Abstract
We evaluated performance of three commercial Japanese encephalitis virus (JEV) IgM antibody capture enzyme-linked immunosorbent assay (MAC ELISA) kits with a panel of serological specimens collected during a surveillance project of acute encephalitis syndrome in India and acute meningitis and encephalitis syndrome in Bangladesh. The serum and cerebral spinal fluid specimens had been referred to the Centers for Disease Control and Prevention (CDC) for confirmatory testing. The CDC results and specimen classifications were considered the reference standard. All three commercial kits had high specificity (95–99.5%), but low sensitivities, ranging from 17–57%, with both serum and cerebrospinal fluid samples. Specific factors contributing to low sensitivity compared with the CDC ELISA could not be determined through further analysis of the limits and dilution end points of IgM detection.
Introduction
Japanese encephalitis virus (JEV) is the leading cause of pediatric viral encephalitis in Asia with an estimated 50,000 cases and 10,000 deaths per year.1 Most JEV infections are asymptomatic or cause an undifferentiated fever and serosurveys in JEV endemic areas have shown that the majority of adults have been exposed to JEV.1 In neuroinvasive JEV infections, patients may experience a prodromal period of 1–2 weeks followed by 1–3 days of altered sensorium.1 Many viral and bacterial pathogens cause similar clinical symptoms, which makes laboratory-based diagnosis essential for accurate elucidation of disease etiology.
Isolation of JEV is not a sensitive method of laboratory diagnosis in clinical specimens because the low-level, transient viremia is cleared soon after onset of illness. In contrast, anti-JEV immunoglobulin M (IgM) is produced soon after infection and is detectable in 90% of cases in cerebrospinal fluid (CSF) by 4 days and in serum by 7–9 days following the development of clinical illness.2–4 The JEV-specific IgM antibody capture enzyme-linked immunosorbant assay (MAC ELISA) has become the first-line serological assay recommended by the World Health Organization (WHO) to diagnose acute JEV infections.5
There are a variety of commercially available and “in-house” laboratory produced JEV IgM detection assays used in diagnostic laboratories, with varying or unknown sensitivities and specificities. Limited performance assessments have been done and these have generally consisted of testing the assays against a panel of well-characterized archived specimens collected from a variety of patients, many of whom did not have clinical symptoms of JEV neurological infections.6–9 An evaluation with “field” specimens that were more representative of the type of samples that would typically be collected in Japanese encephalitis (JE) surveillance and routinely tested in public health laboratories was needed. In this study, we evaluated three kits using serological specimens collected during surveillance of acute encephalitis syndrome (AES) in India and an acute meningitis and encephalitis syndrome (AMES) project in Bangladesh. The specimens were collected primarily at admission to a hospital or clinic and initially tested in the laboratories designated for the project (Featherstone DF and others, unpublished data). All samples were referred to the global specialized laboratory (GSL) at the Centers for Disease Control and Prevention, Division of Vector-Borne Infectious Diseases (CDC/DVBID) for confirmatory testing using a battery of CDC in-house assays. Using the CDC test results and interpretations as the reference standard, a panel of 226 CSF and 294 serum specimens were used to evaluate the performance of three commercially available JEV MAC ELISA kits: Panbio JE-Dengue IgM combo ELISA, XCyton JEV CheX, and InBios JE Detect. Information on the performance of these commercial JEV IgM ELISA kits will be useful to diagnostic laboratories involved in JE surveillance programs, allowing evidence-based decisions on choice of kits to be used and weight of the data generated in such studies.
Materials and Methods
Samples.
A panel of 520 acute-phase serum and CSF samples without personal identifiers were selected from 1,589 specimens referred to CDC for confirmatory JE serological testing from Bangladesh (438 serum and 86 CSF) and India (551 serum and 534 CSF). The specimens had been collected from patients meeting the acute encephalitis clinical case definition10 as part of a WHO South East Asian Region/Centers for Disease Control and Prevention (SEAR/CDC) AES/AMES surveillance project. Samples were referred to the microbiology laboratories associated with the hospital in each of the cities when a case was identified by a surveillance medical officer (SMO) who periodically goes to the hospital to identify AES or AMES cases. After the specimens were tested, the aliquots were referred to the regional reference laboratory and then on to the GSL at the CDC in Fort Collins, Colorado. The samples were collected from 0 to 60 days post onset of illness. Samples that were collected ≤ 7 days post onset accounted for 43.2% (127/294); 23.1% (68/294) were 8–14 days; 7.8% (23/294) were ≥ 15 days; 76 samples had unknown days post onset of disease. All samples were tested at CDC for recent evidence of either JEV or dengue virus (DENV) infection; serum samples were also tested for West Nile virus (WNV) infection. The assessment panel was composed of 294 serum samples, of which 61 (21%) were classified as JEV IgM positive and 233 (79%) were classified as JEV IgM negative (36 JEV and WNV negative/DENV IgM positive, 16 JEV and DENV negative/WNV IgM positive, and 181 JEV/DENV/WNV IgM negative) at CDC. The 226 CSF samples consisted of 30 (13%) JEV IgM positives and 196 (87%) JEV IgM negatives (16 JEV IgM negative/DENV IgM positive and 180 JEV/DENV IgM negative). The InBios JE Detect kit was evaluated after the testing on the Panbio and XCyton kits was completed and 24 serum and 42 CSF samples had been depleted. Therefore, a subset of 454 samples was used to evaluate the InBios JE Detect kit. It was composed of 270 serum specimens, including 51 (19%) JEV IgM positives and 219 (81%) JEV IgM negatives (33 JEV and WNV IgM negative/DENV IgM positive, 16 JEV and DENV IgM negative/WNV IgM positive, and 170 JEV/DENV/WNV IgM negative) and 184 CSF specimens, including 17 (9%) JEV IgM positives and 167 (91%) JEV IgM negatives (10 JEV IgM negative/DENV IgM positive and 157 JEV/DENV IgM negative). The panel stock was stored at −70°C and thawed to aliquot a sample (150 μL CSF and 80 μL serum), which was heat inactivated in a water bath at 56°C for 30 min and stored at −20°C until required for kit evaluation.
Test methods.
One laboratory technician conducted all CDC ELISA, plaque-reduction neutralization test (PRNT), and commercial kit testing, with the exception that confirmatory JEV IgM ELISA testing using suckling mouse brain (SMB) antigen on a subset of positive samples was completed by another laboratory technician, who routinely tests diagnostic specimens in the serological laboratory at CDC. Both technicians are Certified Laboratory Improvement Amendments (CLIA) accredited to conduct ELISA and PRNT.
CDC AES/AMES virology confirmatory testing protocols.
All referred AES/AMES specimens were tested at CDC contingent on sample volume availability and classified according to the CDC/DVBID arbovirus serological testing algorithm (Figure 1). Briefly, samples were tested by JEV and DENV IgM ELISA11–13 using JE and DEN COS-1 antigen.14,15 The CSF was diluted 1:10 and serum samples were diluted 1:400.
Figure 1.

Centers for Disease Control and Prevention (CDC) testing algorithm and results interpretation for Japanese encephalitis (JE) classification. aCerebrospinal fluid (CSF) were treated in the same manner with the exception that they were not tested for West Nile virus (WNV). bSpecimens without adequate volume remaining to conduct the plaque-reduction neutralization test (PRNT) were re-tested by Japanese encephalitis virus (JEV) and dengue virus (DENV) IgM antibody capture enzyme-linked immunosorbent assay (MAC ELISA) with the COS-1 antigen or JE suckling mouse brain (SMB) antigen. Those with concordant results were considered JE presumptive positive and classified as JE positive; specimens with discordant results were excluded from the sample set. cSamples with positive IgM results to more than one of the antigens tested (JEV, DEN, and/or WNV) and without virus-specific antibody response by PRNT (titers to more than one of the viruses without a 4-fold or greater difference) were classified as inconclusive and excluded from the evaluation specimen panel.
If sufficient sample remained after ELISA testing, JEV and/or DENV IgM positive or equivocal results were confirmed by DENV and JEV 90% specimen dilution endpoint plaque reduction neutralization assay (PRNT90) in Vero cells using a 0.5% agarose double overlay, visualized with neutral red staining in the second overlay.16,17 ChimeriVax-JEV (Sanofi Pasteur, France, formerly Acambis Inc., Cambridge, MA) and ChimeriVax- DEN 2 virus (Sanofi Pasteur) were used as challenge viruses in the PRNT.18–21 The initial serum and CSF dilutions were 1:10 and 1:4, respectively, resulting in lower limit of quantitation (LLOQ) titers of 10 and 4. Samples were tested simultaneously to JEV and DENV.22,23 Specimens without adequate volume for the PRNT were re-tested by JEV and DENV MAC ELISA with the COS-1 antigen or JEV SMB antigen. Serum samples were also tested for WNV IgM with confirmatory PRNT90 as described for JEV and DENV.
The CDC results were interpreted as JE positive, JE presumptive, dengue (DEN) positive, DEN presumptive, West Nile (WN) positive, WN presumptive, or negative as outlined in Figure 1. A sample that was JE IgM positive or equivocal with a 4-fold or higher neutralizing antibody titer to JEV compared with the titer to DENV or WNV in the PRNT was considered virus specific and classified as JE confirmed positive. A sample with a JEV IgM positive or equivocal result but which did not have a 4-fold or higher difference between neutralizing antibody titers to JEV over DENV or WNV was classified as JE presumptive. The DEN and WN positive and presumptive samples were classified in the same manner. For evaluation purposes, JE positive and presumptive samples are considered to be positive. The JE-negative samples were those that were classified as DEN or WN positive or presumptive or were IgM negative for all antigens tested. JEV IgM equivocal results with no neutralizing antibody titer in the PRNT were also classified as JE negative. Samples with positive IgM results for more than one of the antigens tested (JEV, DEN, and/or WNV) and without virus-specific antibody response by PRNT (titer to more than one of the viruses without a 4-fold or greater difference) were classified as inconclusive and excluded from the evaluation sample panel.
Commercial kits.
All kits were stored as prescribed by the manufacturers and the tests were conducted following the manufacturers' instructions with two exceptions: First, the InBios JE Detect kit format prescribes that the samples be tested in duplicate and all samples in this evaluation were tested as singles. Second, Panbio recommends against sample heat inactivation and all the samples were heat inactivated before testing. Samples were diluted according to instructions in the buffers provided in the kits; CSF and sera were diluted 1:10 and 1:100, respectively. Kit results were calculated and classified per the manufacturer's instructions. Qualitative parameters such as time, number of samples that can be tested, and ease of use were assessed in a manner similar to that which is presented by Jacobson and colleagues.6
Panbio JE-Dengue IgM combo ELISA.
The Panbio JE-Dengue IgM combo ELISA test (Inverness Medical, Brisbane, Australia) classifies specimens as JE IgM positive, JE IgM equivocal, DEN IgM positive, DEN IgM equivocal, or JE and DEN IgM negative. Results are calculated with separate calibrators for JE and DEN, run in triplicate on each plate. For the purposes of this evaluation DEN IgM positive and JE IgM equivocal results were considered to be JE negative. This assay uses proprietary recombinant antigens and conjugated monoclonal antibodies (Mab).
This kit was recently evaluated on a panel of CSF samples,24 during which a modification of the cutoffs for calculations was recommended.24 This modification is included in the present evaluation. In addition, the kit protocol recommends that the samples should not be heat inactivated, but in the CDC protocol samples are heated at 56°C for 30 minutes to inactivate adventitious viruses before serological testing. Therefore, to determine if heat inactivation would affect results with the Panbio kit, a panel of samples (32 JEV positive sera and 13 JEV positive CSF samples), both normal and heat inactivated, were tested with the same lot of kit before testing the larger sample panel. As there was no difference in results (data not shown) heat-inactivated samples were used to evaluate the Panbio kit.
XCyton JEV CheX.
The XCyton JEV CheX assay (XCyton LLC, Bangalore, India) results were calculated according to the kit instructions, which used a negative control and weak positive controls for both serum and CSF.7 This assay classifies specimens as JE positive, flavivirus positive, and JE negative. For evaluation purposes the flavivirus-positive samples were considered to be JE negative. This assay uses inactivated cell culture virus antigen and a conjugated JEV-specific Mab.
InBios JE Detect.
The InBios JE Detect (Seattle, WA) format and reagents are similar to those of CDC ELISA.11–13 Both positive and negative antigens are included to validate the test and control for background nonspecific reactivity with the antigen. The JE COS-1 antigen14,15 and flavivirus-reactive conjugated monoclonal antibody are licensed by Inbios. Results are interpreted as JE positive, JE equivocal, or negative. For evaluation purposes equivocal samples were considered to be JE negative.
Statistical methods.
The CDC/DVBID test results were considered the reference standard for this evaluation and samples were thereby classified as either JEV IgM positive or JEV IgM negative. All DENV and WNV IgM positive results were considered as JEV IgM negative for calculations. Kit results were compared with CDC/DVBID results for calculation of agreement (i.e., proportion of samples correctly classified), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Individual confidence intervals (CI) for proportions were computed using the Wilson score interval.25 Comparisons of performances between kits in terms of both sensitivity and specificity were made using the Quesenberry and Hurst CI for the difference of correlated proportions recommended by May and Johnson.26 For all statistical analyses, 95% CI were used. These samples are a representative subset from the AES/AMES project, in which prevalence within the population was determined so that predictive values of tests could be calculated.
Ethical considerations.
The patient's serum and CSF specimens were collected by the sentinel hospitals as part of routine diagnostic testing and testing performed at reference laboratories was on deidentified specimens. The CSF specimens were collected in hospitals with the patient's or parent's informed consent. Because this study was for surveillance and used standard clinical specimens, informed consent was not required.
Results
A total of 520 samples, 294 sera, and 226 CSF were used in the Panbio and XCyton evaluations; 454 samples, 270 sera and 184 CSF specimens, were used to evaluate the InBios kit. Kit testing results compared with the CDC testing results are shown in Tables 1 and 2. Sensitivity, specificity, PPV, NPV, and agreement with 95% CI were calculated from the CDC results (Figure 2). Qualitative parameters of using these kits were identical to those observed by Jacobson and colleagues.6
Table 1.
Summary results of three JEV IgM ELISA kits compared with the CDC classifications*
| Kit | Kit result | CDC | ||||||
|---|---|---|---|---|---|---|---|---|
| Serum | CSF | |||||||
| JE Pos | DEN Pos | WNV Pos | Neg | JE Pos | DEN Pos | Neg | ||
| Panbio (Adjusted results) | JE pos | 24 | 0 | 0 | 4 | 6 (9) | 1 (1) | 0 (1) |
| DEN pos | 0 | 9 | 3 | 9 | 0 (0) | 14 (0) | 3 (3) | |
| JE equiv | 6 | 0 | 1 | 3 | 1 (5) | 0 (3) | 0 (20) | |
| DEN equiv | 1 | 1 | 1 | 5 | 0 (0) | 1 (0) | 0 (0) | |
| Neg | 30 | 26 | 11 | 160 | 23 (16) | 0 (12) | 177 (156) | |
| Total | 61 | 36 | 16 | 181 | 30 | 16 | 180 | |
| InBios JE Detect | JE pos | 29 | 2 | 2 | 6 | 9 | 1 | 1 |
| JE equiv | 1 | 2 | 1 | 6 | 0 | 0 | 0 | |
| Neg | 21 | 29 | 13 | 158 | 8 | 9 | 156 | |
| Total | 51 | 33 | 16 | 170 | 17 | 10 | 157 | |
| XCyton JEV CheX | JE pos | 12 | 1 | 0 | 6 | 5 | 1 | 4 |
| Flavivirus pos | 23 | 6 | 4 | 23 | 7 | 2 | 7 | |
| Neg | 26 | 29 | 12 | 152 | 18 | 13 | 169 | |
| Total | 61 | 36 | 16 | 181 | 30 | 16 | 180 | |
Based on CDC serological testing algorithm (see Figure 1). JEV = Japanese encephalitis virus; ELISA = enzyme-linked immunosorbent assay; CDC = Centers for Disease Control and Prevention; CSF = cerebrospinal fluid; JE = Japanese encephalitis; DEN = dengue.
Table 2.
Summary of results from 3 JEV IgM ELISA kits compared to CDC JE classifications*
| Test | CDC | ||||||
|---|---|---|---|---|---|---|---|
| All samples | Serum | CSF | |||||
| JE+ | JE− | JE+ | JE− | JE+ | JE− | ||
| Panbio | JE+ | 30 | 5 | 24 | 4 | 6 | 1 |
| JE− | 61 | 424 | 37 | 229 | 24 | 195 | |
| No. Samples tested (N = 520) | 91 | 429 | 61 | 233† | 30 | 196‡ | |
| InBios JE Detect | JE+ | 38 | 12 | 29 | 10 | 9 | 2 |
| JE− | 30 | 374 | 22 | 209 | 8 | 165 | |
| No. Samples tested (N = 454) | 68 | 386 | 51 | 219§ | 17 | 167¶ | |
| XCyton JEV CheX | JE+ | 17 | 12 | 12 | 7 | 5 | 5 |
| JE− | 74 | 417 | 49 | 226 | 25 | 191 | |
| No. Samples tested (N = 520) | 91 | 429 | 61 | 233† | 30 | 196‡ | |
Based on CDC serological testing algorithm (see Figure 1). JEV = Japanese encephalitis virus; ELISA = enxyme-linked immunosorbent assay; CDC = Centers for Disease Control and Prevention; JE = Japanese encephalitis.
36 JE and West Nile (WN) IgM negative/dengue (DEN) IgM positive, 16 JE and DEN IgM negative/WN IgM positive, and 181 JE/DEN/WN IgM negative.
16 JE negative/DEN IgM positive, and 180 JE/DEN IgM negative.
33 JE and WNV IgM negative/DEN IgM positive, 16 JE and DEN IgM negative/WNV IgM positive, and 170 JE/DEN/WNV IgM negative.
10 JE IgM negative/DEN IgM positive and 157 JE/DEN IgM negative.
Figure 2.
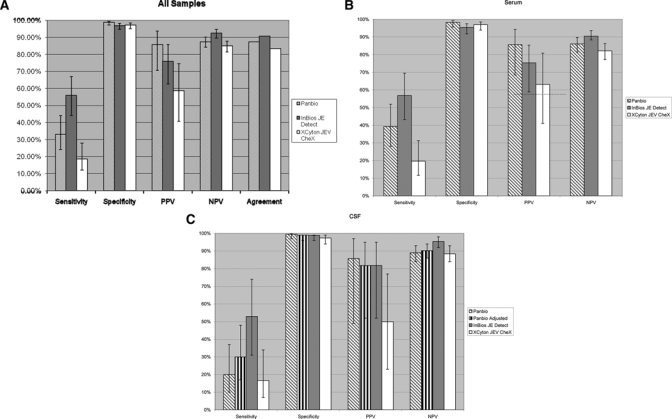
Performance of three Japanese encephalitis (JE) IgM enzyme-linked immunosorbent assay (ELISA) kits compared with Centers for Disease Control and Prevention (CDC) results as reference standard. Error bars represent the 95% confidence interval (CI) for each value. (A) Cerebrospinal fluid (CSF) and serum samples combined, (B) serum samples only, and (C) CSF samples only.
The kits had sensitivities ranging from 17–53% for CSF and 20–57% for serum compared with the CDC standard: Panbio 33% (CI = 24–44%) [CSF, 20% (CI = 10–37%); serum, 39% (CI = 28–52%)]; InBios JE Detect 56% (CI = 44–67%) [CSF, 53% (CI = 31–74%); serum, 57% (CI = 43–70%)]; XCyton JEV CheX 19% (CI = 12–28%) [CSF, 17% (CI = 7–34%); serum, 20% (CI = 12–31%)] (Figure 2). Test sensitivity was also calculated with CDC-confirmed JE-positive serum samples, excluding the JE presumptive results, which had not been confirmed by a 4-fold increase in PRNT titer. In this subgroup of 25 samples (23 for InBios JE Detect kit) Panbio, InBios JE Detect, and XCyton JEV CheX had sensitivities of 52% (CI = 33–70%), 74% (CI = 58–90%), and 24% (CI = 11–43%), respectively. However, the difference was not significant at the 95% confidence level from the sensitivity that included JE presumptive samples (data not shown). The Panbio kit had increased sensitivity, to 30% (CI = 17–48%) for CSF samples, if the modified cutoff values were used in the test result calculations, but this difference was not significant from the non-adjusted value (Figure 2). To ensure that the low IgM detection was not caused by heat inactivation of the samples, a subset of 45 samples was tested by the Panbio kit using normal and heat inactivated JEV-positive samples. There were no differences in test results (data not shown).
In a direct comparison of performance between the kits calculated by Quesenberry and Hurst CI construction, the InBios JE Detect assay had significantly higher sensitivity than XCyton JEV CheX with both CSF and serum sample sets and the Panbio for the serum set (Table 3). Using the same comparison parameters, the InBios JE Detect assay also had significantly higher overall sensitivity than the Panbio and XCyton assays with the combined CSF and serum sample set (Table 3). The Panbio kit also detected JEV IgM in serum samples at a significantly higher rate than the XCyton kit (Table 3).
Table 3.
Comparison of sensitivity* between kits
| Kit comparisons | Overall | CSF | Sera |
|---|---|---|---|
| Panbio-XCyton JEV CheX | 14 (−4, 27) | 3 (−16, 22) | 19 (0.4, 35) |
| Panbio-InBios JE Detect | −24 (−36, −7) | −18 (−39, 13) | −25 (−40, −6) |
| Panbio-Panbio Adjusted | N/D† | −10 (−20, 4) | N/D† |
| XCyton JEV CheX-InBios JE Detect | −40 (−51, −22) | −35 (−52, −1) | −41 (−54, −20) |
| XCyton JEV CheX-Panbio Adjusted | N/D† | −13 (−28, 6) | N/D† |
| InBios JE Detect-Panbio Adjusted | N/D† | 6 (−25, 16) | N/D† |
Percent difference (95% confidence interval [CI]). Percentages shown in bold are significantly different in sensitivity from each other (P < 0.05). Percentages with CIs that do not contain zero are considered to be significantly different. Sensitivity comparisons were computed by the first kit listed versus the second kit; a positive number indicates better performance in the first kit and a negative number indicates better performance in the second kit. Kit results were classified as correct or incorrect based on the CDC reference standard results. For numbers of samples used in these calculations refer to Table 2.
ND = not done; adjusted calculation for CSF only.
As a further estimate of sensitivity, endpoint dilutions of JEV IgM detection were determined in a subset of five samples (Table 4); three sera diluted 4-fold from 1:100 to 1:12800 and two CSF diluted 2-fold from 1:10 to 1:1280. Serum samples 1–3, with high CDC JEV IgM-positive ELISA results (P/N > 20), did not reach endpoint detection at the final 1:12800 dilution in either the CDC ELISA or InBios tests (Table 4). In the Panbio assay the JEV IgM detection endpoint dilution was at 1:3200 for samples 1 and 3, and 1:200 for sample 2. The endpoint dilutions in the XCyton kit were 1:6400 in sample 1 and 1:1600 in sample 2; in sample 3 JEV IgM was detected at the final 1:12800 dilution. The CSF samples 4 and 5 had high CDC JEV IgM positive results and low CDC JEV IgM positive results (P/N < 10), respectively (Table 4). JEV IgM was detected to the final 1:1280 dilution in sample 4 and to 1:80 in sample 5 with the CDC ELISA. In the InBios kit the JEV IgM detection endpoint of sample 4 was at 1:320 dilution; sample 5 had a negative result in the lowest dilution tested (1:10). In the XCyton kit the endpoint dilution was 1:20 in sample 4; no JEV IgM was detected in sample 5 at the initial test dilution (1:10). Compared with the CDC results, with the Panbio kit the endpoint dilutions were 4-fold lower in samples 1 and 3; 64-fold lower in samples 2 and 4; and 8 fold lower in sample 5. Using the adjusted cutoff calculation for CSF sample 4, the endpoint dilution was 32-fold lower. The InBios kit had a 4-fold reduction in titer in sample 4. The XCyton kit had endpoint dilutions 2-, 8-, and 64-fold lower in samples 1, 2, and 4, respectively.
Table 4.
Endpoint dilutions* of IgM detection on three sera and two CSF
| Sample | Type† | CDC | Panbio | XCyton | InBios | Panbio Adjusted |
|---|---|---|---|---|---|---|
| 1 | High serum | > 12,800 | 3,200 | 6,400 | > 12,800 | N/A |
| 2 | High serum | > 12,800 | 200 | 1,600 | > 12,800 | N/A |
| 3 | High serum | > 12,800 | 3,200 | > 12,800 | > 12,800 | N/A |
| 4 | High CSF | > 1,280 | 40 | 20 | 320 | 20 |
| 5 | Low CSF | 80 | 10 | < 10 | < 10 | 10 |
Endpoints are given as the reciprocal of the highest dilution at which Japanese encephalitis virus (JEV) IgM was detected by the assay from an initial serum dilution of 1:100 diluted 4-fold to 1:12800 and initial cerebrospinal fluid (CSF) dilution of 1:10 diluted 2-fold to 1:1280. Endpoints of > 12,800 (sera) or > 1,280 (CSF) indicate that JEV IgM was detected by the assay to the final dilution of the analysis.
Based on initial Centers for Disease Control and Prevention (CDC) Japanese encephalitis virus (JEV) IgM antibody capture enzyme-linked immunosorbent assay (MAC ELISA) results at 1:400 (sera) and 1:10 (CSF) screening dilutions. High serum or CSF, P/N > 20; low CSF, P/N < 10.
To determine if the kit sensitivity was low with samples near the CDC positive cutoff, kit results were compared with CDC JEV IgM positive results, separated into low (P/N < 10), medium (10 ≤ P/N < 20), and high (P/N ≥ 20) P/N ratios. There was no significant difference in performance of any of the kits at any of the levels (Figure 3), however the InBios kit detected significantly more low (P/N < 10) and medium (10 ≤ P/N < 20) CDC JEV IgM-positive samples than the XCyton kit. Sensitivity of the kits was also analyzed by comparing kit results to CDC JEV IgM positive results, categorized by the number of days from illness onset to sample collection. There was no significant difference in performance between the days post onset or the assays evaluated (Figure 4).
Figure 3.

Comparison of kit results to Low (P/N < 10), Medium (P/N = 10–20), and High (P/N > 20) Centers for Disease Control and Prevention (CDC) Japanese encephalitis virus (JEV) positive IgM antibody capture enzyme-linked immunosorbent assay (MAC ELISA) results.
Figure 4.

Comparison of kit results to Centers for Disease Control and Prevention (CDC) Japanese encephalitis virus (JEV) IgM positive results by number of days from illness onset to specimen collection.
Compared with the CDC reference standard, the Panbio kit had overall specificity of 98.8% (CI = 97–99.5%) [CSF, 99.5% (CI = 97–99.7%); serum, 98.2% (CI = 96–99.3%)]; the InBios JE Detect kit had specificity of 96.9% (CI = 95–98%) [CSF, 98.8% (CI = 96–99%); serum, 95.4% (CI = 92–98%)]; and the XCyton JEV CheX kit had specificity of 97.2% (CI = 95–98%) [CSF, 97.4% (CI = 94–99%); serum, 97.0% (CI = 94–99%)] (Figure 2). False JEV IgM positive results caused by cross-reactivity with DENV IgM-positive samples were as follows (Table 1): Panbio 2% (CI = 0.4–10%) [CSF (1/16), 6.3% (CI = 0–28%); serum (0/36), 0% (CI = 0–10%)]; InBios JE Detect 7.0% (CI = 2–19%) [CSF (1/10), 11.1% (CI = 0–40%); serum (2/33), 6.1% (CI = 2–20%)]; and XCyton JEV CheX 3.8% (CI = 1–13%) [CSF(1/16), 6.3% (CI = 0–28%); serum (1/36), 2.8% (CI = 0–14%)]. In addition (2/16) 12.5% (CI = 4–36%) of the WNV IgM-positive samples had positive JE results in the InBios JE Detect kit (Table 1); (3/16), 18.8% (CI = 6–43%) of WNV IgM-positive samples were classified as DENV IgM positive by the Panbio assay; there were no false JEV positive results with the XCyton kit from the WNV-positive samples. There were no significant differences in kit performance on direct comparison between kits based on the CI for the differences of proportions (data not shown).27
Overall PPVs were Panbio 86% (CI = 78–97%) [CSF, 86% (CI = 49–97%); serum, 86% (CI = 69–94%)]; InBios JE Detect 76% (CI = 63–86%) [CSF, 82% (CI = 52–95%); serum, 75% (CI = 59–85%)]; and XCyton JEV CheX 59% (CI = 41–74%) [CSF, 50% (CI = 23–77%); serum, 63% (CI = 41–81%)] (Figure 2). The NPVs for the kits were: Panbio 87% (CI = 84–90) [CSF, 89% (CI = 84–93%); serum, 86% (CI = 81–90%)]; InBios JE Detect 93% (CI = 90–95%) [CSF 95% (CI = 92–98%); serum 91% (CI = 86–94%)]; and XCyton JEV CheX 85% (CI = 94–93%) [CSF, 88% (CI = 83–92%); serum, 82% (CI = 77–86%)] (Figure 2).
Discussion
An ideal field-based diagnostic test should meet the ASSURED criteria: affordable to diagnostic laboratories with limited resources, sensitive, specific, user-friendly, rapid, equipment-free, and delivered to those who need it.28 The JEV IgM ELISA format has the potential to fulfill most of these criteria. The JEV IgM ELISA is relatively sensitive, with detection of JEV IgM in 95% of patients within 5 days of illness onset.29 IgM is less cross-reactive than IgG to other flavivirus antigens, resulting in higher specificity of the IgM ELISA compared with the IgG ELISA. It requires very little equipment or technical expertise to run the assay, and can be carried out rapidly, particularly with the commercial kits that contain all the components and clear easy-to-understand instructions. Most laboratories have staff with technical experience in running similarly formatted assays.
Meningoencephalitis may result from a number of bacterial and viral etiologies so laboratory confirmation of disease etiology is of utmost importance. Although both sensitivity and specificity are important, one must consider the scope of testing to be done. In diagnostic and surveillance activities, specificity of assays is of primary importance as some etiologies of meningoencephalitis have available treatment and/or vaccines for prevention. If assays have low specificity and samples are falsely identified as JEV infections there are several potential consequences. These consequences may arise because of a lack of further testing after the JEV diagnosis is made. This would preclude patients infected with treatable diseases, such as bacterial meningitis, from receiving needed treatment. In addition, false diagnosis of JEV can cause overestimation of disease burden leading to extensive and possibly ineffective vaccination campaigns, which can be quite costly and may put unnecessary burdens on already weak public health systems.
These kits were evaluated previously with panels of well-characterized sera by Jacobson and colleagues.6 The serum panel was composed of JE IgM-positive, DEN-IgM positive, and JE/DEN IgM-negative samples that were archived at the study site, thus the samples did not necessarily fit the AES/AMES case definition. With these selected specimens the kits had high sensitivity (89.3–99.2%). Because of high cross-reactivity with the DENV IgM-positive sera, the InBios and XCyton kits had low specificity (56.1% and 65.3%, respectively) compared with the Panbio JE/DEN duo IgM ELISA kit (99.2%) against the Armed Forces Research Institute of Medical Science (AFRIMS) in-house reference assay.30,31 Specificity of both kits increased to 96.1% if the DENV IgM-positive sera were excluded from the analysis.6 Following this evaluation InBios modified the threshold cutoff to increase the specificity (Raychaudhuri S, personal communication). The low specificity identified by the Jacobson study in the InBios and XCyton assays was not apparent in this field evaluation of AMES samples collected from India and Bangladesh, over the study period.
CSF is the preferred specimen to test in AMES cases as an IgM positive result in CSF is indicative of a nervous system infection. In contrast, flavivirus cross-reactive antibodies in sera from patients who may have recent asymptomatic DENV or WNV infections, or individuals with recent JE vaccination could be misdiagnosed as having a current JEV infection. Ravi and colleagues24 showed that the Panbio and XCyton kits had high specificity and sensitivity in a panel of CSF samples from patients with encephalitis syndrome, which included JE, herpes simplex, SSPE, and encephalitis cases for which the etiology was not identified. Although these results were encouraging, a field evaluation with specimens collected from an AMES patient population was needed.
The CSF and serum specimens comprising the test panel in this evaluation were collected in an AES and AMES surveillance project in India and Bangladesh, respectively. A single CSF or serum specimen was collected on hospital entry in 88% (857/975) of the cases in India and in 76% (583/756) of cases in Bangladesh, and diagnosis was made from testing this single acute specimen. Convalescent samples were not obtained for most of the cases and the dates of illness onset and sample collection were not available for many of the samples, such that a final diagnosis could not be made for 81% of the JEV IgM-positive specimens, and could not be confirmed by PRNT because of a lack of detectable neutralizing titer. These were classified as presumptive JEV infections, and considered JE positives in the analyses. As patients typically develop neutralizing antibodies by 7 days post onset, if the IgM-positive sample was collected less than 7 days post onset with a negative PRNT, the sample would be classified as an acute unconfirmed positive sample and a convalescent sample would be requested. Conversely, if the IgM-positive sample was collected more than 7 days post onset with a negative PRNT, the sample would be classified as having non-specific reactivity.
All the kits had high specificity, ranging from 96–98%. The JEV IgM false–positive rate among the DENV IgM-positive specimens was 3.9% with the XCyton kit, 2% with the Panbio kit, and 7.1% with the InBios kit. The InBios JE Detect kit also had 12.5% (2/16) cross-reactivity with WNV-positive samples. These false–positive rates are much lower than those seen from previous evaluations by Jacobson6 (70–100%) and A-Nuegoonpipat (9%)6,30 and the 10–40% cross-reactivity reported using the CDC ELISA.6,11,24 A large difference in cross-reactivity rates, in DEN-positive sera, between the evaluations was noted, 70% in the XCyton JEV CheX in the Jacobson evaluation,6 using the AFRIMS assay as the reference standard,30 versus 2.8% in serum samples presented here. As both reference standards have similar levels of IgM cross-reactivity on DEN-positive samples, 9% cross-reactivity reported in the AFRIMS assay30 versus approximately 20% cross-reactivity in the CDC assay on this serum sample set, the difference in the cross-reactivity rates between the two evaluations is likely caused by the types of samples used. This assessment used AES field survey cases versus well-characterized samples, including dengue fever cases, in the Jacobson assessment.6 It is likely that the higher cross-reactivity in the Jacobson6 assessment is caused by the inclusion of the samples from dengue fever patients, rather than AES cases that would not typically be assessed when undertaking JEV surveillance. The NPV were calculated to be 85–93%, therefore use of these kits in surveillance would result in an accurate assessment of non-JE cases.
Overall, the kits had low sensitivities, ranging from 20–60%. The rate of false–negative results was so high that if these kits were used as the only diagnostic test in a population of meningoencephalitis patients in which the true JE prevalence was 13–35%, 40–83% of JEV infections would not be detected. For example, in a surveillance project if 1,000 specimens were tested in a population in which the true JE prevalence was 13.2%, the calculated prevalence using the Panbio, XCyton, or InBios kit would be 4.35%, 2.47%, and 7.38%. However, in routine surveillance rarely are 1,000 specimens tested. If fewer than 100 samples were tested with these kits, most of the JE cases could be missed. Underestimation of JE cases during an epidemic could significantly impact the public health response, such as vaccination campaigns.
In an attempt to determine the factors that accounted for the low sensitivity of the kits, CDC JEV IgM positive results were analyzed three different ways. The kit threshold of IgM detection was measured by endpoint serum and CSF dilution assays (Table 4). In serum specimens the Panbio kit had the lowest sensitivity and InBios the highest sensitivity and equivalent to that of the CDC ELISA. The initial CSF dilution of 1:10 was near the endpoint level of detection by the three kits, with one exception. JEV IgM was detected in CSF sample 4 to dilution 1:300 with the InBios kit (Table 4). These results suggest that modifying the instructions to test CSF undiluted or at a higher concentration could increase sensitivity of the kits with CSF specimens. The second analysis was designed to determine if there was an association between IgM concentration and kit detection. Although the CDC MAC-ELISA is not quantitative, P/N ratios are indicative of IgM levels. The CDC JEV IgM-positive sera were categorized as low (P/N < 10), medium (10 ≤ P/N < 20), or high (P/N ≥ 20) and compared with the kit test results. As shown in Figure 3 there was no association between CDC P/N ratio and kit JEV IgM-positive rate; all kits failed to detect high, medium, and low CDC JEV IgM positives. In a third analysis, we wanted to determine if IgM in specimens collected in the very acute phase of illness, when the IgM levels were just beginning to rise, may have been below the kit threshold of detection (Figure 4). The CDC and kit test results were plotted by number of days post onset of illness to specimen collection (Figure 4). No significant correlation was observed between the interval of illness onset to specimen collection and the JEV IgM positivity rate by the kits. Thus, the factors contributing to the significantly lower sensitivities of the kits with sera remain unclear and require further analysis.
All of the three kits are based on a standard ELISA format and results calculated, although the reagents and components vary among them. The Panbio kit contains JE and DEN recombinant antigens produced in an insect cell expression system. Whether the antigen is a complete envelope protein or not is proprietary information; however, antigen conformation has been shown to affect reactivity to IgM.32 The DENV and JEV Mab conjugates used in the Panbio kit also have not been disclosed, but the antigenic sites that they recognize and their avidity may factor into the sensitivity of the assay. The XCyton JEV CheX kit contains inactivated cell-culture JEV antigen from the JEV Indian prototype (NIVP20778, 1956, Vellore, India). The Mab conjugate used in the XCyton kit was developed against the Indian prototype as well.7 Thus, the XCyton kit might be expected to have the highest sensitivity in this group of specimens from India and Bangladesh, as the JEV Indian prototype is probably antigenically the closest to the JEV strain presently circulating there. The reactivity of the JEV Mab has not been determined and it is possible that the Mab has high specificity but low avidity for the JE antigen, which could account for the low sensitivity. The InBios JE kit format is most closely modeled to that of the CDC IgM ELISA. In addition to the plate set-up, the same reagents are used in both assays.33 The JEV and DENV antigens, derived from virus-like particles expressed from stably transformed COS-1 cells, and the flavivirus group reactive Mab conjugate 6B6C-1/HRP are the same reagents used in the MAC ELISA at CDC/DVBID.14,34,35 Not surprisingly, the InBios kit had the highest sensitivity compared with the CDC ELISA.
All of these assays contain components to control for nonspecific- and cross-reactivity. The Panbio and XCyton assays incorporate low positive samples that are used in calculation of cutoff to reduce the likelihood of false–positive results. Similar to the CDC ELISA format, in the InBios assay each sample is tested against a viral antigen and normal, negative antigen. Background or nonspecific reactivity of the sample with the normal control antigen invalidates the test. This reduces the likelihood of false–positive results caused by reactivity of the serological samples with the components used in antigen production.
Factors such as lot-to-lot variability and reagent degradation during shipping or storing also may have contributed to kit performance. There is some evidence of lot-to-lot variation in the XCyton JEV CheX kits. The XCyton JEV CheX kit uses phenol red as a pH indicator in the dilution buffer. In a side-by-side comparison of three unexpired XCyton JEV CheX kits, difference in dilution buffer color intensity between the lots was observed. Changes in pH can lead to conformational changes in protein structure,32 which could potentially change the interactions between the coat antibodies and the antibodies in solution and change the structures of the antigens used in the assays, altering the Mab affinities and thus reducing sensitivity.
Laboratory diagnosis of flavivirus infections, particularly from a single acute specimen, is challenging even at a reference laboratory. Specificity of the IgM ELISA may be a problem in areas where flaviviruses co-circulate or in vaccinated populations, because of the cross-reactivity of IgM to other flavivirus antigens used in the ELISA, such as between JEV and DENV. False–positives can be resolved by PRNT in primary flavivirus infections, ideally with a convalescent specimen collected 7–10 days following collection of the first specimen. However, in secondary flavivirus infections neutralizing antibody from the first flavivirus infection may be more reactive, or cross-reactive, to the challenge viruses in the PRNT than antibodies elicited in the current infection, again reducing the specificity.
Sensitivity of the assay can also present a problem in patients with encephalitis who present to the hospital soon after onset of illness, which may be before the rise of the neutralizing antibodies, or even IgM, to detectable levels.2 The IgM ELISA may have low sensitivity in this population of acutely ill patients.
However, given the problems with flavivirus diagnosis, the low sensitivity of the kits compared with the CDC ELISA under field conditions is still of concern. Further analyses of the factors contributing to the low sensitivities of the kits are needed. To further evaluate the performance of these and other JE ELISA kits, a standardized reference panel of well-characterized serum and CSF specimens is in preparation, coordinated by the WHO. This will enable reference laboratories to provide recommendations on the use and improvements of assays for use in surveillance and point-of-care diagnostic situations.
Acknowledgments
This project was possible through extensive collaboration with the South-East Asia Regional Office of the World Health Organization (WHO-SEARO), the national laboratories in India and Bangladesh, and National Institute of Mental Health and Neuro Sciences (NIMHANS). Support for the kit evaluations at the U.S. Centers for Disease Control and Prevention (CDC), Department of Health and Human Services, was provided by the Acute Meningitis-Encephalitis Syndrome Surveillance project of CDC's Global Disease Detection and Response Initiative. The authors acknowledge the manufacturers of Panbio, XCyton, and InBios diagnostic kits for submitting their assays for evaluation. The technical support of Robert Lanciotti, Amanda Panella, Janeen Laven, and Olga Kosoy at the CDC/DVBID Arboviral Diseases Diagnostic Laboratory is much appreciated.
Footnotes
Authors' addresses: Jaimie S. Robinson, Brad J. Biggerstaff, and Barbara W. Johnson, Centers for Disease Control and Prevention, Division of Vector-Borne Infectious Diseases, Fort Collins, CO, E-mails: fxa3@cdc.gov, BBiggerstaff@cdc.gov, and bfj9@cdc.gov. David Featherstone, Expanded Programme on Immunization/Immunization, Vaccines and Biologicals Department/Family and Child Health Cluster/World Health Organization, Geneva, Switzerland, E-mail: featherstoned@who.int. Ravi Vasanthapuram and Anita Desai, Department of Neurovirology, National Institute of Mental Health and Neuro Sciences (NIMHANS), Bangalore, India, E-mails: virusravi@gmail.com and adesai@gmail.com. Nalini Ramamurty, World Health Organization - South-East Asia Regional Office I.P. Estate, New Delhi, India, E-mail: Ramamurtyn@SEARO.WHO.INT. Anwarul Haque Chowdhury, National Polio and Measles Laboratory, IPH, Dhaka, Bangladesh, E-mail: banplab@gmail.com. Hardeep S. Sandhu and Kathleen F. Cavallaro, CORP Bldg, Atlanta, GA, E-mails: hsandhu@cdc.gov and kfc1@cdc.gov.
References
- 1.Vaughn DW, Hoke CH., Jr The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol Rev. 1992;14:197–221. doi: 10.1093/oxfordjournals.epirev.a036087. [DOI] [PubMed] [Google Scholar]
- 2.Burke DS, Nisalak A, Ussery MA, Laorakpongse T, Chantavibul S. Kinetics of IgM and IgG responses to Japanese encephalitis virus in human serum and cerebrospinal fluid. J Infect Dis. 1985;151:1093–1099. doi: 10.1093/infdis/151.6.1093. [DOI] [PubMed] [Google Scholar]
- 3.Chanama S, Sukprasert W, Sa-ngasang A, An A, Sangkitporn S, Kurane I, Anantapreecha S. Detection of Japanese encephalitis (JE) virus-specific IgM in cerebrospinal fluid and serum samples from JE patients. Jpn J Infect Dis. 2005;58:294–296. [PubMed] [Google Scholar]
- 4.Solomon T, Ooi MH, Beasley DW, Mallewa M. West Nile encephalitis. BMJ. 2003;326:865–869. doi: 10.1136/bmj.326.7394.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . Manual for the Laboratory Diagnosis of Japanese Encephalitis Virus Infection. Geneva: World Health Organization; 2007. [Google Scholar]
- 6.Jacobson JA, Hills SL, Winkler JL, Mammen M, Thaisomboonsuk B, Marfin AA, Gibbons RV. Evaluation of three immunoglobulin M antibody capture enzyme-linked immunosorbent assays for diagnosis of Japanese encephalitis. Am J Trop Med Hyg. 2007;77:164–168. [PubMed] [Google Scholar]
- 7.Ravi V, Desai A, Balaji M, Apte MP, Lakshman L, Subbakrishna DK, Sridharan G, Dhole TN, Ravikumar BV. Development and evaluation of a rapid IgM capture ELISA (JEV-Chex) for the diagnosis of Japanese encephalitis. J Clin Virol. 2006;35:429–434. doi: 10.1016/j.jcv.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Cuzzubbo AJ, Endy TP, Vaughn DW, Solomon T, Nisalak A, Kalayanarooj S, Dung NM, Warrilow D, Aaskov J, Devine PL. Evaluation of a new commercially available immunoglobulin M capture enzyme-linked immunosorbent assay for diagnosis of Japanese encephalitis infections. J Clin Microbiol. 1999;37:3738–3741. doi: 10.1128/jcm.37.11.3738-3741.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrivastva A, Tripathi NK, Parida M, Dash PK, Jana AM, Lakshmana Rao PV. Comparison of a dipstick enzyme-linked immunosorbent assay with commercial assays for detection of Japanese encephalitis virus-specific IgM antibodies. J Postgrad Med. 2008;54:181–185. doi: 10.4103/0022-3859.40959. [DOI] [PubMed] [Google Scholar]
- 10.WHO . WHO-recommended standards for surveillance of selected vaccine preventable diseases. Geneva: WHO; 2006. [Google Scholar]
- 11.Martin DA, Biggerstaff BJ, Allen B, Johnson AJ, Lanciotti RS, Roehrig JT. Use of immunoglobulin M cross-reactions in differential diagnosis of human flaviviral encephalitis infections in the United States. Clin Diagn Lab Immunol. 2002;9:544–549. doi: 10.1128/CDLI.9.3.544-549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol. 2000;38:1823–1826. doi: 10.1128/jcm.38.5.1823-1826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanciotti RS, Tsai TF. In: Manual of Clinical Microbiology. Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Washington, DC: ASM Press; 2007. pp. 1486–1500. (Arboviruses). [Google Scholar]
- 14.Chang GJ, Hunt AR, Holmes DA, Springfield T, Chiueh TS, Roehrig JT, Gubler DJ. Enhancing biosynthesis and secretion of premembrane and envelope proteins by the chimeric plasmid of dengue virus type 2 and Japanese encephalitis virus. Virology. 2003;306:170–180. doi: 10.1016/s0042-6822(02)00028-4. [DOI] [PubMed] [Google Scholar]
- 15.Purdy DE, Chang GJ. Secretion of noninfectious dengue virus-like particles and identification of amino acids in the stem region involved in intracellular retention of envelope protein. Virology. 2005;333:239–250. doi: 10.1016/j.virol.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 16.Lindsey HS, Calisher CH, Mathews JH. Serum dilution neutralization test for California group virus identification and serology. J Clin Microbiol. 1976;4:503–510. doi: 10.1128/jcm.4.6.503-510.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaty BJ, Calisher CH, Shope RE. In: Arboviruses. Lennette EH, Lennette DL, Lennette ET, editors. Washington, DC: American Public Health Association; 1995. pp. 465–478. (Diganostic procedures for viral, rickettsial and chlamydial infections). [Google Scholar]
- 18.Chambers TJ, Nestorowicz A, Mason PW, Rice CM. Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J Virol. 1999;73:3095–3101. doi: 10.1128/jvi.73.4.3095-3101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guirakhoo F, Arroyo J, Pugachev KV, Miller C, Zhang ZX, Weltzin R, Georgakopoulos K, Catalan J, Ocran S, Soike K, Ratterree M, Monath TP. Construction, safety, and immunogenicity in nonhuman primates of a chimeric yellow fever-dengue virus tetravalent vaccine. J Virol. 2001;75:7290–7304. doi: 10.1128/JVI.75.16.7290-7304.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugachev KV, Guirakhoo F, Mitchell F, Ocran SW, Parsons M, Johnson BW, Kosoy OL, Lanciotti RS, Roehrig JT, Trent DW, Monath TP. Construction of yellow fever/St. Louis encephalitis chimeric virus and the use of chimeras as a diagnostic tool. Am J Trop Med Hyg. 2004;71:639–645. [PubMed] [Google Scholar]
- 21.Johnson BW, Kosoy O, Hunsperger E, Beltran M, Delorey M, Guirakhoo F, Monath T. Evaluation of chimeric Japanese encephalitis and dengue viruses for use in diagnostic plaque reduction neutralization tests. Clin Vaccine Immunol. 2009;16:1052–1059. doi: 10.1128/CVI.00095-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 23.WHO . Guidelines for Plaque Reduction Neutralizing Testing of Human Antibodies to Dengue Viruses. Geneva: WHO; 2007. [Google Scholar]
- 24.Ravi V, Robinson JS, Russell BJ, Desai A, Ramamurty N, Featherstone D, Johnson BW. Evaluation of IgM antibody capture enzyme-linked immunosorbent assay kits for detection of IgM against Japanese encephalitis virus in cerebrospinal fluid samples. Am J Trop Med Hyg. 2009;81:1144–1150. doi: 10.4269/ajtmh.2009.09-0144. [DOI] [PubMed] [Google Scholar]
- 25.Wilson EB. Probable inference the law of succession and statistical inference. J Am Stat Assoc. 1927;22:209–212. [Google Scholar]
- 26.May WL, Johnson WD. Confidence intervals for differences in correlated binary proportions. Stat Med. 1997;16:2127–2136. doi: 10.1002/(sici)1097-0258(19970930)16:18<2127::aid-sim633>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 27.May WL, Johnson WD. Hypothesis testing and confidence interval construction in 2 × 2 tables of correlated proportions. J Biopharm Stat. 1998;8:115–130. doi: 10.1080/10543409808835226. [DOI] [PubMed] [Google Scholar]
- 28.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nat Rev Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 29.Burke DS, Nisalak A, Ussery MA, Laorakpongse T, Chantavibul S. Kinetics of IgM and IgG responses to Japanese encephalitis virus in human serum and cerebrospinal fluid. J Infect Dis. 1985;151:1093–1099. doi: 10.1093/infdis/151.6.1093. [DOI] [PubMed] [Google Scholar]
- 30.A-Nuegoonpipat A, Panthuyosri N, Anantapreecha S, Chanama S, Sa-Ngasang A, Sawanpanyalert P, Kurane I. Cross-reactive IgM responses in patients with dengue or Japanese encephalitis. J Clin Virol. 2008;42:75–77. doi: 10.1016/j.jcv.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 31.Burke DS, Nisalak A, Ussery MA. Antibody capture immunoassay detection of Japanese encephalitis virus immunoglobulin M and G antibodies in cerebrospinal fluid. J Clin Microbiol. 1982;16:1034–1042. doi: 10.1128/jcm.16.6.1034-1042.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roehrig JT, Johnson AJ, Hunt AR, Bolin RA, Chu MC. Antibodies to dengue 2 virus E-glycoprotein synthetic peptides identify antigenic conformation. Virology. 1990;177:668–675. doi: 10.1016/0042-6822(90)90532-v. [DOI] [PubMed] [Google Scholar]
- 33.Martin DA, Biggerstaff BJ, Allen B, Johnson AJ, Lanciotti RS, Roehrig JT. Use of immunoglobulin M cross-reactions in differential diagnosis of human flaviviral encephalitis infections in the United States. Clin Diagn Lab Immunol. 2002;9:544–549. doi: 10.1128/CDLI.9.3.544-549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. doi: 10.1016/s0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- 35.Roehrig JT, Mathews JH, Trent DW. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology. 1983;128:118–126. doi: 10.1016/0042-6822(83)90323-9. [DOI] [PubMed] [Google Scholar]


