Abstract
We report Trypanosoma cruzi infection rates of the native kissing bug Triatoma protracta in southern California. The rates are within the historically reported range, but differ significantly between the two sites (19% in Escondido and 36% in Glendora). Identification of T. cruzi in T. protracta was conducted for the first time by using partial 18S ribosomal RNA and 24Sα ribosomal RNA sequences. Incongruence of 24Sα ribosomal RNA phylogeny with current T. cruzi genotype classification supports non-clonality of some T. cruzi genotypes.
Chagas disease is caused by the protozoan parasite Trypanosoma cruzi, which is transmitted by Triatominae or kissing bugs (Insecta: Hemiptera: Reduviidae). An estimated 9.8 million persons are infected with this disease, and 40 million are at risk in Latin America.1 Transmission, although apparently rare, also occurs in the southwestern United States, as indicated by six autochthonous transmission cases since 1955, including one in California. Surveillance of T. cruzi infection rates among local populations of Triatominae is therefore critical for assessing the public health risk.
Several species of Triatominae are native to the United States, with Triatoma protracta (Uhler) being the most common of the three species endemic to California. Triatoma protracta is widespread in California and host specific to woodrats (Neotoma spp.), which nest in middens found in a wide range of natural habitats. Human bites by T. protracta in houses occur when T. protracta is attracted to lights on warm evenings.2 The low incidence of autochthonous T. cruzi transmission may be explained by the transmission inefficacy of endemic kissing bugs, especially T. protracta,3 but also by limited contact of humans with the vectors, or low infection rates of kissing bugs with T. cruzi. Autochthonous transmission in the southwestern United States is rare, although studies based on blood bank screenings and serosurveys suggest undiagnosed cases.4,5 Such cases remain unverified but suggest the need of constant monitoring. A recent study in Arizona showed that a much higher infection rate was found in T. rubida (Uhler) compared with rates in historical surveys, which suggests temporal variability with regards to infection rates.6
Large-scale surveys of T. cruzi infection rates of local T. protracta populations in California have not been conducted since the 1980s. To test for the potential of autochthonous Chagas disease transmission in southern California, we measured infection rates with T. cruzi among two populations of T. protracta bordering the large population centers of Los Angeles and San Diego.
We additionally developed a polymerase chain reaction (PCR)–based method primarily to identify T. cruzi and its genotypes in Triatominae. Several genotypes (also referred to as natural clones or major lineages) of T. cruzi are associated with different host lineages. In the United States, genotypes TcI and TcIV occur exclusively in marsupials (opossums) and placental mammals (e.g., raccoons), respectively.7 Different genotypes are also known to result in differences in clinical manifestations of the disease.8 Genotyping of trypanosome isolates is therefore an important endeavor, and we use the most recent T. cruzi genotype nomenclature.9
Specimens were collected using light traps (ultraviolet and mercury vapor) in Escondido (San Diego County; 33°12¢44.9994²N, 117°5′36.9954²W, elevation = 405 meters) and Glendora (Los Angeles County; 34°9¢59.364²N, 117°50¢18.204²W, elevation = 391 meters), California. The sites are located at a suburban residence (Escondido) or less than 1 km from a suburban residence (Glendora). A total of 161 specimens (Escondido = 139 and Glendora = 22) were collected over an eight-week period (June–September 2008). All specimens were identified as T. protracta and entered into a database (collecting and dissection data). Voucher specimens were submitted to the Entomology Research Museum at the University of California, Riverside.
Abdominal contents of live specimens were examined according to the protocols of Westenberger and others.10 Microscopic examination served as the basis for determining the infection rates. The average infection rate of T. protracta with T. cruzi was 21.1%, and there were a significant difference between the two populations (Glendora = 36.4% and Escondido = 18.7%) (Table 1).
Table 1.
Infection rates for Triatoma protracta, southern California
| Characteristic | Glendora | Escondido | P |
|---|---|---|---|
| No. T. protracta | 22 | 139 | |
| No. (%) Trypanosoma cruzi infected | 8 (36.4)* | 26 (18.7) | 0.000911 |
Significantly different with respect to the mean at the 5% level by chi-square test.
We used a rapid method to identify T. cruzi based on amplification and sequencing of partial 18S and 24Sα ribosomal genes.11 Instead of identifying the trypanosomes only on the basis of size polymorphisms of the amplicons, we sequenced the PCR products and phylogenetically analyzed the sequences. DNA was extracted by using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). The PCR parameters were those of Souto and others.12 The PCR amplification used primer pairs Tcz18Sf (5′-TTAACGGGAATATCCTCAGC, which was designed during this study) and TczS829r13 and yielded a 440-basepair fragment of the 18S ribosomal RNA. Primers Tcz24S-D71 and Tcz24S-D7212 were used to amplify a 110-basepair fragment of the 24Sα ribosomal RNA gene. Twenty 18S DNA sequences and two 24Sα DNA sequences were analyzed.
Sequences were aligned with sequences from GenBank by using MAFFT (G-INS-i default settings).14 With each set of sequences, all phylogenetic approaches used (parsimony, maximum likelihood, and Bayesian) identified parasites from both localities as T. cruzi (Figures 1 and 2 represent the Bayesian and maximum likelihood analyses, respectively; trees generated by other methods are not shown). The 18S rRNA gene fragment was too conserved to differentiate between T. cruzi genotypes (Figure 1) and showed only seven single nucleotide polymorphisms from 427 positions (variability = 1.64%). The 24Sα fragment was more informative for genotyping (63 polymorphisms from 103 positions, variability = 61.17%). The 24Sα gene phylogeny (Figure 2) places the isolates from southern California in a clade that comprises members of the TcII and TcVI genotypes, although the relationships within this clade were not resolved.
Figure 1.
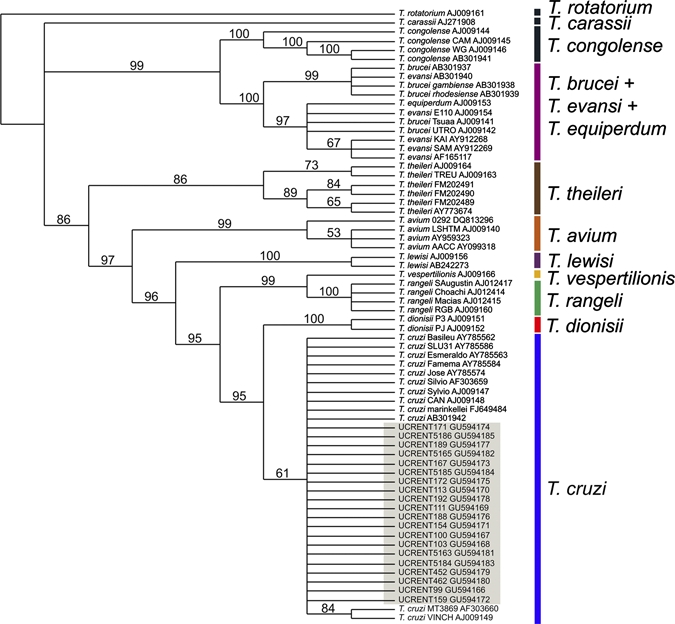
Bayesian 50% majority-rule consensus phylogeny of Trypanosoma cruzi in southern California based on partial 18S ribosomal RNA sequences. A total of 20 sequences (GenBank accession numbers GU594166–59416685) were obtained in this project. The remaining sequences were retrieved from GenBank with accession numbers listed. Numbers above branches indicate posterior probabilities. The analysis was performed by using MrBayes (CIPRES portal) with generalized time reversible + I + Γ model parameters. A total of 2 million generations were conducted with temperature settings at 0.2. A sampling frequency of 1,000 and a burn-in at 25% of the sampled trees were set for final tree production. This figure appears in color at www.ajtmh.org.
Figure 2.

24Sα ribosomal RNA phylogram of Trypanosoma cruzi in southern California generated by RaxML (CIPRES portal) by using generalized time reversible + I + Γ model parameters (default settings). Abbreviations of current T. cruzi genotype classification are listed on the right with known hybrids noted. Two sequences were generated in this study (GenBank accession numbers GU594186 and GU594187). The remaining 15 sequences were retrieved from GenBank. This figure appears in color at www.ajtmh.org.
Our study presents an updated report of infection rates of two T. protracta populations with T. cruzi in southern California and provides the first molecular identification of T. cruzi in this region. Infection rates differ significantly between the two populations, but overall are comparable with those in historical data. Previous studies of infection rates in California were predominantly conducted by Sherwin F. Wood (1930s–1960s); these infection rates ranged typically from 20% to 30%.15 A recent study of infection rates of T. protracta from Escondido in southern California was based on only 20 specimens, four (20%) of which were positive by PCR detection.3 Our study, which was based on a larger sample size, shows virtually identical prevalence for Escondido (19%), but a much higher infection rate for Glendora (36%), which indicated that infection levels of T. protracta may show significant geographic variation.
All phylogenetic analyses of the 24Sα dataset found the isolates from California to be closely related to the TcII and TcVI group members. This result was unexpected because neither of these groups has been previously reported in North America. In addition, none of the TcII, TcIV, TcV, and TcVI groups was recovered as monophyletic in our study (Figure 2), which indicated that the current classification may not fully reflect relationships of the T. cruzi genotypes. Our results lend support to the prevailing view that different T. cruzi genotypes may not be strictly clonal, especially with the recognition that several genotypes are actually hybrids (Figure 2) or suspected to be hybrids.9 Further investigations are needed to establish the relationships of T. cruzi in southern California with the remaining major lineages of this species. We recommend use of identification techniques using DNA sequences for the added advantage of providing nucleotide information that is valuable for documenting genetic variation within T. cruzi from different geographic regions.
Acknowledgments
We thank Chris Conlan (Escondido) and the Gomer family (Glendora) for donating specimens, Kyle Risser for providing field and laboratory assistance, and Jan Walter for critically reading of the manuscript.
Footnotes
Financial support: This study was supported by the California Desert Research Fund (Wei Song Hwang and Guanyang Zhang), the Department of Entomology at the University of California, Riverside, and the National Science Foundation grant PEET DEB-0933853 (Christiane Weirauch).
Authors' addresses: Wei Song Hwang, Guanyang Zhang, Christiane Weirauch, Department of Entomology, University of California, Riverside, CA. Dmitri Maslov, Department of Biology, University of California, Riverside, CA.
References
- 1.Schofield CJ, Jannin J, Salvatella R. The future of Chagas disease control. Trends Parasitol. 2006;22:583–588. doi: 10.1016/j.pt.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Ryckman R. The kissing bug problem in western North America. Bulletin of the Society of Vector Ecologists. 1981;6:3. [Google Scholar]
- 3.Klotz SA, Dorn PL, Klotz JH, Pinnas JL, Weirauch C, Kurtz JR, Schmidt J. Feeding behavior of triatomines from the southwestern United States: an update on potential risk for transmission of Chagas disease. Acta Trop. 2009;111:114–118. doi: 10.1016/j.actatropica.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Navin TR, Roberto RR, Juranek DD, Limpakarnjanarat K, Mortenson EW, Clover JR, Yescott RE, Taclindo C, Steurer F, Allain D. Human and sylvatic Trypanosoma cruzi infection in California. Am J Public Health. 1985;75:366–369. doi: 10.2105/ajph.75.4.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bern C, Montgomery SP, Katz L, Caglioti S, Stramer SL. Chagas disease and the US blood supply. Curr Opin Infect Dis. 2008;21:476–482. doi: 10.1097/QCO.0b013e32830ef5b6. [DOI] [PubMed] [Google Scholar]
- 6.Reisenman CE, Lawrence G, Guerenstein PG, Gregory T, Dotson EM, Hildebrand JG. Infection of kissing bugs with Trypanosoma cruzi, Tucson, Arizona, USA. Emerg Infect Dis. 2010;16:400–405. doi: 10.3201/eid1603.090648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roellig DM, Brown EL, Barnabe C, Tibayrenc M, Steurert FJ, Yabsley MJ. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg Infect Dis. 2008;14:1123–1125. doi: 10.3201/eid1407.080175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, Vargas J, Torrico F, Diosque P, Valente V, Valente SA, Gaunt MW. Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog. 2009;5:9. doi: 10.1371/journal.ppat.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- 10.Westenberger SJ, Sturm NR, Yanega D, Podlipaev SA, Zeledon R, Campbell DA, Maslov DA. Trypanosomatid biodiversity in Costa Rica: genotyping of parasites from heteroptera using the spliced leader RNA gene. Parasitology. 2004;129:537–547. doi: 10.1017/s003118200400592x. [DOI] [PubMed] [Google Scholar]
- 11.Souto RP, Zingales B. Sensitive detection and strain classification of Trypanosoma cruzi by amplification of a ribosomal RNA sequence. Mol Biochem Parasitol. 1993;62:45–52. doi: 10.1016/0166-6851(93)90176-x. [DOI] [PubMed] [Google Scholar]
- 12.Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi. Mol Biochem Parasitol. 1996;83:141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 13.Maslov DA, Lukes J, Jirku M, Simpson L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol Biochem Parasitol. 1996;75:197–205. doi: 10.1016/0166-6851(95)02526-x. [DOI] [PubMed] [Google Scholar]
- 14.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood SF. Trypanosoma cruzi; new foci of enzootic Chagas disease in California. Exp Parasitol. 1975;38:153–160. doi: 10.1016/0014-4894(75)90017-x. [DOI] [PubMed] [Google Scholar]


