from Lawnicki et al.12 The percentages do not add up to 100% because JγP rearrangements are not included in the calculations as JγP is not part of the TCRG Gene Clonality Assay.
Despite this design limitation, no specific guidelines for interpretation of clonality in TRG@ assays were given in the initial BIOMED-2 report for the two-tube system. Instead, the authors suggested that in questionable cases, subsequent studies using heteroduplex analysis should be undertaken, but no percentage of resolution of cases was provided.11 However, adding a gel may not be a practical or economical approach for many laboratories. Other BIOMED-2 reports in T-cell malignancies16 and reactive tissues17 have similarly provided no rule set or ratio definition of a clonal result, other than to describe it to be “clear” or “unequivocal.”17 In a follow-up BIOMED report, comments on unclear results stated that “repeat analysis and consulting experienced labs was preferred over speculation about peak height, surface-under-curve, …”18
Other authors have tried to fill the need and defined clonality in various ways for the BIOMED-2 system. For example, in a study detecting circulating clonal T cells in patients with nephrogenic systemic fibrosis, a predominant peak was considered clonal if it was three times the amplitude of the third highest peak in the polyclonal background distribution.19 In a study of anaplastic large cell and peripheral T-cell lymphomas, clonality was assigned if the height of at least one, but not more than two, distinct peak(s) exceeded that of the polyclonal background by at least twofold.20 Likewise, in a study evaluating cutaneous T-cell lymphomas by comparing DGGE and capillary electrophoresis of BIOMED-2 products, a similar rule was used.21 The authors also recommended duplicate analyses for each case to reproduce the result and to resolve ambiguous or oligoclonal cases.21 Unexpectedly, the authors observed that 14% of patients with benign inflammatory disorders had a positive or clonal Genescan result using the BIOMED-2 primers. These “pseudomonoclonal” peaks were thought by the authors to result from small populations of T cells containing rare rearrangements or a small amount of T cell-derived DNA.21 Interestingly, three cases of the benign inflammatory disorders demonstrated a polyclonal result when analyzed by DGGE. They surmised that the BIOMED-2 protocol was more sensitive because of the use of two separate tubes, thereby preventing competition between rearrangements.21 We would argue that competitive amplification is necessary within a large polyclonal population to prevent false positive results. Compared to capillary electrophoresis, DGGE is known to be more robust in separating polyclonal rearrangements using the unique sequences present, something capillary electrophoresis cannot do, as it separates sequences only by length.3,7
Due to the recognized need for standardization of interpretation of T cell rearrangements using the BIOMED-2 system, a new computerized, statistical method was developed using a best-fit curve analysis.22 When outlier peaks were compared to the best-fit curve of a polyclonal distribution, a χ2 error value of ≥1 suggested a clonal population was present when using Vγ1-8 primers. However, the authors cautioned that higher cut-offs may be necessary for different primer combinations using more rarely rearranged genes such as Vγ9.22 We believe this would be equivalent to saying that one may need different peak ratios to determine a positive result when multiple distributions of uncommonly used gene segments are present in an assay. In a series of 80 samples, the authors then compared their computational method to previously-suggested analysis methods including relative peak height, relative peak ratio, and peak height ratio; and compared the results to DGGE and/or Southern blot. Based on their results, relative peak height, which was defined similar to our RPB formula, was most concordant with their computational method. However, there were many false positives (compared to DGGE/SB) with the BIOMED-2 system, generating a false positive percentage that ranged between 10 to 24% for the various analysis methods.22 This software method represents a second method and perhaps the most objective solution to help maximize specificity.
The need for additional rules for ratio of peak heights is increased with the number of fluorochromes and polyclonal size distributions that are designed in an assay. If all TCRγGR are amplified in a single distribution it is easy to define a positive result using a singular ratio of two or three times the height of the polyclonal background. However, the inclusion of multiple size distributions forces a decision as to whether to define a positive result using a common peak height ratio or whether it is necessary to use a different ratio for different sized fluorochrome labeled distributions. Recently, Patel et al compared the performance of the BIOMED-2 assay with a four-tube, four-distribution, laboratory-developed system with fluorescent labeled variable region primers.23 The variable region primers, which produce four different product sizes, were previously described in our report of a DGGE assay3 and differ from the variable primers used in our current report. Using a 3× RPB threshold for a positive result for both the BIOMED-2 and the four-distribution system, similar performance was observed between the two systems in sensitivity (75% vs. 76% respectively), however, specificity was not calculated.23 We have shown that a lower peak height ratio (RPB = 2×), that is designed for a tri-color or one-color assay, cannot be used for an assay with a separate single joining segment distribution, nor can it be used with the BIOMED-2 design and maintain specificity. Therefore, a low ratio of 2× is not recommended for the BIOMED-2-based TCRG Gene Clonality Assay, as the specificity will be too low.
We developed a rule of using five times the height of the polyclonal background to define a positive result with the BIOMED-2 assay, with the intent of minimizing false positive calls and maximizing specificity. Laboratorians need to determine what the appropriate ratio would be for clonality for each of Tubes A and B in their own laboratory when using the TCRG Gene Clonality Assay, whether it be 3×, 4×, or 5× the polyclonal background. A table of the variable and joining segments used in cell lines is provided to determine sensitivity within each distribution. (Table 4).36
Table 4.
Gene Segments Used in TCRγGR in the Cell Lines
| Variable | Cell line | Joining | Cell line |
|---|---|---|---|
| V1-8 | MOLT4 | Jγ1 & 2 | HSB2 |
| CEM | CEM | ||
| 8402 | JURKAT | ||
| JURKAT | SUPB15 | ||
| Peer | Peer | ||
| V-9 | HSB2 | JγP1/2 | MOLT4 |
| Peer | SUPB15 | ||
| SNT-836 | |||
| SNT-1336 | |||
| SNT-1536 | |||
| V-10 | HSB2 | JγP | SNT-836 |
| 8402 | SNT-1336 | ||
| MOLT4 | SNT-1536 | ||
| V-11 | Jurkat |
The gene usage has been verified for each cell line in the authors’ lab with the exception of SNT-8, 13, and 15, which are referenced.
In our laboratory, we use primers that are designed in such a way so as to use one size distribution centered around 190 nucleotides, thereby including all T-cell receptor gamma gene rearrangements within the tube in a three-color polyclonal background. More small peaks may be eliminated by the use of a single tube reaction and one polyclonal distribution in a one-color approach. Use of multiple primer sets in a single tube, with either the tri-color or one-color approach, which amplify products in a single size distribution, allows competitive amplification of all gene rearrangements to determine whether a significant population is present, while maintaining high specificity.7 This is an advantage of using a complete multiplex assay, as a rare peak “identified” by a separate single Vγ or Jγ distribution may not be found to be a true clonal peak if compared to the entire population of T-cells in one distribution of rearrangements.
Conclusions
Multiplex assays designed to detect products with multiple fluorochomes and multiple different size ranges may lead to false positive interpretations due to formation of pseudoclonal peaks in the presence of inadequate low-amplitude polyclonal distributions for comparison. The high incidence of pseudoclonal spikes in the TCRG Gene Clonality Assay is directly attributed to the design of the BIOMED-2 assay in that there are eight potential normal distributions of TCRγGR that are produced within the two tubes. Because of a lack of a sufficient number of T-cells to form all of the normal distributions, it makes it extremely difficult to assess the importance of small spikes, especially in tube B, without an optimized RPB ratio. The use of a complete multiplex reaction using three joining primers in a one-tube method, with all primers competing to form one overlapping normal distribution, is easier to interpret than the BIOMED-2 platform.
Acknowledgements
We thank Debra Lytle for assistance with the laboratory experiments and sequence alignments. We thank Lisa Roseland for preparation of the manuscript.
Footnotes
Supported by the Department of Pathology and Microbiology, University of Nebraska Medical Center, Omaha, Nebraska.
This work was presented at the AMP 2008 Annual Meeting October 29–November 2, 2008 in Grapevine, TX.
T.C.G. is a consultant for InVivoScribe Technologies, LLC.
References
- 1.Lamberson C, Hutchison RE, Shrimpton AE. A PCR assay for detecting clonal rearrangement of the TCR-gamma gene. Mol Diagn. 2001;6:117–124. doi: 10.1054/modi.2001.25321. [DOI] [PubMed] [Google Scholar]
- 2.Benhattar J, Delacretaz F, Martin P, Chaubert P, Costa J. Improved polymerase chain reaction detection of clonal T-cell lymphoid neoplasms. Diagn Mol Pathol. 1995;4:108–112. doi: 10.1097/00019606-199506000-00006. [DOI] [PubMed] [Google Scholar]
- 3.Greiner TC, Raffeld M, Lutz C, Dick F, Jaffe ES. Analysis of T cell receptor-gamma gene rearrangements by denaturing gradient gel electrophoresis of GC-clamped polymerase chain reaction products. Correlation with tumor-specific sequences. Am J Pathol. 1995;146:46–55. [PMC free article] [PubMed] [Google Scholar]
- 4.Bottaro M, Berti E, Biondi A, Migone N, Crosti L. Heteroduplex analysis of T-cell receptor gamma gene rearrangements for diagnosis and monitoring of cutaneous T-cell lymphomas. Blood. 1994;83:3271–3278. [PubMed] [Google Scholar]
- 5.Guitart J, Kaul K. A new polymerase chain reaction-based method for the detection of T-cell clonality in patients with possible cutaneous T-cell lymphoma. Arch Dermatol. 1999;135:158–162. doi: 10.1001/archderm.135.2.158. [DOI] [PubMed] [Google Scholar]
- 6.Lynas C, Howe D. Simple, reliable detection of T cell clones by PCR-LIS-SSCP analysis of TCRgamma rearrangement. Mol Cell Probes. 1998;12:41–48. doi: 10.1006/mcpr.1997.0146. [DOI] [PubMed] [Google Scholar]
- 7.Greiner TC, Rubocki RJ. Effectiveness of capillary electrophoresis using fluorescent-labeled primers in detecting T-cell receptor gamma gene rearrangements. J Mol Diagn. 2002;4:137–143. doi: 10.1016/s1525-1578(10)60694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier VS, Rufle A, Gudat F. Simultaneous evaluation of T- and B-cell clonality, t(11;14) and t(14;18), in a single reaction by a four-color multiplex polymerase chain reaction assay and automated high-resolution fragment analysis: a method for the rapid molecular diagnosis of lymphoproliferative disorders applicable to fresh frozen and formalin-fixed, paraffin-embedded tissues, blood, and bone marrow aspirates. Am J Pathol. 2001;159:2031–2043. doi: 10.1016/S0002-9440(10)63055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega F, Medeiros LJ, Jones D, Abruzzo LV, Lai R, Manning J, Dunmire V, Luthra R. A novel four-color PCR assay to assess T-cell receptor gamma gene rearrangements in lymphoproliferative lesions. Am J Clin Pathol. 2001;116:17–24. doi: 10.1309/5WFQ-N12E-DT05-UX1T. [DOI] [PubMed] [Google Scholar]
- 10.Luo V, Lessin SR, Wilson RB, Rennert H, Tozer C, Benoit B, Leonard DG. Detection of clonal T-cell receptor gamma gene rearrangements using fluorescent-based PCR and automated high-resolution capillary electrophoresis. Mol Diagn. 2001;6:169–179. doi: 10.1054/modi.2001.27056. [DOI] [PubMed] [Google Scholar]
- 11.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 12.Lawnicki LC, Rubocki RJ, Chan WC, Lytle DM, Greiner TC. The distribution of gene segments in T-cell receptor gamma gene rearrangements demonstrates the need for multiple primer sets. J Mol Diagn. 2003;5:82–87. doi: 10.1016/s1525-1578(10)60456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SC, Berg KD, Racke FK, Griffin CA, Eshleman JR. Pseudo-spikes are common in histologically benign lymphoid tissues. J Mol Diagn. 2000;2:145–152. doi: 10.1016/S1525-1578(10)60630-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shadrach B, Warshawsky I. A comparison of multiplex and monoplex T-cell receptor gamma PCR. Diagn Mol Pathol. 2004;13:127–134. doi: 10.1097/01.pdm.0000126419.92931.a3. [DOI] [PubMed] [Google Scholar]
- 15.Dippel E, Assaf C, Hummel M, Schrag HJ, Stein H, Goerdt S, Orfanos CE. Clonal T-cell receptor gamma-chain gene rearrangement by PCR-based GeneScan analysis in advanced cutaneous T-cell lymphoma: a critical evaluation. J Pathol. 1999;188:146–154. doi: 10.1002/(SICI)1096-9896(199906)188:2<146::AID-PATH334>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Bruggemann M, White H, Gaulard P, Garcia-Sanz R, Gameiro P, Oeschger S, Jasani B, Ott M, Delsol G, Orfao A, Tiemann M, Herbst H, Langerak AW, Spaargaren M, Moreau E, Groenen PJ, Sambade C, Foroni L, Carter GI, Hummel M, Bastard C, Davi F, Delfau-Larue MH, Kneba M, van Dongen JJ, Beldjord K, Molina TJ. Powerful strategy for polymerase chain reaction-based clonality assessment in T-cell malignancies. Report of the BIOMED-2 Concerted Action BHM4 CT98-3936. Leukemia. 2007;21:215–221. doi: 10.1038/sj.leu.2404481. [DOI] [PubMed] [Google Scholar]
- 17.Langerak AW, Molina TJ, Lavender FL, Pearson D, Flohr T, Sambade C, Schuuring E, Al Saati T, van Dongen JJ, van Krieken JH. Polymerase chain reaction-based clonality testing in tissue samples with reactive lymphoproliferations: usefulness and pitfalls. A report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2007;21:222–229. doi: 10.1038/sj.leu.2404482. [DOI] [PubMed] [Google Scholar]
- 18.van Krieken JH, Langerak AW, Macintyre EA, Kneba M, Hodges E, Sanz RG, Morgan GJ, Parreira A, Molina TJ, Cabecadas J, Gaulard P, Jasani B, Garcia JF, Ott M, Hannsmann ML, Berger F, Hummel M, Davi F, Bruggemann M, Lavender FL, Schuuring E, Evans PA, White H, Salles G, Groenen PJ, Gameiro P, Pott C, Dongen JJ. Improved reliability of lymphoma diagnostics via PCR-based clonality testing: report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21:201–206. doi: 10.1038/sj.leu.2404467. [DOI] [PubMed] [Google Scholar]
- 19.Kreuter A, Hoxtermann S, Gambichler T, Tigges C, Hahn SA, Schieren G. Detection of clonal T cells in the circulation of patients with nephrogenic systemic fibrosis. Arch Dermatol. 2009;145:1164–1169. doi: 10.1001/archdermatol.2009.225. [DOI] [PubMed] [Google Scholar]
- 20.Tan BT, Seo K, Warnke RA, Arber DA. The frequency of immunoglobulin heavy chain gene and T-cell receptor gamma-chain gene rearrangements and Epstein-Barr virus in ALK+ and ALK− anaplastic large cell lymphoma and other peripheral T-cell lymphomas. J Mol Diagn. 2008;10:502–512. doi: 10.2353/jmoldx.2008.080054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goeldel AL, Cornillet-Lefebvre P, Durlach A, Birembaut P, Bernard P, Nguyen P, Grange F. T-cell receptor gamma gene rearrangement in cutaneous T-cell lymphoma: comparative study of polymerase chain reaction with denaturing gradient gel electrophoresis and GeneScan analysis. Br J Dermatol. 2010;162:822–829. doi: 10.1111/j.1365-2133.2009.09575.x. [DOI] [PubMed] [Google Scholar]
- 22.Kuo FC, Hall D, Longtine JA. A novel method for interpretation of T-cell receptor gamma gene rearrangement assay by capillary gel electrophoresis based on normal distribution. J Mol Diagn. 2007;9:12–19. doi: 10.2353/jmoldx.2007.060032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel KP, Pan Q, Wang Y, Maitta RW, Du J, Xue X, Lin J, Ratech H. Comparison of BIOMED-2 versus laboratory-developed polymerase chain reaction assays for detecting T-Cell receptor-gamma gene rearrangements. J Mol Diagn. 2010;12:226–237. doi: 10.2353/jmoldx.2010.090042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukowsky A, Richter S, Dijkstal K, Sterry W, Muche JM. A T-cell receptor gamma polymerase chain reaction assay using capillary electrophoresis for the diagnosis of cutaneous T-cell lymphomas. Diagn Mol Pathol. 2002;11:59–66. doi: 10.1097/00019606-200206000-00001. [DOI] [PubMed] [Google Scholar]
- 25.Munro NJ, Snow K, Kant JA, Landers JP. Molecular diagnostics on microfabricated electrophoretic devices: from slab gel- to capillary- to microchip-based assays for T- and B-cell lymphoproliferative disorders. Clin Chem. 1999;45:1906–1917. [PubMed] [Google Scholar]
- 26.Ng SB, Lai KW, Murugaya S, Lee KM, Loong SL, Fook-Chong S, Tao M, Sng I. Nasal-type extranodal natural killer/T-cell lymphomas: a clinicopathologic and genotypic study of 42 cases in Singapore. Mod Pathol. 2004;17:1097–1107. doi: 10.1038/modpathol.3800157. [DOI] [PubMed] [Google Scholar]
- 27.Chang TL, Salto-Tellez M, Thamboo TP, Lee YS, Koay ES. Diagnostic validation of capillary electrophoresis analysis of T-cell receptor gamma-chain gene rearrangements: prediction of malignant transformation of cutaneous T-cell lymphoproliferative disorders. Clin Chem. 2003;49:513–515. doi: 10.1373/49.3.513. [DOI] [PubMed] [Google Scholar]
- 28.Nga ME, Tan SH, Teh M, Koay ES, Chong SM, Putti TC, Salto-Tellez M. Lymphocytic gastritis-like T cell lymphoma: molecular evidence of an unusual recurrence. J Clin Pathol. 2004;57:1222–1224. doi: 10.1136/jcp.2004.017350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakirevich E, Jackson CL, Meitner PA, MacKenzie D, Tavares R, Robinson-Bostom L, DeLellis RA, Resnick MB. Analysis of T-cell clonality using laser capture microdissection and high-resolution microcapillary electrophoresis. J Mol Diagn. 2007;9:490–497. doi: 10.2353/jmoldx.2007.070006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon M, Kind P, Kaudewitz P, Krokowski M, Graf A, Prinz J, Puchta U, Medeiros LJ, Sander CA. Automated high-resolution polymerase chain reaction fragment analysis: a method for detecting T-cell receptor gamma-chain gene rearrangements in lymphoproliferative diseases. Am J Pathol. 1998;152:29–33. [PMC free article] [PubMed] [Google Scholar]
- 31.Beaubier NT, Hart AP, Bartolo C, Willman CL, Viswanatha DS. Comparison of capillary electrophoresis and polyacrylamide gel electrophoresis for the evaluation of T and B cell clonality by polymerase chain reaction. Diagn Mol Pathol. 2000;9:121–131. doi: 10.1097/00019606-200009000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Sprouse JT, Werling R, Hanke D, Lakey C, McDonnel L, Wood BL, Sabath DE. T-cell clonality determination using polymerase chain reaction (PCR) amplification of the T-cell receptor gamma-chain gene and capillary electrophoresis of fluorescently labeled PCR products. Am J Clin Pathol. 2000;113:838–850. doi: 10.1309/02M7-5JCC-YRTK-MGDR. [DOI] [PubMed] [Google Scholar]
- 33.Juarez T, Isenhath SN, Polissar NL, Sabath DE, Wood B, Hanke D, Haycox CL, Wood GS, Olerud JE. Analysis of T-cell receptor gene rearrangement for predicting clinical outcome in patients with cutaneous T-cell lymphoma: a comparison of Southern blot and polymerase chain reaction methods. Arch Dermatol. 2005;141:1107–1113. doi: 10.1001/archderm.141.9.1107. [DOI] [PubMed] [Google Scholar]
- 34.Tang MB, Chong TK, Tan ES, Sun YJ, Tan SH. A comparative study of polymerase chain reaction detection of clonal T-cell receptor gamma chain gene rearrangements using polyacrylamide gel electrophoresis versus fluorescence capillary electrophoresis. Ann Acad Med Singapore. 2008;37:27–31. [PubMed] [Google Scholar]
- 35.Ponti R, Fierro MT, Quaglino P, Lisa B, Paola FC, Michela O, Paolo F, Comessatti A, Novelli M, Bernengo MG. TCRgamma-chain gene rearrangement by PCR-based GeneScan: diagnostic accuracy improvement and clonal heterogeneity analysis in multiple cutaneous T-cell lymphoma samples. J Invest Dermatol. 2008;128:1030–1038. doi: 10.1038/sj.jid.5701109. [DOI] [PubMed] [Google Scholar]
- 36.Oyoshi MK, Nagata H, Kimura N, Zhang Y, Demachi A, Hara T, Kanegane H, Matsuo Y, Yamaguchi T, Morio T, Hirano A, Shimizu N, Yamamoto K. Preferential expansion of Vgamma9-JgammaP/Vdelta2-Jdelta3 gammadelta T cells in nasal T-cell lymphoma and chronic active Epstein-Barr virus infection. Am J Pathol. 2003;162:1629–1638. doi: 10.1016/s0002-9440(10)64297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


