Abstract
With rapid improvements in medical treatment and health care, many datasets dealing with time to relapse or death now reveal a substantial portion of patients who are cured (i.e., who never experience the event). Extended survival models called cure rate models account for the probability of a subject being cured and can be broadly classified into the classical mixture models of Berkson and Gage (BG type) or the stochastic tumor models pioneered by Yakovlev and extended to a hierarchical framework by Chen, Ibrahim, and Sinha (YCIS type). Recent developments in Bayesian hierarchical cure models have evoked significant interest regarding relationships and preferences between these two classes of models. Our present work proposes a unifying class of cure rate models that facilitates flexible hierarchical model-building while including both existing cure model classes as special cases. This unifying class enables robust modeling by accounting for uncertainty in underlying mechanisms leading to cure. Issues such as regressing on the cure fraction and propriety of the associated posterior distributions under different modeling assumptions are also discussed. Finally, we offer a simulation study and also illustrate with two datasets (on melanoma and breast cancer) that reveal our framework’s ability to distinguish among underlying mechanisms that lead to relapse and cure.
Keywords: Bayesian hierarchical model, Cure fraction, Cure rate model, Latent activation scheme, Markov chain Monte Carlo algorithm, Moment-generating functions, Survival analysis
1. INTRODUCTION
With significant progress in the medical and health sciences, scientists and health professionals increasingly encounter datasets in which some patients are expected to be cured. This concept is subtle because a subject surviving the time window of the experiment is considered “censored,” whereas he or she is cured if he or she will never fail from the event under study. We can never “observe” a cure because of a finite monitoring time, yet interest in formulating models accounting for cure is ripe for evaluating prognosis in potentially fatal diseases (e.g., cancers). Here traditional parametric survival models, such as Weibull or gamma (Cox and Oakes 1984), which do not account for possibility of cure, may prove inadequate.
To address this conceptually challenging problem, cure rate models that incorporate a cure fraction have been proposed, starting with the mixture models by Berkson and Gage (1952) (the BG model), with subsequent investigations by Farewell (1982, 1986), Goldman (1984), and Ewell and Ibrahim (1997) among others. More recent developments include cancer models (Yakovlev et al. 1993; Yakovlev 1996; Yakovlev and Tsodikov 1996) that propose mechanisms that bring about metastasis. Being motivated from mechanistic considerations of the particular disease, these are referred to as mechanistic models. Based on these, Chen, Ibrahim, and Sinha (1999) proposed an alternative class of cure models, which we call the YCIS models (see also Ibrahim, Chen, and Sinha 2001; Tsodikov, Ibrahim, and Yakovlev 2003; Banerjee and Carlin 2004). These models assume that an individual is at risk of failure if he or she is exposed to at least one (perhaps several) of the so-called latent factors or latent risks. Otherwise, the individual is not at risk and is considered cured. Failure is observed when one (or some) of these latent factors becomes activated.
Existing cure models are almost exclusively modifications of either the BG (see, e.g., Kuk and Chen 1992; Taylor 1995; Sy and Taylor 2000; Li and Taylor 2002) or the YCIS models (see, e.g., Tsodikov et al. 2003). Although insight into the biological processes leading to disease manifestation often sheds some light on appropriate model assumptions, controversy persists on whether to use the BG model or the YCIS model (see, e.g., Yin and Ibrahim 2005; Tsodikov et al. 2003; Tucker and Taylor 1996; Tucker, Thames, and Taylor 1990). The problem is exacerbated only in realistic settings, because survival datasets are unlikely to identify (and hence validate) underlying mechanistic model parameters. In this vein, both the BG and YCIS models are somewhat inflexible, because they do not account for the uncertainty in the transition from the underlying process to disease manifestation. This in turn can lead to poorer fits to the data.
Our current work proposes a class of hierarchical models that address the aforementioned issues by considering stochastically modeling of the process of disease manifestation. We achieve greater modeling flexibility by stochastically modeling the ordered sequence of latent events that lead to disease manifestation. We show that this framework leads to identifiable models, of which the BG and YCIS are special instances (Sec. 2). We also consider the identifiability of regression parameters and the resulting hazard structures (Sec. 3). Although assumptions made on the latent event distributions cannot be verified a priori, we assess their relative merits a posteriori using a posterior predictive criterion (Sec. 4). In Section 5 we illustrate model performance with a simulated experiment and then with survival data on two forms of cancer (melanoma and breast), indicating potential advantages of our approach. The melanoma data come from a clinical trial by the Eastern Cooperative Oncology Group (ECOG) (see Kirkwood et al. 1996) and has been analyzed by, for instance, Chen et al. (1999). The breast cancer dataset is a sample from the SEER databases (www.seer.cancer.gov). Finally, in Section 6 we offer a summary and indicate areas for future research.
2. MODELS WITH LATENT FACTORS AND ACTIVATION SCHEMES
Cure models based on latent activation schemes assume that an observed failure time T (when the individual fails) is generated by the latent event times (activation times for the N latent factors) Y1, …, YN. If N = 0, then the individual is not exposed to any of the latent factors and is considered cured (not at risk of failure) and T = ∞. For a given N, the are assumed to be independently distributed with a common survival function, P(Y > t) = S(t) = 1 − F(t), independent of N.
2.1 Hierarchical-Activation Schemes
A crucial issue for further development is relating the latent to the observed T. Generally, if we assume that r out of N latent factors need to activate for the subject to fail, then T = Y(r) for 1 ≤ r ≤ N where Y(1) < ··· < Y(N) are the ordered Yk. Thus r is a threshold variable, the biological interpretation of which is addressed later. It can be a fixed constant or a function of N or can even be treated as random by specifying a conditional distribution for r given N (denoted as r|N).
The conditional distribution of T, given N and r, can be written as
| (1) |
where 1(·) is the indicator function and
is the incomplete beta function. Derivation of (1) follows easily from a standard result on order statistics using the binomial theorem (see, e.g., Rao 1973, p. 215). The unconditional survival function of T, say S*(t), is given in terms of the latent distribution as
| (2) |
where the expectation EN,r is taken over the joint distribution of (N, r). Note that S*(t) exists and is bounded between 0 and 1 for any valid distribution of (N, r) restricted to N ≥ r ≥ 1. In addition because limt → ∞ S(t) = 0, we have that S*(t) is improper whenever limt → ∞ S*(t) = P(N = 0) > 0. Indeed, then P(N = 0) is the probability of a person being cured, called the cure fraction, which depends only on the distribution of N irrespective of the value of r. Although S*(t) is improper, we can still consider the hazard h*(t) such that h*(t) dt ≈ P(T ∈ [t, t + dt)|T > t), and the corresponding improper density,
| (3) |
where f (t) is the proper density of F(t). The variable N can never be observed and must be modeled using a probabilistic assumption. On the other hand, the scientific context can sometimes suggest a fixed value or a function of N for r in (2).
A flexible hierarchical modeling approach specifies the joint distribution of r and N through a marginal specification for N and a conditional distribution for r given N. The conditional distribution of T given N and r remains as in (1), and S*(t) is easily derived from (2) to yield
| (4) |
Practical modeling further benefits by characterizing cure models that are identifiable from the data (see, e.g., Li, Taylor, and Sy 2001). In our hierarchical context, we seek conditions in which θ is identifiable under a noninformative prior g(θ). This is relevant only when some individuals experience the event (i.e., not all are censored) and amounts to the finiteness of . Identifiability permits regression parameters with flat priors through a link on θ. For instance, assuming the marginal specification N ~ Po(θ) with mean θ, we obtain the cure fraction P(N = 0) = exp(−θ). The objective (noninformative) scale-invariant prior sets g(θ) = 1/θ, which amounts to a flat prior on log(θ). Under this setup, we can prove (see item 1 in the App.) that θ is identifiable whenever r = 1 or r = N. (In fact, we prove this identifiability more generally whenever r is fixed or N − r is fixed.) Setting r = 1 implies that activating any one of the N latent events brings about the observed failure, that is, T = min1≤k≤N Yk (the first-activation, or FA, scheme). On the other hand, setting r = N implies that T = max1≤k≤N Yk and delivers a different scheme (the last-activation, or LA, scheme). A well-identified mixture scheme is obtained immediately with r|N having positive mass on {1, N} with probabilities 1 − π and π. Indeed, this model is identifiable for any π ∈ (0, 1) with a posterior distribution that indicates the data’s support for first or last activation.
More generally, with r|N having positive mass on values other than 1 and N, θ may no longer be identifiable. A particularly interesting result follows by specifying r|N as DiscreteUnif (1, N) (discrete uniform). Then (4) simplifies to (see item 2 in the App.)
| (5) |
which is a classical BG-type model with cure fraction P(N = 0) depending on the distribution of N. With N ~ Po(θ) and g(θ) = 1/θ, we find that θ is not identifiable. The same is true for μ with g(μ) ∝ 1 in the BG model, where N ~ Ber(eμ/(1 + eμ)) with cure fraction 1/(1 + eμ).
The identifiability of the cure fraction is determined by the nature of the mixing in (2). The question of characterizing non-degenerate probability distribution for r given N ~ Po(θ) on the support of {1, …, N} that leads to an identifiable θ is more challenging (see item 3 in the App.). For instance, with N ~ Po(θ), we may specify r − 1|N as Bin(N − 1, π) (the HA–Bin model). Although θ is weakly identifiable for π ∈ (0, 1), the hyperparameter π can be assigned, at least conceptually, a suitable hyperprior (e.g., a uniform or beta distribution) with Markov chain Monte Carlo (MCMC) techniques used for their estimation. In fact, this is a clear advantage of the Bayesian mechanism, because a richer framework is obtained without impairing model identifiability. Note that the hierarchical model includes the first- and last-activation schemes as special cases when π = 0 and π = 1, and its posterior distribution may indicate favoring first or last activation. However, the data often do not inform as much about this hyperparameter, and the posterior of π might be inconclusive. A more practical approach fits separate models (say, fixing π = 0, 1) and uses statistical model comparison metrics to assess “better” performance. We discuss such a practical strategy in Section 4. Alternatively, as discussed earlier, the mixing probabilities in the mixture model are better identified and can be used to test for activation schemes (see Sec. 5.1).
2.2 First-Activation Scheme
The first-activation scheme sets r = 1 in the hierarchical setting assuming a single activation leads to observed failure. A biological model for patients diagnosed with cancer assumes that N is the number of metastasis-competent cells (clonogens) that are in an irreversible process toward metastasis and that Yk is the time needed for the kth clonogen to produce “detectable” tumor. This occurs as soon as any one of the clonogens metastasizes, so that T = min1≤k≤N Yk, and (1) simplifies to P(T ≥ t|N) = 1(N = 0) + [S(t)]N1(N ≥ 1). The YCIS models of Chen et al. (1999) are a special instance of this scheme with N ~ Po(θ). Hanin, Tsodikov, and Yakovlev (2001) have provided arguments supporting this assumption for postradiation clonogens in a patient’s body.
Several authors, including Tucker et al. (1990) and Tucker and Taylor (1996), have questioned the universal validity of the Poisson assumption and this scheme. From a modeling standpoint, N can have any finite-mean integer-valued distribution with moment-generating function (mgf) m(t) = E[exp(tN)] and a cure fraction given by P(N = 0) = m(−∞). The marginal distribution of T is then obtained as
| (6) |
For example, in the YCIS model with N ~ Po(θ), we have m(t) = exp[− θ (1 − et)], which yields S*(t) = exp(−θ(1 −S(t))), with cure fraction exp(−θ). Note that (6) offers flexibility to go beyond the YCIS and BG models and model the biology of disease occurrence/relapse suitable for the application at hand.
In the BG-type models, N is binary with only one latent dominant event (say, one dominant metastasis-competent tissue mass), so N ~ Ber(θ), where θ is the probability of activation and m(t) = 1 − θ (1 − et). Here S*(t) = 1 − θ (1 − S(t)), with a cure fraction of 1 − θ. An alternative K-site cancer model discussed by Gail, Santner, and Brown (1980) suggests that for each patient, an unknown N out of K dominant mutation sites within a disease location become mutated, with K fixed a priori based on scientific considerations. This implies a BG(K) model in which N ~ Bin(K, θ) and m(t) = (1 − θ (1 − et))K, yielding S*(t) = (1 − θ (1 − S(t)))K with a cure fraction of (1 − θ)K. Ideally, K also could be stochastically modeled, although this would be computationally burdensome and likely would entail reversible-jump MCMC (see, e.g., Carlin and Louis 2000, sec. 6.4.3). Although the choice of K is subjective, and the model’s performance can be sensitive to it, the flexible hierarchical schemes add robustness to the model’s performance.
Yet another specification of a mechanistic model of cancer considers a biological model (see Moolgavkar, Luebeck, and de Gunst 1990) in which exposure to genetic damage causes the patient’s body to produce N mutated cells/tissues before the immune system is activated. If every new mutation (initiation) produces with probability 1 − θ an effective immune response capable of destroying the last mutated tissue/cell to halt the mutation process, then N ~ Geo(θ), a geometric distribution with mean θ/(1 − θ) and mgf m(t) = (1 − θ)/(1 − θet). With now being the iid promotion times of N mutated cells remaining in the body, a first-activation scheme results in S*(t) = (1 − θ)/(1 − θS(t)) with a cure fraction of 1 − θ.
2.3 Last-Activation Scheme
The mechanistic framework of the first-activation scheme, originally proposed by Yakovlev et al. (1993), can be questioned for certain diseases. Alternatively, N can be the number of latent factors that must all be activated for failure. For instance, for certain situations (e.g., types of cancer), the underlying mechanism involves metastasis-competent mutation of the tissue mass that generates the first primary. During the short interval of this mutation, the patient’s immune response also becomes activated to initiate N immune responses to this mutation. When there is no mutation, there is no immune response (i.e., N = 0). Each immune response is a latent factor capable of resisting disease manifestation or death until its promotion (destruction) time, Yk. Failure (detectable disease/death) occurs after all of the N factors have been activated, so the observed failure time is T = max Yk, k = 1, …, N, a special case of (1) with r = N. Here we also discuss Poisson, Bernoulli, binomial, and geometric specifications for N.
The conditional distribution of T given N is now easily expressed in terms of F(t) = 1 − S(t) as
| (7) |
with cure fraction S*(∞) = m(−∞). Analogous to (6), (7) characterizes S*(t) for last activation in terms of m(t) and helps provide insight into regression (see Sec. 3). In addition, S*(t) in (7), although different from (6), tends to have the same cure fraction, m(−∞) = P(N = 0). In particular, when N ~ Po(θ), we have S*(t) = 1 + exp(−θ)(1 − exp(θF(t))), which is different from that under first activation, but approaches the same cure fraction, exp(−θ). For N ~ Geo(θ), using the mgf of the geometric distribution, we obtain, after some algebra, S*(t) = (1 − θ) + [θ2F(t)/(1 − θF(t))], with 1 − θ as the cure fraction for 0 < θ < 1. The biological motivation for the geometric model is similar to that described for the first activation, except that now failure occurs after the last promotion time for the mutated cell.
3. REGRESSION IN CURE MODELS
Equation (3) shows f*(t) in a generally complex relationship with the latent density function, but gives the hazard function h*(t) for the first and last activation schemes in terms of as
and
Forming , we see that g(t) depends on both S(t) and θ. With N ~ Po(θ) we find that g(t) = (1+e−θ)eθS(t) −1, a decreasing function in t satisfying limt→ ∞ g(t) = e−θ (the cure fraction) and limt→0 g(t) = eθ. This behavior occurs for any latent survival distribution and suggests that activation schemes are identifiable from data with specific hazard structures.
In what follows, we specify the latent survival function S(t) using a two-parameter Weibull distribution, Weib(ρ, η), with survival function S(t) = exp(−tρeη). Cure rate models can incorporate a regressor vector x in the Weibull scale parameter η = η(x), ensuring both a proportional hazards structure and an accelerated failure time model for the latent activation time. But this does not necessarily translate to S*(t|x). Whenever the regressor x is modeled through the activation time using an accelerated life model structure as S(t|x) = S0(tφ (x)), the corresponding cure rate model under the first-activation scheme becomes an accelerated life model with , with for any m(u) that is free of x. Using the uniqueness of the mgf, we can show that when S(t) does not depend on x, the cure model cannot have an accelerated failure time structure. Similarly, when S(t|x) = S0(tφ (x)) and S*(t|x) has an accelerated failure time distribution, the mgf m(u|x) = E[exp(uN)|x] is free of x. These results characterize the activation time distribution and the distribution of N when the cure rate model under the first activation scheme follows an accelerated failure time structure.
For the special case of the YCIS model in which N is Poisson with mean θ = θ(x) and S(t) is free of x, we obtain a proportional hazards structure for h*(t|x) = θ (x)f(t). On the other hand, a model with either N ~ Bin(K, θ) or N ~ Geo(θ) does not render a proportional hazards structure regardless of whether we regress through θ (x) or η (x). In fact, using the uniqueness property of the Poisson mgf, we can show that when S(t) is free of x and h*(t|x) has a proportional hazards structure, then N|x ~ Po(θ (x)) with mean θ (x) (a function of x). Therefore, when the available observed data inform about the form of S*(t|x), we can deduce the distributional structures of the corresponding latent activation times and N.
Under the last-activation scheme, we do not have a proportional hazards structure with Poisson, binomial, or geometric mgf’s, and the hazard structures are more complex (and perhaps less intuitive) than in the first-activation setting. Nevertheless, our earlier discussions on identifiability (see Sec. 4 and the App., item 1) show that with a Poisson mgf, we have identifiable regression in log(θ) under fairly general conditions, whereas for a Bernoulli or binomial mgf, regressing on θ with flat priors on regression coefficients produces improper posteriors. On the other hand, regressing through η in the latent Weibull distribution is always valid, yielding proper posteriors even with improper priors with an appropriate link.
To see how the foregoing assumptions lead to flexible classes of models, consider the setting with I subjects, where the ith subject has a vector of risk factors (covariates) xi. Letting ηi and θi be the respective analog of the foregoing η and θ for subject i, we introduce regression as either or , where g is a suitable link mapping onto the positive real line (recall that θi has positive support). We investigate cure rate models based on the activation scheme, the distribution of N, and the regression, in particular considering each of the following models under different activation schemes:
Models 1(a)–1(d) regress on the latent Weibull mean, ensuring proper models irrespective of the distribution of N, but maintain a constant cure parameter θ. Model 2 regresses on the cure parameter θ and renders proper models for N ~ Po(θ), but restricts the latent survival distribution to a common Weibull mean η. Note that all activation schemes coincide for model 1(b) (N binary). In models 1(a)–1(d), higher values of ηi indicate longer latent event times and hence prolonged survival, so covariates influence the latent factors that cause failure, whereas in model 2 they influence the cure fraction directly. In our subsequent analysis, we compare performances of these models under first-, last-, and hierarchical-activation schemes.
The covariates that affect the latent event times also may affect the probability of cure. In fact, a natural question that arises here relates to the feasibility and identification of regressions in both η and θ. Such settings will not necessarily yield proportional hazards, proportional odds, or accelerated failure structures in S*(t) and in fact might lead to nonidentifiable regression models for the marginal survival function. In a different context, this is a more relevant issue for developing spatial models for geographically referenced cure data (see, e.g., Banerjee and Carlin 2004) with spatially informative priors, and it merits separate investigation.
4. BAYESIAN ESTIMATION AND MODEL COMPARISONS
4.1 The Likelihood and Posterior
For the ith individual, our observed data consist of Di = (ti, νi, xi), where ti is the observed failure time, νi is the failure indicator, and xi is a set of covariates. We collect the model-specific parameters (and hyperparameters) into Ωi. Generally, we write Ωi = (ηi(β), θi(β), ρ, ψ) to denote regression in either ηi or θi (but not both) and let ψ denote set of other hyperparameters that may arise in specific models. Suppressing the dependence of the latent distributions on ηi and ρ and implicitly assuming N ≥ r ≥ 1, subject i’s contribution to the data likelihood (in a right-censored setting) is
where P(T ≥ ti|Ni, ri)as in (1) and . The posterior distribution of {Ωi} (marginalized over Ni and ri) can be now be written generically as
| (8) |
where P(Ωi, Ni, ri) are the joint prior probabilities. Note that for models 1(a)–1(c), we have P(θi(β)) ≡ P(θ) and P(ηi(β)) ≡ P(β), whereas the reverse is true for model 2. In addition, for the first- and last-activation schemes, P(ri|Ni) is degenerate, and we need consider only P(Ni|θi).
In general, marginalization in (8) is analytically intractable and performed using a MCMC algorithm (see, e.g., Carlin and Louis 2000). Implementation of the MCMC may be simplified considerably using a marginalized likelihood. In fact, marginalizing the (Ni, ri)’s out of (8) significantly reduces the estimation space and amounts to evaluating
for each i. Using the facts that νi equals 1 or 0 and that the derivative can be interchanged with the expectation, we can rewrite this as (h*(ti))νiS*(ti), yielding the data likelihood as
| (9) |
Thus we may sample P({Ωi}|{Di}) in (8) using, say, Metropolis, and avoid sampling the (Ni, ri).
4.2 Model Comparison
With the broad range of models that the foregoing formulation encompasses, model evaluation and selection become important issues. We believe that the model selection issue involves scientific considerations that might favor certain assumptions and assess the models with respect to the data at hand. Regarding this, note that the data likelihood in (9) can distinguish between the models and the activation schemes based on the differing hazard and survival structures [h*(t) and S*(t)] that they imply. As a practical guideline for model selection, we adopt a measure that rewards goodness of fit but penalizes complexity. In practice, when several models fit the data adequately well, scientific considerations (e.g., knowledge about the disease) often guide the final model choice(s). In the absence of such knowledge, our measure will favor the simpler models unless the complex model offers substantially better fits.
To assess goodness of fit, we perform posterior predictive comparisons. With Ω as the parameter set, we generate (future) data replicates from the posterior predictive distribution,
| (10) |
where P(t*|Ω, {νi, xi}) is the underlying probability distribution in the data likelihood. More precisely, given postconvergence posterior samples of size M from MCMC output, say (Ω1, …, ΩM) from P(Ω|{Di}), for each Ωj we generate a predictive data replicate (an I × 1 vector) by drawing from P(t*|Ωj, {νi, xi}). This yields an I × M posterior predictive ensemble matrix, say , whose ith row is a sample from the posterior predictive distribution of the survival time for the ith individual.
Note that, unlike in usual survival models, here the impropriety of S*(t) in the likelihood complicates these computations somewhat, in particular, simulating from P(t*|Ω, {νi, xi}). However, by simulating our latent event schemes, we can generate posterior predictive samples for the ith individual by plugging in the posterior sample values Ωj, j = 1, …, M, for the required parameter estimates. For instance, if νi = 1, then subject i has failed, so we first generate Ni from the truncated Po(θi(β))1(Ni ≥ 1) distribution, ensuring that Ni ≥ 1, and set ri = 1 (first activation), ri = Ni (last activation) or generate ri ~Bin(Ni, πi) (hierarchical activation) for the respective schemes. Next, we generate Y1, …, YN from the Weibull family and set Ti = Y(ri). On the other hand, if νi = 0, then the subject has a positive probability of being cured, so we generate Ni from a Po(θi(β)) distribution, admitting the possibility that Ni = 0. If Ni = 0, then we set Ti = ∞. (Operationally, what is actually assigned as ∞ will depend on the computing environment, but for practical purposes, any number much larger than the observed data will suffice.) Otherwise, we follow the procedures for Ni ≥ 1 as for νi = 1, but truncate Ti to exceed the observed censoring time ti.
Although several models may provide an adequate fit to the data, it is beneficial to have a framework for choosing among them. Theoretically, due to the presence of the point mass at Ti = ∞, the moments of the posterior predictive distribution do not exist for cure models, but in practice we replace ∞ with a large number and effectively build model comparison procedures based on these. Here we adopt the posterior predictive L-measure (Laud and Ibrahim 1995; Gelfand and Ghosh 1998), computed as the sum of a goodness-of-fit measure and a penalty term,
| (11) |
where μ* = Et*|{Di}[t*] is the posterior predictive mean of the replicated data and t is a vector of the observed time points {ti}. Letting P = tr(var(t*|t))) [which equals the first term in (11)] and G = (t − μ*)T (t − μ*), we see that L tends to P + G as δ → ∞. This approach has decision-theoretic justifications for treating model choice as a minimizing decision rule for a squared-error loss function. To compute G, P, and L, we first compute and then insert it to compute and G = (t − μ*)T(t − μ*). Operationally, we often work on a transformed scale, replacing ti and by log(ti) and , to ensure greater numerical stability and a better scale for comparison. Smaller L scores suggest better models in terms of predictive fit, and simulation draws from different likelihoods can be compared fairly. Alternative model comparison measures such as the deviance information criterion (DIC) (Spiegelhalter, Best, Carlin, and van der Linde 2002), although computationally simpler, are unsuitable here, because our likelihoods are not log-concave. Furthermore, because the L-measure is computed purely from predictive samples, we can compare models not only within a given activation scheme, but also across different activation schemes—something that is precluded in DIC without computation of normalizing constants, which greatly detracts from its computational simplicity.
5. ILLUSTRATIONS
We illustrate our methods first through a simulation study for assessing model performance, and then using datasets for melanoma and breast cancer. The melanoma data, where failure is melanoma relapse, was previously analyzed using YCIS modeling (Chen et al. 1999) to capture a pronounced “plateau” effect in its survival curve. The breast cancer data, for which failure is defined as death from breast cancer (here deaths from many aggressive cancers are attributed to disease relapse, and relapse is practically indistinguishable from death), does not reveal a dominant plateau effect in the study time frame. Nonetheless, cure modeling for breast cancer is relevant, especially with prognosis often showing marked improvement where cure fraction estimates are better measures of treatment efficacy.
We always assigned a N(0, 103) prior to the regression slopes, whereas we assigned a relatively weak Gamma(.001,. 001) prior to ρ. For model 2, we set η ~ N(0, 100), with θ ~ Gamma(.001, .001) in model 1(a) and U(0, 1) in models 1(b) and 1(c). These priors were sufficiently weak for the data to drive the posterior inference and yield acceptable MCMC convergence. Experimenting with different hyper parameter values revealed very robust posteriors. For the HA–Bin scheme, we selected r − 1|N ~ Bin(N − 1, π) and experimented with a beta or uniform prior on π and also with π = 1/2. The resulting inferences did not differ much, so we present only the π = 1/2 cases. When fitting the mixture model (Mix) in which r ∈ {1, N} with probability 1 − π and π, we assigned π ~ U(0, 1). For each analysis, we ran two initially dispersed parallel MCMC chains for 30,000 iterations each. Convergence diagnostics suggested discarding the first 5,000 iterations from each chain as a preconvergence burn-in, yielding 2 × 25,000 = 50,000 samples for posterior analysis.
5.1 Simulation Study
To assess the performance of the activation schemes, we generated datasets from each set of known model specifications and activation schemes that were subsequently analyzed by fitting models 1(a), 1(c), 1(d), and 2. Model 1(b), the classical BG-type model, does not distinguish among the different activation schemes and is not considered here. For each of models 1(a), 1(c), 1(d), and 2 and each activation scheme, FA, LA, HA–Bin, and (Mix), we generated 10,000 datasets: 2,000 each with cure fractions of 20%, 25%, 30%, 35%, and 40%. The regressors comprised an intercept and a continuous covariate generated from a N(0, 1) distribution. We fixed β= (−5, 2) for models 1(a) and 1(c) and (−3, 2) for model 1(d), with ρ = 1.5 for all of the models, and let the censoring rate vary between 20% and 60%. For model 2, we set η = −3 and β1 = 1, with β0 determined from β1 and the respective cure fraction. MCMC iterations subsequently produced consistent estimates of all of the model parameters with the 95% credible interval including the true values 95–100% of the time.
The first two columns in Table 1 indicate the model and scheme generating the data, with each cell displaying the means of G, P, and L over the 10,000 simulations. The bold diagonal entries for the L measure indicate, quite expectedly, the better performance of the true model on average. This pattern is very consistent, barring only the Mix scheme in model 1(c), where HA–Bin’s L score mean is marginally lower. The table also reveals superior performance of the hierarchical schemes, both HA–Bin and Mix; even when not the truth, they perform very robustly, with Ḡ, P̄, and L̄ appreciably closer to those of the true model and substantially lower than those of the misspecified nonhierarchical scheme. Note that the differences between the HA–Bin and the Mix schemes were usually minor, although the former’s performance under the true Mix model seems better than the reverse [especially for models 1(c) and 1(d)]. Model 1(c) in Table 1 corresponds to K = 10; we also conducted analysis for other values of K (e.g., 5, 15, and 20) and found activation scheme performance very consistent with that presented here.
Table 1.
Model Performance in Simulation Examples
| Simulated data |
Ḡ |
P̄ |
L̄ |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Simulation scheme | FA | LA | HA–Bin | Mix | FA | LA | HA–Bin | Mix | FA | LA | HA–Bin | Mix |
| 1(a) | FA | 50.1 | 57.3 | 51.4 | 53.6 | 55.1 | 68.2 | 57.8 | 57.1 | 105.2 | 125.5 | 109.2 | 110.7 |
| LA | 23.1 | 17.9 | 18.1 | 18.9 | 37.8 | 20.7 | 25.8 | 30.6 | 60.9 | 38.6 | 43.9 | 49.5 | |
| HA–Bin | 44.2 | 43.7 | 29.5 | 31.5 | 39.9 | 38.7 | 31.6 | 35.1 | 84.1 | 82.4 | 61.1 | 66.6 | |
| Mix | 62.7 | 58.6 | 53.1 | 50.2 | 69.4 | 68.7 | 63.8 | 64.9 | 132.1 | 127.3 | 116.9 | 115.1 | |
| 1(c) | FA | 51.1 | 64.1 | 52.2 | 51.9 | 53.9 | 59.8 | 55.8 | 57.4 | 105.0 | 123.9 | 108.0 | 109.3 |
| LA | 27.7 | 19.5 | 19.8 | 19.7 | 27.5 | 21.6 | 24.0 | 24.4 | 55.2 | 41.1 | 43.8 | 44.1 | |
| HA–Bin | 45.5 | 43.9 | 35.8 | 35.9 | 48.6 | 40.0 | 36.5 | 39.3 | 94.1 | 83.9 | 72.3 | 75.2 | |
| Mix | 49.8 | 49.5 | 43.7 | 43.4 | 62.3 | 60.8 | 59.3 | 60.1 | 112.1 | 110.3 | 103.0 | 103.5 | |
| 1(d) | FA | 71.6 | 72.7 | 72.0 | 71.2 | 68.7 | 83.1 | 70.7 | 78.1 | 140.3 | 155.8 | 142.7 | 149.3 |
| LA | 26.7 | 18.5 | 20.3 | 20.5 | 35.5 | 22.7 | 26.5 | 26.1 | 62.2 | 41.2 | 46.8 | 46.6 | |
| HA–Bin | 43.7 | 47.8 | 29.6 | 33.5 | 43.5 | 43.3 | 31.3 | 35.6 | 87.2 | 91.1 | 60.9 | 68.1 | |
| Mix | 67.3 | 69.8 | 60.6 | 61.9 | 86.9 | 91.7 | 82.3 | 80.6 | 154.2 | 161.5 | 142.9 | 142.5 | |
| 2 | FA | 45.9 | 53.4 | 48.0 | 48.6 | 55.9 | 67.4 | 58.8 | 61.1 | 101.8 | 120.8 | 106.8 | 109.7 |
| LA | 31.6 | 25.1 | 26.1 | 25.2 | 34.4 | 29.9 | 35.1 | 34.5 | 66.0 | 55.0 | 61.2 | 59.7 | |
| HA–Bin | 59.7 | 62.6 | 47.6 | 49.5 | 69.4 | 69.6 | 60.6 | 63.5 | 129.1 | 132.2 | 108.2 | 113.0 | |
| Mix | 57.8 | 56.2 | 46.0 | 46.2 | 67.5 | 65.5 | 63.4 | 60.5 | 125.3 | 121.7 | 109.4 | 106.7 | |
NOTE: The first and second columns denote the underlying model and mechanism that generated the data. The remaining columns show means of goodness of fit (Ḡ), the penalty (P̄), and the L measure (L̄) under first-, last-, and hierarchical-activation schemes. The cells with the lowest means for each measure are highlighted.
To assess the mixture model’s ability to distinguish between fast activation and last activation, we also monitored the mixing probability P(r = 1). This parameter appeared to be well identified in our MCMC computations. For example, with a randomly selected simulated dataset, the Mix scheme with P(r = 1) = P(r = N) = 1/2 and model 2 yielded 95% posterior credible interval for P(r = 1) about (.394, .791). With fast activation and last activation as the true underlying schemes, they were consistently estimated as (.845, .997) and (.029, .444), whereas with HA–Bin, the interval was (.017, .557).
These simulations demonstrate the advantages for accounting for the uncertainty in activation schemes. Fixed-activation schemes seem to perform inferiorly to our hierarchical schemes, whose comparatively robust performance should make them invaluable modeling tools for more realistic settings in which the “true” underlying mechanisms generating the data are rarely known.
5.2 Analysis of Melanoma Data
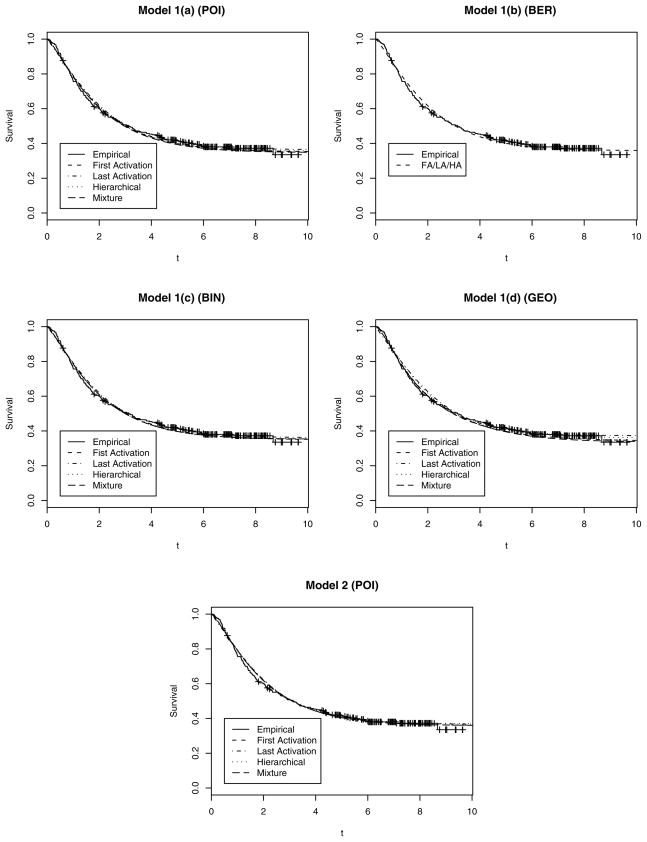
Melanoma incidence rates are among the highest of all solid tumors; despite of earlier detection and screening, high-risk melanoma patients continue to have mortality rates between 60% and 75% (Kirkwood et al. 2000). The data that we consider come from two recent Phase three clinical trials (Kirkwood et al. 1996) in which subjects were given postoperative chemotherapy with interferon alpha-2b (IFN). The dataset comprises 284 subjects, of whom 113 are female, with 174 who relapsed and the remainder who were censored. In addition, an indicator covariate (fully active, other) representing the performance status (PS) of each subject is also available. Basic descriptive analysis through a Kaplan–Meier curve (see the solid line in Fig. 2) reveals a typical “plateau,” indicating a significant proportion of patients who appear to be cured. Table 2 provides the posterior predictive L measures for the different models for the melanoma dataset. Recall that model 1(b) is the BG model, in which the activation schemes coincide; thus, the L measures are same across this row. Model 1(d) (regressing on the Weibull mean) for the Mix scheme has the lowest L-measure score among all of the different models under any activation scheme with a marginal improvement over the YCIS model (model 2, FA) and more pronounced improvements over the BG-type model.
Figure 2.
Posterior Estimates (median) of the Survival Plots for the Melanoma Data Under the Different Activation Schemes Showing Adequate Fits Under the Different Schemes.
Table 2.
The Goodness-of-Fit, Penalty, and L-Measure Values for Various Models Under First-, Last-, and Hierarchical-Activation Schemes for the Melanoma Dataset
| Model | Melanoma |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First |
Last |
HA–Bin |
Mixture |
|||||||||
| G | P | L | G | P | L | G | P | L | G | P | L | |
| 1(a) | 206.98 | 421.93 | 628.91 | 206.71 | 463.99 | 670.70 | 210.16 | 438.00 | 648.16 | 209.15 | 419.40 | 628.55 |
| 1(b) | 206.29 | 438.30 | 644.59 | 206.29 | 438.30 | 644.59 | 206.29 | 438.30 | 644.59 | 206.29 | 438.30 | 644.59 |
| 1(c) | 207.93 | 423.87 | 631.80 | 206.87 | 459.21 | 666.08 | 210.09 | 437.96 | 648.05 | 207.78 | 422.63 | 630.41 |
| 1(d) | 209.35 | 409.10 | 618.45 | 206.81 | 492.75 | 699.56 | 206.05 | 436.08 | 642.13 | 210.75 | 405.57 | 616.32 |
| 2 | 206.59 | 414.22 | 620.81 | 205.43 | 459.62 | 665.05 | 203.50 | 425.25 | 628.75 | 205.65 | 413.49 | 619.14 |
NOTE: The model with the best L-score in each column is highlighted.
Table 3 presents results for the L-best model under the different activation schemes. The covariates included are age at diagnosis, sex (male or female), and performance status (fully active or not). Recall from Section 3 that for model 2, the parameters affect the cure probability, with positive estimates implying lower cure probabilities, whereas for models 1(a), 1(b), and 1(c) they affect the Weibull link, now with positive estimates implying greater hazard of failing. The lack of significance in the covariate estimates is somewhat disappointing, but these findings agree with the results reported by Chen et al. (1999) (although they discussed only the FA scheme). The Weibull scale parameter ρ is significantly greater than 1, consistent with the marked slope in the hazard curve. We also remark that the Mix model 1(d) estimated of P(r = 1) as .859 with a 95% confidence interval of (0.651, 0.989), indicating strong support for first activation.
Table 3.
Posterior Quantiles for Different Models (corresponding to the models with best L-measure) Under the Three Activation Schemes for the Melanoma Dataset
| Parameter | First [model 1(d)] |
Last [model 1(b)] |
HA–Bin (model 2) |
Mixture [model 1(d)] |
||||
|---|---|---|---|---|---|---|---|---|
| Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | |
| Intercept | −1.835 | (−2.242, −1.479) | −.924 | (−1.196, −.675) | .089 | (−.130, .255) | −1.665 | (−2.156, −1.236) |
| Age | −.019 | (−.244, .215) | −.023 | (−.186, .144) | .118 | (−.033, .283) | −.024 | (−.250, .220) |
| Sex (male = 0) | −.041 | (−.568, .441) | .025 | (−.385, .396) | −.164 | (−.472, .164) | −.014 | (−.582, .472) |
| Performance status | −.283 | (−1.151, .472) | −.246 | (−1.023, .372) | −.239 | (−.732, .308) | −.362 | (−1.226, .407) |
| η (Weibull link) | −.929 | (−1.136, −.749) | ||||||
| Cure fraction (exp(− θ)) | .342 | (.270, .408) | .359 | (.297, .419) | .338 | (.250, .404) | ||
| ρ | 1.426 | (1.232, 1.622) | 1.193 | (1.038, 1.353) | 1.192 | (1.050, 1.356) | 1.477 | (1.283, 1.670) |
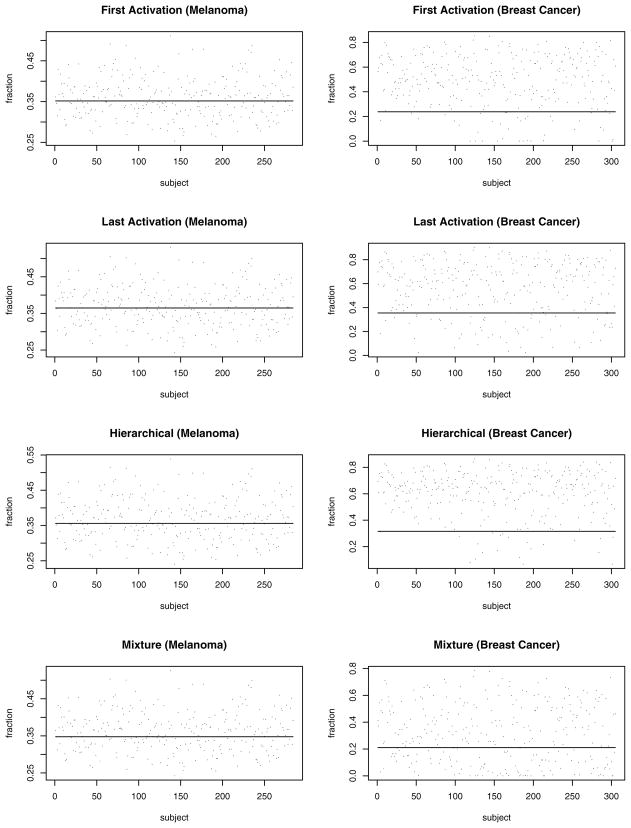
The first column of Figure 1 plots the individual-specific cure fraction estimates (posterior medians) for the different activation schemes as provided by model 2 for the melanoma dataset. Each plot also shows a solid line corresponding the constant cure fraction estimated from model 1(a) (regression in the Weibull link). These plots reveal the sensitivity in the cure fractions to individual characteristics, somewhat corroborating the results in Table 2 that indicated model 2 as one of the best models. Finally, Figure 2 overlays the median of the posterior predictive survival plots (dashed and dotted lines) with Kaplan–Meier plots (solid lines) from the raw data. Almost all of the models seem to provide, adequate fits to the empirical curve, capturing the plateau quite effectively. This is noteworthy given that a smooth parametric family (Weibull) is modeling a latent, rather than an observed, survival distribution.
Figure 1.
Plots of the Median Estimates of the Cure Fractions for the Melanoma and Breast Cancer Datasets Under Different Activation Schemes. The horizontal solid line corresponds to the constant cure fraction estimate from model 1(a), whereas the dots are the subject-specific cure fraction estimates provided by model 2.
5.3 Analysis of Breast Cancer Data
The National Cancer Institute’s Surveillance, Epidemiology, and End Results (SEER) database (available at http://seer.cancer.gov) provides a national cohort of women who have been progressively monitored for assessing breast cancer prognosis. In addition, available individual-level covariates include racial information, age at diagnosis, the number of primary cancers diagnosed, and the stage of the disease [local, regional (95 patients), or distant (12 patients), with local as baseline]. We consider a sample of 305 patients diagnosed with breast cancer in January 1992 and monitored until 1998. The response here is time to death from breast cancer; only those who have been identified as having died from metastasis of nodes in the breast (there were 102 such deaths) are considered having failed, whereas the rest (including those who might have died from metastasis of other types of cancer or other causes) are considered censored. With the time unit “month,” the longest observation time is 84 months. Note that with such an endpoint definition, a cure rate model is essentially mandatory, because we know that some positive fraction will die from causes other than breast cancer.
Table 4 provides the L scores for the different models for the breast cancer dataset. Clearly, the FA scheme (YCIS model) does not perform well here. Indeed, the hierarchical and LA frameworks with regression in the cure fraction (model 2) seem to be more desirable, with the HA–Bin scheme in model 2 having the lowest L score. Table 5 presents parameter inference results from model 1(d) with LA along with those from model 2 for FA and the hierarchical schemes (the best models under each scheme). For all three models, we see a significant positive impact for age at diagnosis (with later diagnosis corresponding to greater hazard). The number of primary nodes has a positive estimate, significantly so for the LA [model 1(d)] scheme. Expectedly, regional or distant stages yield significant hazard increases relative to local. The Weibull scale parameter ρ is estimated robustly across the models. The mixture model 2 still seems to slightly favor the FA scheme, although much less than the melanoma data, estimating P(r = 1) with .745 and a 95% confidence interval of (.495, .928).
Table 4.
The Goodness-of-Fit, Penalty, and L-Measure Values for Various Models Under First-, Last-, and Hierarchical-Activation Schemes for the Breast-Cancer Dataset
| Model | Breast cancer |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First |
Last |
HA–Bin |
Mixture |
|||||||||
| G | P | L | G | P | L | G | P | L | G | P | L | |
| 1(a) | 172.28 | 274.40 | 446.68 | 129.42 | 223.62 | 353.04 | 141.42 | 219.83 | 361.25 | 177.80 | 270.42 | 448.22 |
| 1(b) | 149.44 | 242.15 | 391.59 | 149.44 | 242.15 | 391.59 | 149.44 | 242.15 | 391.59 | 149.44 | 242.15 | 391.59 |
| 1(c) | 180.26 | 283.63 | 463.89 | 137.89 | 235.04 | 372.93 | 149.71 | 246.21 | 395.92 | 180.90 | 288.43 | 469.33 |
| 1(d) | 188.19 | 306.76 | 494.95 | 117.49 | 204.24 | 321.73 | 175.87 | 199.06 | 374.93 | 188.20 | 295.28 | 483.48 |
| 2 | 98.54 | 261.03 | 359.57 | 98.71 | 231.43 | 330.14 | 91.63 | 171.11 | 262.74 | 98.71 | 217.25 | 315.96 |
NOTE: The model with the best L-score in each column is highlighted.
Table 5.
Posterior Quantiles for Different Models (corresponding to best L-measures) Under the Three Activation Schemes for the Breast-Cancer Dataset
| Parameter | First (model 2) |
Last [model 1(d)] |
HA–Bin (model 2) |
Mixture (model 2) |
||||
|---|---|---|---|---|---|---|---|---|
| Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | Median | (2.5%, 97.5%) | |
| Intercept | −2.907 | (−4.306, −.439) | −7.789 | (−9.776, −6.018) | −2.650 | (−2.705, −2.552) | −3.105 | (−4.211, −1.963) |
| Age | .034 | (.018, .050) | .025 | (.013, .037) | .024 | (.018, .029) | .032 | (.018, .052) |
| Primaries | .196 | (−.191, .545) | .621 | (.089, 1.119) | .062 | (−.088, .273) | .351 | (−.172, 1.037) |
| Stage (local = 0) | ||||||||
| Regional | .538 | (.103, .955) | .737 | (.286, 1.201) | .788 | (.532, .891) | .700 | (.142, 1.318) |
| Distant | 2.484 | (1.782, 3.124) | 2.531 | (1.882, 3.150) | 1.647 | (1.429, 1.763) | 3.380 | (2.609, 4.063) |
| η (Weibull link) | −6.403 | (−7.835, −5.406) | −6.002 | (−6.061, −5.810) | −6.763 | (−8.093, −5.930) | ||
| Cure fraction (exp(− θ)) | .398 | (.246, .520) | ||||||
| ρ | 1.358 | (1.052, 1.737) | 1.262 | (.992, 1.577) | 1.528 | (1.426, 1.612) | 1.512 | (1.285, 1.822) |
Turning to the median cure fraction estimates in the second column of Figure 1, the variation in the cure fraction estimates appears to be less tightly centered about the fixed cure fraction estimate than for the melanoma data. Unlike in the melanoma data, here the differences in performance across models are quite evident. The lack of a “plateau” in the observed time frame induces the downward shift of cure fraction for the poorly performing models, so model 1(a) tends to move the cure fraction bar off the center. This reveals greater sensitivity to the modeling assumptions and a bias in the models with a constant cure fraction.
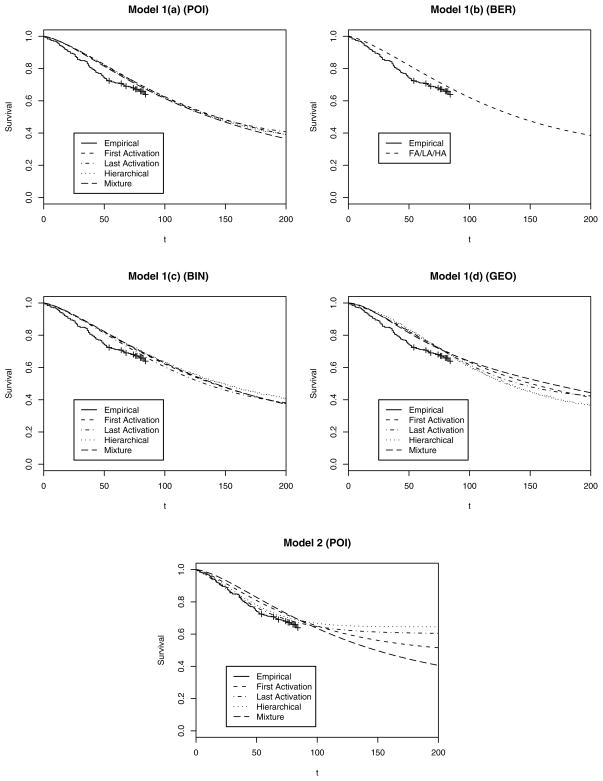
This message carries over to the posterior-predictive survival plots show in Figure 3, where the Kaplan–Meier plot for the survival data runs up to only 84 months, but we extend our fitted curves up to 200 months to show the “plateau.” Generally, the fit of models 1(a)–1(d) (including BG-type models) is less satisfactory than that of model 2, demonstrating the inadequacy of a constant cure fraction. In model 2, the LA and HA–Bin schemes offer better fits than FA (YCIS model), nicely capturing the flattening of the survival curve for the observed data after 50 months. These plots also reveal the differences in the plateaus more distinctly than the melanoma data. Generally, we find that LA reaches the plateau fastest and FA reachest it the slowest, whereas HA–Bin and Mix have intermediate rates. This is expected due to the more stringent conditions for failure in LA than in FA. Finally, we remark that these predictive fits seem to be consistent with the goodness-of-fit measures (G) in Table 4, although they do not inform about formal model penalty (P), which is accounted for by the L measure. In any case, the breast cancer data demonstrate the weaker performances of the existing BG-type and YCIS models and the improvements available using the flexible hierarchical methods.
Figure 3.
Posterior Estimates (median) of the Survival Plots for the Breast Cancer Data Under the Different Activation Schemes. The Kaplan–Meier plot of the observed data is the solid line up to 84 months, whereas the fitted curves are extended up to 200 months to better reveal their shape. The estimated curves are much more sensitive to the modeling assumptions than for the melanoma data.
6. SUMMARY AND DISCUSSION
We have proposed a general class of cure models motivated from latent-activation mechanisms and outlined a hierarchical framework with MCMC inference. Our models generalize existing ones, offer robust inference, and should be especially beneficial in realistic settings where the disease’s mechanistic nature can be only surmised. In fact, statistical estimation of when the survival function hits a plateau is particularly pertinent in cancer studies (e.g., of the breast), where cure is believed although short-term monitoring data might conceal such a plateau. Our melanoma analysis reveals that our models perform consistently with existing models, whereas our breast cancer example demonstrates the need for greater flexibility.
Extensions of our work could explore further possibilities beyond N ~ Po(θ). In fact, interesting hazard structures (e.g., proportional odds) arise with N ~ Geo(θ) in model 2, leading to new classes of identifiable cure models. In addition, whereas the mixing distribution is unidentifiable in semiparametric formulations, joint modeling of cure rate survival data and multivariate longitudinal data (e.g., Ibrahim et al. 2001; Taylor 1995) involves characterizing the mixing structure through observed multivariate longitudinal data. We envision adapting our richer framework for such data. Taking a different tack, we could consider spatial cure models extending the simpler BG-type structures (e.g., Banerjee and Carlin 2004) by incorporating random effects or frailties that are correlated. Our framework can certainly accommodate such modeling, but unresolved issues arise regarding associations in both the latent link and the cure fraction.
Acknowledgments
The work of the first three authors was supported in part by National Institutes of Health grants 1-R01-CA95955 and 1-R01-CA112444. The work of Dr. Sinha was supported by National Cancer Institute grant 9-R01-CA69222. The authors thank the joint editor, associate editor, and three referees for several suggestions, and also Minghui Chen, University of Connecticut, for useful discussions.
APPENDIX: TECHNICAL DETAILS
-
We discuss the identifiability of θ under the prior g(θ) with respect to the density f*(t|θ), which amounts to being finite for all t > 0. We could work with the likelihood , but the general idea of the proof would remain the same. Note that if N ~ Po(θ) and a(N) is any function on the integers, then we have the following simple relationship:
(A.1) First, suppose that r is fixed at w + 1 for some nonnegative integer w. Then f *(t|θ) = f (t)EN [N1(N ≥ w + 1)P(W = w|N)], where W|N ~ Bin(N − 1, F(t)), which equals, by (A.1), θf (t)EN [1(N ≥ w)P(W* = w|N)], where W*|N ~ Bin(N, F(t)). This can be further simplified to givewhich equals θf(t)P(V = w), where V ~ Po(θF(t)). Now we haveWith g(θ) = 1/θ, the foregoing further simplifies to If*(t) = f(t)/F(t) < ∞. This proves the identifiability of θ under the scale-invariance prior whenever r is degenerate at a single point. Even when r and N both vary in the foregoing setting but the difference N − r remains constant, calculations analogous to the previous reveal that If*(t) = f(t)/S(t) = h(t) < ∞ under g(θ) = 1/θ. This proves in particular the identifiability of the cure fraction for the last activation models (N − r = 0).
-
Here we derive (5), which demonstrates how the hierarchical framework leads to a BG-type model. Taking r|N ~ DiscreteUnif (1, N) in (4) and setting r* = r − 1, we obtain
For N ~ Po(θ), it is easily seen by differentiating S*(t|θ) above [or directly by using (A.1)] that f*(t|θ) = f(t)(1 − e−θ), and thus . However, with g(θ) = 1/θ, we immediately see that If*(t) = ∞, proving the nonidentifiability of θ. The classical BG-type model assumes that N ~ Ber(θ), with 1 − θ as the cure fraction. With a logistic link, we have θ = eμ/(1 + eμ), which gives f*(t|μ) = eμf(t)/(1 + eμ) and . With the noninformative flat prior g(μ) ∝ 1, it is straightforward to show that If* (t) = ∞.
-
Let PN (·) denote the conditional probability distribution of r − 1 given N with support {0, …, N − 1}. Then, with N ~ Po(θ), using (A.1) we can write f*(t|θ) = θf (t)E(N,V*)[P(W* = V*|N)], where W*|N ~ Bin(N, F(t)) (as in item 1) and V*|N ~ PN+1(·). Therefore, for θ to be identifiable under g(θ) = 1/θ, the family PN(·) must satisfy
For general PN(·) (i.e., one without all its mass at a single point), this condition may be difficult to verify.
Contributor Information
Freda Cooner, Division of Biostatistics, Office of Surveillance and Biometrics, Center for Devices and Radiological Health, Food and Drug Administration.
Sudipto Banerjee, Email: sudiptob@biostat.umn.edu, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455.
Bradley P. Carlin, Public Health, Division of Biostatistics, School of Public Health, University of Minnesota, Minneapolis, MN 55455.
Debajyoti Sinha, Department of Biostatistics and Bioinformatics, Medical University of South Carolina, Charleston, SC 29425.
References
- Banerjee S, Carlin BP. Parametric Spatial Cure Rate Models for Interval-Censored Time-to-Relapse Data. Biometrics. 2004;60:268–275. doi: 10.1111/j.0006-341X.2004.00032.x. [DOI] [PubMed] [Google Scholar]
- Berkson J, Gage RP. Survival Curve for Cancer Patients Following Treatment. Journal of the American Statistical Association. 1952;47:501–515. [Google Scholar]
- Carlin BP, Louis TA. Bayes and Empirical Bayes Methods for Data Analysis. 2. Boca Raton, FL: Chapman & Hall/CRC Press; 2000. [Google Scholar]
- Chen M-H, Ibrahim JG, Sinha D. A New Bayesian Model for Survival Data With a Surviving Fraction. Journal of the American Statistical Association. 1999;94:909–919. [Google Scholar]
- Cox DR, Oakes D. Analysis of Survival Data. London: Chapman & Hall; 1984. [Google Scholar]
- Ewell M, Ibrahim JG. The Large Sample Distribution of the Weighted Log-Rank Statistic Under General Local Alternatives. Lifetime Data Analysis. 1997;3:5–12. doi: 10.1023/a:1009690200504. [DOI] [PubMed] [Google Scholar]
- Farewell VT. The Use of Mixture Models for the Analysis of Survival Data With Long-Term Survivors. Biometrics. 1982;38:1041–1046. [PubMed] [Google Scholar]
- Farewell VT. Mixture Models in Survival Analysis: Are They Worth the Risk? Canadian Journal of Statistics. 1986;14:257–262. [Google Scholar]
- Gail MH, Santner TJ, Brown CC. An Analysis of Comparative Carcinogenesis Experiments Based on Multiple Times to Tumor. Biometrics. 1980;36:255–266. [PubMed] [Google Scholar]
- Gelfand AE, Ghosh SK. Model Choice: A Minimum Posterior Predictive Loss Approach. Biometrika. 1998;85:1–11. [Google Scholar]
- Goldman AI. Survivorship Analysis When Cure Is a Possibility: A Monte Carlo Study. Statistics in Medicine. 1984;3:153–163. doi: 10.1002/sim.4780030208. [DOI] [PubMed] [Google Scholar]
- Hanin L, Tsodikov A, Yakovlev A. Optimal Schedules of Cancer Surveillance and Tumor Size at Detection. Mathematical and Computer Modelling. 2001;33:1419–1430. [Google Scholar]
- Ibrahim JG, Chen M-H, Sinha D. Bayesian Survival Analysis. New York: Springer-Verlag; 2001. [Google Scholar]
- Kirkwood JM, Ibrahim JG, Sondak VK, Richards J, Flaherty LE, Ernstoff MS, Smith TJ, Rao U, Steele M, Blum RH. High- and Low-Dose Interferon Alfa-2b in High-Risk Melanoma: First Analysis of Intergroup Trial E1690/S9111/C9190. Journal of Clinical Oncology. 2000;19:1226–1228. doi: 10.1200/JCO.2000.18.12.2444. [DOI] [PubMed] [Google Scholar]
- Kirkwood JM, Strawderman MH, Ernstoff MS, Smith TJ, Borden EC, Blum RH. Interferon Alfa-2b Adjuvant Therapy of High-Risk Resected Cutaneous Melanoma: The Eastern Cooperative Oncology Group Trial EST 1684. Journal of Clinical Oncology. 1996;14:7–17. doi: 10.1200/JCO.1996.14.1.7. [DOI] [PubMed] [Google Scholar]
- Kuk AYC, Chen CH. A Mixture Model Combining Logistic Regression With Proportional Hazards Regression. Biometrika. 1992;79:531–541. [Google Scholar]
- Laud P, Ibrahim J. Predictive Model Selection. Journal of the Royal Statistical Society, Ser B. 1995;57:247–262. [Google Scholar]
- Li CS, Taylor JMG. A Semiparametric Accelerated Failure Time Cure Model. Statistics in Medicine. 2002;21:3235–3247. doi: 10.1002/sim.1260. [DOI] [PubMed] [Google Scholar]
- Li CS, Taylor JMG, Sy JP. Identifiability of Cure Models. Statistics and Probability Letters. 2001;54:389–395. [Google Scholar]
- Moolgavkar SH, Luebeck EG, de Gunst M. Two-Mutation Model for Carcinogenesis: Relative Roles of Somatic Mutations and Cell Proliferation in Determining Risk. In: Moolgavkar SH, editor. Scientific Issues in Quantitative Cancer Risk Assessment. Boston: Birkhäuser; 1990. pp. 136–152. [Google Scholar]
- Rao CR. Linear Statistical Inference and Its Applications. 2. New York: Wiley; 1973. [Google Scholar]
- Spiegelhalter DJ, Best N, Carlin BP, van der Linde A. Bayesian Measures of Model Complexity and Fit (with discussion) Journal of the Royal Statistical Society, Ser B. 2002;64:583–639. [Google Scholar]
- Sy JP, Taylor JMG. Estimation in a Cox Proportional Hazards Cure Model. Biometrics. 2000;56:227–236. doi: 10.1111/j.0006-341x.2000.00227.x. [DOI] [PubMed] [Google Scholar]
- Taylor JMG. Semiparametric Estimation in Failure Time Mixture Models. Biometrics. 1995;51:899–907. [PubMed] [Google Scholar]
- Tsodikov A, Ibrahim J, Yakovlev A. Estimating Cure Rates From Survival Data: An Alternative to Two-Component Mixture Models. Journal of the American Statistical Association. 2003;98:1063–1078. doi: 10.1198/01622145030000001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SL, Taylor JMG. Improved Models of Tumor Cure. International Journal of Radiational Biology. 1996;70:539–553. doi: 10.1080/095530096144743. [DOI] [PubMed] [Google Scholar]
- Tucker SL, Thames HD, Taylor JMG. How Well Is the Probability of Tumor Cure After Fractionated Irradiation Described by Poisson Statistics? Radiation Research. 1990;124:273–282. [PubMed] [Google Scholar]
- Yakovlev AY. Threshold Models of Tumor Recurrence. Mathematical and Computer Modelling. 1996;23:153–164. [Google Scholar]
- Yakovlev AY, Asselain B, Bardou VJ, Fourquet A, Hoang T, Rochefordiere A, Tsodikov AD. A Simple Stochastic Model of Tumor Recurrence and Its Application to Data on Premenopausal Breast Cancer. In: Asselain B, Boniface M, Duby C, Lopez C, Masson JP, Tranchefort J, editors. Biometrie et Analyse de Donnees Spatio-Temporelles. Vol. 12. Rennes, France: Société Française de Biométrie; 1993. pp. 66–82. [Google Scholar]
- Yakovlev AY, Tsodikov AD. Stochastic Models of Tumor Latency and Their Biostatistical Applications. Singapore: World Scientific; 1996. [Google Scholar]
- Yin GS, Ibrahim JG. Cure Rate Models: A Unified Approach. The Canadian Journal of Statistics. 2005;33:559–570. [Google Scholar]