Abstract
The high sterol concentration in eukaryotic cell membranes is thought to influence membrane properties such as permeability, fluidity and microdomain formation. Drosophila cannot synthesize sterols, but do require them for development. Does this simply reflect a requirement for sterols in steroid hormone biosynthesis, or is bulk membrane sterol also essential in Drosophila? If the latter is true, how do they survive fluctuations in sterol availability and maintain membrane homeostasis? Here, we show that Drosophila require both bulk membrane sterol and steroid hormones in order to complete adult development. When sterol availability is restricted, Drosophila larvae modulate their growth to maintain membrane sterol levels within tight limits. When dietary sterol drops below a minimal threshold, larvae arrest growth and development in a reversible manner. Strikingly, membrane sterol levels in arrested larvae are dramatically reduced (dropping sixfold on average) in most tissues except the nervous system. Thus, sterols are dispensable for maintaining the basic membrane biophysical properties required for cell viability; these functions can be performed by non-sterol lipids when sterols are unavailable. However, bulk membrane sterol is likely to have essential functions in specific tissues during development. In tissues in which sterol levels drop, the overall level of sphingolipids increases and the proportion of different sphingolipid variants is altered. These changes allow survival, but not growth, when membrane sterol levels are low. This relationship between sterols and sphingolipids could be an ancient and conserved principle of membrane homeostasis.
Keywords: Drosophila, Ecdysone, Growth, Membranes, Sphingolipids, Sterols
INTRODUCTION
The ability to synthesize sterols evolved in the earliest microbial eukaryotes (Volkman, 2003), coincident with rising atmospheric oxygen levels (Chen et al., 2007). The synthesis of one cholesterol molecule requires 11 molecules of oxygen (Espenshade and Hughes, 2007) and, unlike other lipids, no energy is produced by sterol catabolism (Haines, 2001). Sterols make up a significant fraction of the lipids in cell membranes, and high concentrations of membrane sterol are thought to confer the biophysical properties that are essential for membrane function. Studies of model membranes containing simple lipid mixtures show that cholesterol influences lipid chain order, thereby preventing the formation of gel-phases at low temperatures and affecting the balance of liquid disordered and liquid ordered phases at higher temperatures. It has been proposed that cholesterol-dependent phase separation within lipid bilayers contributes to the spatial segregation of membrane proteins in vivo (Mouritsen and Zuckermann, 2004; Simons and Vaz, 2004). A cholesterol-dependent increase in lipid packing order also leads to a decrease in ion permeability in model membranes, suggesting that membrane cholesterol might help to maintain membrane potential in vivo (Haines, 2001). Results of cholesterol depletion experiments support the idea that bulk membrane cholesterol has an important role in vivo. Lowering membrane cholesterol levels by cyclodextrin extraction or by inhibiting cholesterol biosynthesis enzymes disturbs protein sorting and signaling. This has been attributed to perturbation of liquid ordered membrane microdomains (Brown and London, 2000; Lucero and Robbins, 2004; Pike, 2004; Pike, 2005; Schuck and Simons, 2004; Sengupta et al., 2007; Simons and Ikonen, 1997; Simons and Vaz, 2004). Thus, cholesterol has important functions in the membrane that justify the large energetic investment in its synthesis.
In this context, it is surprising that the ability to synthesize sterols has been lost in many invertebrates, such as insects and nematodes (Clayton, 1964; Hobson, 1935b). It has been known for over 70 years that insects are sterol auxotrophs, requiring dietary sources of sterols for their development (Hobson, 1935a). However, the exact nature of this requirement has been unclear. Could an animal that does not synthesize sterols reliably accumulate sufficiently high levels to maintain membrane biophysical properties? Vertebrate cells sense membrane sterol levels in the endoplasmic reticulum (ER) via the sterol regulatory element-binding protein (SREBP) pathway. When activated by low sterol levels, SREBP increases sterol biosynthesis and the uptake of sterols from lipoproteins (Chang et al., 2006; Espenshade and Hughes, 2007). Flies cannot activate sterol biosynthesis in response to low membrane sterol levels, and the Drosophila SREBP pathway does not respond to sterols, but to phosphatidylethanolamine (Dobrosotskaya et al., 2002; Kunte et al., 2006; Seegmiller et al., 2002). If sterols do have important functions in fly membranes, how are its levels controlled and protected from fluctuations in dietary availability? Another sterol auxotroph, the nematode Caenorhabditis elegans, has apparently solved this problem by evolving other lipids to replace the biophysical functions of sterols. These animals do not accumulate sterol in the membranes of most tissues, even when they are present in the diet (Matyash et al., 2001) (see also Fig. S1 in the supplementary material), and require them only for steroid hormone biosynthesis (Kurzchalia and Ward, 2003; Matyash et al., 2004). However, sterols do make up a significant fraction of Drosophila cell membranes (Rietveld et al., 1999). Are they necessary for membrane function? Or do Drosophila, like C. elegans, require sterols only for steroidogenesis? To answer these questions, we have manipulated the availability of dietary sterol during Drosophila larval development. By feeding with different sterols and with the steroid hormone ecdysone, we distinguish a requirement for bulk membrane sterol from its function in steroidogenesis. We show that Drosophila maintain membrane sterol levels within tight limits, in part by regulating growth. Finally, we show that Drosophila survive extreme dietary sterol depletion by increasing the production of specific sphingolipid variants that compensate for low membrane sterol.
MATERIALS AND METHODS
Cell culture
S2R+ cells were maintained in complete medium [Schneider's complete medium (Invitrogen) containing 10% fetal bovine serum (FBS) and penicillin or streptomycin] or in delipidated medium (delipidated FBS) (Gupta et al., 2009).
Flies
npc1b mutants (Voght et al., 2007) were provided by L. Pallanck. RNAi lines against fa2h (CG30502 – FlyBase) were obtained from the Vienna Drosophila RNAi Center (VDRC) and the National Institute of Genetics (NIG). Both were combined with UAS:Dicer2 (X chromosome) to enhance RNAi efficiency. To produce UAS:fa2h transgenics, we cloned fa2h cDNA (RE68078), mutated to remove an EcoR1 site, into pUhr3 (Marois et al., 2006) cut with EcoR1 and Kpn1. The lace mutant l(2)k05305 has been described previously (Adachi-Yamada et al., 1999). GAL4 lines used were: tubulinGAL4, elavGAL4 and adhGAL4 (Bloomington), npc1bGAL4 (Voght et al., 2007) and lipoGAL4 (Brankatschk and Eaton, 2010).
Feeding experiments
Embryos were collected for 2 hours on apple juice/agar plates, rinsed in PBS containing 0.05% Triton X-100, treated for 30 seconds with 50% NaClO, then rinsed with distilled water. One embryo was placed in each well of a 24-well plate containing lipid-depleted medium [LDM; 10% chloroform-extracted yeast autolysate (Sigma), 10% glucose, 1% chloroform-extracted agarose and 0.015% Nipagen] or yeast medium (YM; 10% live yeast, 10% glucose, 1% chloroform-extracted agarose and 0.015% Nipagen). Isolation of larvae prevents sterol acquisition by cannibalism, ensures identical nutrition and increases developmental synchrony. Unless otherwise specified, panels represent the average of three independent experiments with 48 larvae.
Glucose medium contained 10% glucose, 1% chloroform-extracted agarose and 0.015% Nipagen. Where indicated, LDM was supplemented with either sterol or ecdysone. Sterols were added in a 1 mM ethanolic solution. α-Ecdysone (Fluka), stored as a 5 mM ethanolic solution, was added at intervals to mimic the endogenous bursts of ecdysone synthesis associated with molting and pupariation.
Identification of larval stages and measurement of size
Larval stages were distinguished on the basis of mouth hook and spiracle morphology (Bodenstein, 1994). Larval volume was measured as described previously (Colombani et al., 2005). Adult wing measurements were performed as described previously (Eugster et al., 2007).
Preparation of membrane lipids
Larvae were homogenized, then centrifuged at 1000 g to remove nuclei, unbroken cells and cuticles. Supernatants were centrifuged for 3 hours at 154,000 g to pellet membranes. The procedure was performed at 4°C. Membrane lipids were the extracted as described previously (Bligh and Dyer, 1959).
Thin layer chromatography (TLC)
Phospholipids were quantified as described previously (Rouser et al., 1970). Lipid extracts containing equal amounts of phospholipids were loaded on silica TLC plates (Merck) and run in two sequential solvent systems (Kuerschner et al., 2005). Where indicated, sterol separation was increased by soaking TLC plates in a 12.5% aqueous solution of silver nitrate before loading. Lipids were detected by spraying with 20% sulfuric acid and heating to 150-200°C for 5 minutes.
Saponification
To remove saponifiable lipids, dried lipid extract containing 140 nmol inorganic phosphate was warmed for 1 hour at 80°C in 2 ml 0.3 M methanolic KOH. Non-saponifiable lipids were extracted three times with diethyl ether and run through DEAE Sephadex A-50 columns to remove fatty acids.
Mass spectrometry (MS)
Top-down analysis of the fly total membrane lipidome was performed on an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a NanoMate robotic nanoflow ion source (Advion BioSciences, Ithaca, NJ, USA) as described previously (Schwudke et al., 2007a). MS/MS experiments on the LTQ Orbitrap and the QSTAR Pulsar i (MDS Sciex, Toronto, Canada) mass spectrometers were performed as described previously (Schwudke et al., 2007b). Absolute quantification of ceramides and hexosyl-ceramides was performed by adding 250 nmol of Cer 35:1;2 and GlcCer 30:1;2 (Avanti Polar Lipids. Alabaster, AL, USA) as standards to the lipid extracts. Quantification of sphingolipids relied on extracted intensities of specific structural fragments of the long chain base (LCB): m/z 264.2 (for LCB 18), 236.2 (for LCB 16), 234.2 (for LCB 16:1), 208.2 (for LCB 14) and 206.2 (for LCB 14:1).
To quantify lipids in hemolymph and individual tissues, internal standards were added to the samples prior to extraction: PC 18:3-18:3, 40 pmol; PE 17:0-17:0, 52 pmol; Cer 35:1;2, 20 pmol; GalCer 30:1;2, 20 pmol; LacCer 30:1;2, 20 pmol. Sample amounts were adjusted so that ∼ 2 nmol of total lipid was extracted from each. Samples were extracted as described previously (Zech et al., 2009), then re-suspended and infused in positive ion mode as described previously (Schwudke et al., 2007a). The amount of each species is expressed as mole percent (mol%) with respect to all polar lipids detected (including phospholipids and sphingolipids). CerPE species were quantified using the PE 17:0-17:0 internal standard.
Lipid identification was performed using LipidX software (developed at the Max-Planck Institute of Molecular Cell Biology and Genetics by Ronny Herzog, D.S. and A.S.).
Sterol quantification
Cholesterol was quantified enzymatically using a Cholesterol/Cholesteryl Ester Quantitation Kit (K603-100; Biovision, Mountain View, California, USA) following the manufacturer's instructions.
Filipin staining
Tissues were fixed and stained with the fluorescent sterol-binding compound filipin (Sigma) as described previously (Voght et al., 2007) and mounted using VECTASHIELD mounting medium (Vector Laboratories).
Immunostaining
Second instar fat bodies were removed on ice in PBS containing protease inhibitors (Roche) on ice, immediately transferred to 4% formaldehyde in PBS, then stained as described previously (Colombani et al., 2005). Anti-Drosophila Foxo antibody (Colombani et al., 2005) was used at 1:500 and DAPI at 1:5000.
Microscopy
Tissues were imaged using a Zeiss LSM 510 confocal microscope using a Plan-Neofluor 40×/1.3 Oil Ph3 objective. Tissues from the same experiment were imaged under identical conditions on the same day. Image J and Adobe Photoshop were used for image processing.
RT-PCR
Real-time RT-PCR was performed as described previously (Colombani et al., 2005). Larvae kept in single wells were fed either LDM or LDM + cholesterol and were collected 4 days after egg laying (AEL). Larval RNA (2.5 μg per reaction) was reverse transcribed and 3 ng of the resulting cDNA was used as a template for PCR. Primers were designed using the Primer3: WWW primer tool (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) and selected according to efficiency (100% ± 2%). Primer concentrations were: 200 nM for gapdh1, fa2h and e74b, and 300 nM for 4ebp. e74b was used for normalization as its expression did not change under different feeding conditions. Transcript levels for each of three independent larval collections were measured in triplicate. Relative gene expression was calculated using the comparative CT method as described previously (Schmittgen and Livak, 2008).
For non-quantitative RT-PCR, 135 ng of RNA was reverse transcribed. 10 ng of cDNA was used as a template for fa2h and 5 ng for actin5C in a 35 cycle reaction using HotStarTaq Plus DNA Polymerase (QIAGEN).
Primers are listed in Table S1 in the supplementary material.
RESULTS
Sterol depletion causes reversible arrest of larval growth and development
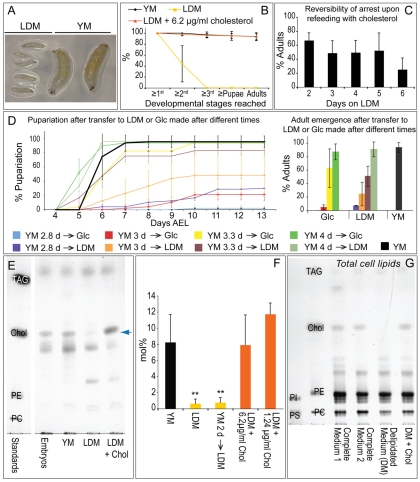
To investigate how Drosophila respond to sterol depletion, we placed single embryos on either live yeast-containing medium (YM) or on lipid-depleted medium (LDM) containing chloroform-extracted yeast lysate, and observed larval development (see Fig. S2 in the supplementary material). Most animals fed YM developed to adults. By contrast, those fed LDM arrested in the first or second larval instar (Fig. 1A,B). Although the second larval instar normally lasts one day, larvae that arrested in the second instar upon lipid depletion live for more than five days without further development, continuing to feed until shortly before death. Strikingly, adding cholesterol to LDM allowed 95% of animals to complete adult development (Fig. 1B). Thus, sterol is the only essential dietary lipid missing from LDM; absence of dietary sterol arrests larval growth and development.
Fig. 1.
Dietary sterol depletion reversibly arrests growth and development and reduces membrane sterol concentration. (A) Larvae 120 hours (5 days) after egg laying (AEL) fed on either lipid-depleted medium (LDM) or yeast medium (YM). (B) Percentage of larvae reaching the indicated developmental stages when fed YM, LDM or LDM + 6.2 μg/ml cholesterol. (C) Percentage of adults emerging when larvae were transferred to LDM + cholesterol after being maintained for different times on LDM. (D) Larvae were transferred from YM to either glucose (Glc) medium or LDM 2.8-4 days AEL (e.g. YM 2.8→Glc means larvae were transferred from YM to Glc medium 2.8 days AEL). Left panel shows the percentage pupariation of these animals over time. Right panel shows the percentage of animals reaching adulthood. (E) Thin layer chromatography (TLC) of non-saponifiable membrane lipids of embryos and larvae fed on different diets. Blue arrow indicates sterols. (F) Quantification of sterol (mol% with respect to phospholipids) present in different larval membrane lipid extracts. Asterisks indicate a statistically significant difference (P≤0.01) when compared with larvae fed YM. Membrane sterol levels in larvae fed 1.24 μg/ml or 6.2 μg/ml cholesterol did not differ significantly from each other or from that of YM-fed animals (P≤0.07). (G) TLC of total lipids from S2R+ cells grown in complete or delipidated medium. Equal amounts of phospholipid were loaded. TAG, triacylglycerides; Chol, cholesterol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PC, phosphatidylcholine; PS, phosphatidylserine. Error bars indicate standard deviations.
To determine whether sterol depletion-induced growth arrest is reversible, larvae fed only LDM for different lengths of time were provided with food containing cholesterol. After 5 days on LDM, at least 50% of larvae resumed growth and completed adult development when cholesterol was provided (Fig. 1C). Thus, growth arrest is reversible; larvae maintained on LDM are viable with no irreversible damage that prevents subsequent adult development.
In order to determine whether dietary sterol is continuously necessary for larval development, or is required only at specific stages, we transferred larvae from YM to LDM at different times and quantified pupariation and adult emergence (Fig. 1D). Larvae transferred to LDM at 4 days AEL pupariated earlier than those left on yeast, and 90% emerged as adults. Fifty percent of larvae fed with sterols until 3.3 days AEL produced viable adults, although pupariation was delayed. When transferred at earlier stages, most larvae arrested in the second or third larval instar. These effects were similar in timing to those caused by transfer to medium containing glucose alone (Fig. 1D). Thus, the times at which dietary sterol no longer influenced pupariation or adult development correspond to well-known insect growth milestones: `critical weight' and `minimal viable weight'. At minimal viable weight, total starvation no longer prevents adult development, and at critical weight it does not delay pupariation (Davidowitz et al., 2003; Edgar, 2006; Mirth and Riddiford, 2007). Continuous sterol availability was necessary for larvae to reach these milestones.
Dramatic reductions in membrane sterol do not affect cell or larval viability
To determine whether dietary sterol depletion reduces membrane sterol concentrations in growth-arrested larvae, we used TLC and enzymatic quantification to determine the mole percentage (mol%) of sterol (with respect to phospholipids) in larval membranes (Fig. 1E,F). Larvae raised on YM or on LDM + cholesterol accumulated 9 mol% membrane sterol. Strikingly, sterol-depleted animals retained on average only 1.5 mol% membrane sterol. A sixfold decrease was also detected by gas chromatography-mass spectrometry (GC-MS) (see Fig. S3 in the supplementary material). As growth-arrested larvae remain viable, this shows that dramatically reducing average membrane sterol concentration does not reduce larval viability.
We depleted sterols from the medium used to grow Drosophila S2R+ cells in order to determine whether Drosophila cell viability requires membrane sterol. These cells grow at a normal rate following adaptation to medium containing delipidated serum (Gupta et al., 2009). Cholesterol was present in membranes of S2R+ cells grown using normal serum; however, it was undetectable in cells grown on delipidated serum (Fig. 1G). This confirms previous results using Kc insect cells (Silberkang et al., 1983) and shows that these cells do not require significant levels of membrane sterol.
Specific tissues and membrane regions retain sterols preferentially
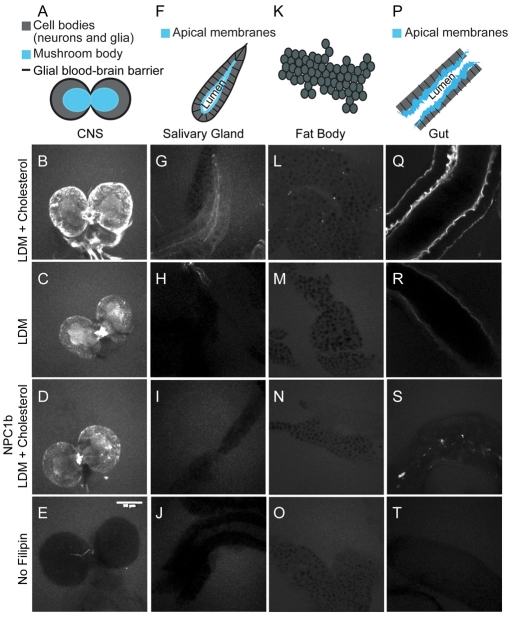
To determine how sterol depletion affects different tissues, we stained a range of tissues with filipin, a fluorescent sterol-binding compound, and imaged them under identical conditions. In cholesterol-fed second instar larvae, filipin staining was strongest in the gut (Fig. 2Q) and central nervous system (CNS) (Fig. 2B). In epithelial tissues, such as the gut and salivary glands, sterols were enriched in apical membranes (Fig. 2G,Q). Surprisingly, filipin staining in fat body cell membranes was low, even when larvae were fed LDM + cholesterol (Fig. 2L). Thus, sterol normally accumulates to different levels in different cell membranes. When sterols were depleted, either by dietary methods (Fig. 2C,H,M,R) or by mutation of the gut sterol transporter NPC1b (Voght et al., 2007) (Fig. 2D,I,N,S), some cell membranes retained sterols preferentially. Although filipin staining was reduced in blood-brain barrier glia surrounding the CNS (Fig. 2A, black) and on neuronal cell bodies (Fig. 2A, gray), it did not decrease in the mushroom body (Fig. 2A, blue), which is composed of fasciculated axon bundles (Fig. 2, compare B with C,D). Sterol also remained in gut apical membranes when animals were fed LDM, although staining disappeared from other regions of gut cells (Fig. 2R). Thus, the ability to retain membrane sterol is tissue specific. Although axonal membranes probably retain more than the larval average of 1.5 mol% sterol upon sterol depletion, membranes in other tissues are likely to retain even less. Thus, whereas the function of membrane sterol is replaceable in most cells, sterols might still have essential tissue-specific functions in neurons or epithelia. Interestingly, depletion of apical membrane sterol in the gut was more dramatic in the npc1b mutant than in wild-type larvae fed with LDM (Fig. 2, compare R with S). Despite this, npc1b mutants absorb glucose normally (Voght et al., 2007), suggesting that the function of the gut is not severely compromised by loss of membrane sterol.
Fig. 2.
Specific tissues and membrane regions retain sterols preferentially. (A-T) Second instar larval tissues (represented diagrammatically in A,F,K,P) stained with filipin. The CNS (A-E), salivary glands (F-J), fat bodies (K-O) and midguts (P-T) from wild-type larvae fed LDM + cholesterol (B,G,L,Q) or LDM alone (C,H,M,R), and from npc1b mutant larvae fed LDM + cholesterol (D,I,N,S) are shown. E, J, O and T are unstained controls. Scale bar: 50 μm.
Adult development requires both bulk membrane sterol and steroid hormones
Why do larvae arrest growth upon dietary sterol depletion? One possibility is that growth requires normal levels of membrane sterol. Another possibility is that sterol depletion might reduce sterol levels below a threshold required for ecdysone biosynthesis. Pulses of the steroid hormone ecdysone regulate the larval molts, the larval to pupal transition and a variety of pupal events including eclosion (Thummel, 2001). In order to test whether ecdysone was the only essential factor missing from sterol-depleted larvae, we supplemented LDM with ecdysone at times that would mimic normal ecdysone pulses. Three additions of 1 μg/ml ecdysone allowed all larvae to molt to the second instar (Fig. 3A). Increasing the third ecdysone pulse to 50 μg/ml allowed 20% to reach the third instar (Fig. 3A). However, although these larvae molt, their size did not exceed that of second instar larvae fed LDM alone (Fig. 3B). Thus, whereas ecdysone is sufficient to allow larval molting, growth requires other sterol functions.
Fig. 3.
Adult development requires both bulk membrane sterol and steroid hormones. (A) Percentage of larvae reaching the indicated developmental stages when fed either LDM, LDM + three additions of 1 μg/ml ecdysone (Ecd), or LDM + two additions of 1 μg/ml ecdysone + one addition of 50 μg/ml ecdysone. (B) Larval volume over time when larvae are fed either LDM or LDM + two additions of 1 μg/ml ecdysone + one addition of 50 μg/ml ecdysone. (C) Structures of different sterols and ecdysone. Green shaded sterols support adult development. Pink shaded sterols do not. Cholesterol is shown with the numbering system indicating different carbons. Red structural regions highlight differences from cholesterol. (D,E) Percentage of larvae reaching the indicated developmental stages when fed LDM supplemented with sterols or ecdysone (6.2 μg/ml in D or the indicated amounts in μg/ml in E). (F) Argentated TLC of membrane lipid extracts of larvae fed LDM + 6.2 μg/ml desmosterol + 0.13 μg/ml cholesterol. Desmosterol (red arrow) is the only detectable membrane sterol. TAG, triacylglycerides; D, desmosterol; Erg, ergosterol; C, cholesterol. Error bars indicate standard deviations.
To more precisely distinguish sterol requirements in cell membranes from those of steroidogenesis, we fed larvae with a wide range of different sterols (Fig. 3C). All had structural features that should allow them to fulfill bulk requirements in membranes (a planar ring, 3′ hydroxyl group and alkyl side chain) (Bloch, 1983; Demel and De Kruyff, 1976). However, some sterols might be unusable as precursors for ecdysteroid biosynthesis. We found that many sterols supported the development of Drosophila larvae to adulthood, consistent with previous reports (Cooke and Sang, 1970). In addition to cholesterol, these included plant sterols (stigmasterol and sitosterol), 7-dehydrocholesterol and cholestanol (Fig. 3D). Thus, these sterols can fulfill any structural function in the membrane and can also be used as substrates for the biosynthesis of any required sterol-derived signaling molecules. By contrast, neither ergosterol nor desmosterol supported adult development (Fig. 3D). These animals reproducibly arrested development in the first (desmosterol) or third (ergosterol) larval instar.
These sterols might fail to support adult development because they do not interact normally with membrane proteins, or because the biophysical properties of membranes containing them are abnormal. Alternatively, these sterols might be unable to support the biosynthesis of specific signaling molecules. Addition of small amounts (1:50 molar ratio) of cholesterol should not affect the bulk properties of membranes containing desmosterol or ergosterol. Indeed, only desmosterol was detected in the membranes of flies fed cholesterol:desmosterol in a molar ratio of 1:50 (0.13 μg/ml cholesterol + 6.2 μg/ml desmosterol) (Fig. 3F). However, small amounts of cholesterol might rescue the production of signaling molecules, which act at much lower concentrations. To distinguish these possibilities, we investigated whether the development of animals fed desmosterol or ergosterol could be rescued to adulthood by the addition of 0.13 μg/ml cholesterol (an amount that alone cannot support adult development). Cholesterol (0.13 μg/ml) allowed 40% of desmosterol-fed animals and 83% of ergosterol-fed animals to complete development (Fig. 3E). This suggests that both ergosterol and desmosterol function fairly normally in cell membranes, but that one or more essential signaling molecules cannot be produced from these sterols. Furthermore, because 0.13 μg/ml cholesterol cannot support adult development in the absence of larger amounts of other sterols, it is likely that significant amounts of bulk membrane sterol are essential for adult development.
In order to determine whether ecdysone was the only sterol-derived signaling molecule missing from animals fed with desmosterol, we provided larvae with exogenous ecdysone at intervals that mimicked the naturally occurring peaks. All larvae fed desmosterol alone arrested in the first instar, whereas 30% of animals supplemented with ecdysone pupariated (Fig. 3, compare D with E), but did not emerge as adults. Because pupae do not feed, they cannot access ecdysone in the food, which might explain why these animals fail to complete adult development. These data show that desmosterol cannot be used to synthesize ecdysone, and indicate that ecdysone is the only essential sterol-derived signaling molecule missing from desmosterol-fed larvae, at least until pupal stages. Taken together, these data show that Drosophila growth and development require a small amount of sterol that is suitable as an ecdysone precursor, and larger amounts of sterols with more flexible structural requirements, again suggesting that sterols might have important functions as bulk components of the membrane.
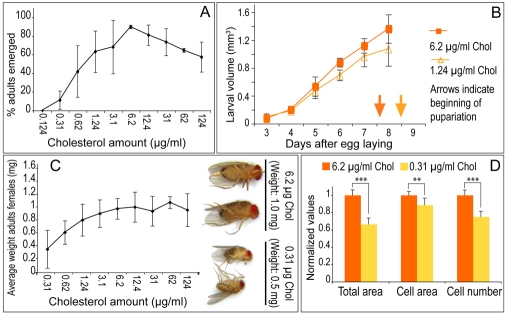
To further assess requirements for bulk sterol in the membrane, we examined whether adult development requires a minimum level of membrane sterol. In order to do this, we first titrated dietary sterol to determine the minimum amount required to complete development (Fig. 4A). Ninety-five percent of larvae grown on plates containing LDM + 6.2 μg/ml cholesterol became adults, similar to larvae fed YM (Fig. 1B, Fig. 4A). Sixty percent emerged as adults when fed LDM + 1.24 μg/ml cholesterol, and no adults emerged when larvae were fed 0.12 μg/ml cholesterol (Fig. 4A). We then compared membrane sterol levels in larvae fed YM, LDM + 6.2 μg/ml cholesterol and LDM + 1.24 μg/ml cholesterol (the minimal amount supporting adult development). Animals fed 1.24 μg/ml cholesterol accumulated as much membrane sterol as did animals fed YM (Fig. 1F). Thus, membrane sterol levels in animals fed minimally sufficient amounts of sterol are equivalent to those of animals fed ad libitum. Furthermore, increasing dietary sterol availability did not elevate membrane sterol beyond this point (Fig. 1F). Thus, membrane sterol concentration is tightly regulated and adult development does not occur until the optimal level is reached.
Fig. 4.
Growth is modulated by dietary sterol availability. (A) Percentage of adults emerging from larvae fed LDM + different amounts of cholesterol. (B) Growth of larvae fed LDM supplemented with different amounts of cholesterol. (C) Average weights of adult females fed LDM + different amounts of cholesterol as larvae. Photos show examples of how females developed on LDM + indicated amounts of cholesterol/ml. (D) Average wing area, cell area and cell number in wings from adult females fed LDM + indicated amounts of cholesterol. ***, P≤0.001, **, P≤0.01. Error bars indicate standard deviations.
Drosophila larvae regulate growth to maintain optimal membrane sterol levels
When exposed to a level of dietary sterol above the threshold required to release growth arrest, Drosophila larvae maintain the same optimal membrane sterol level, even when widely varying amounts are present in the food. What mechanisms ensure proper membrane sterol levels under such different conditions? We examined the idea that larvae might regulate their growth so that they do not exceed their ability to absorb sterols and incorporate them into the membrane. We compared larval growth rate and final adult body size of animals reared on LDM containing between 1.24 μg/ml cholesterol (which allows 60% adult development) and 6.2 μg/ml dietary cholesterol (which allows 95% adult development) (Fig. 4B,C). Within this range, increasing the amount of dietary cholesterol increased larval growth rate, decreased the time required for pupariation (Fig. 4B) and produced larger adults (Fig. 4C). The increase in body size reflected mainly an increase in cell number (Fig. 4D). These data suggest that Drosophila larvae regulate their growth to maintain optimal levels of membrane sterol when dietary availability is limited.
How do sterols regulate growth? Real-time RT-PCR from RNA of larvae grown on LDM and on LDM + cholesterol revealed that sterol depletion reduced mRNA levels for the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase 1 (GAPDH1) by 35% (P≤0.05) and increased mRNA levels of the translation inhibitor 4EBP 5.5-fold (P≤0.05). Thus, sterol depletion might reduce growth, in part, by inhibiting glycolysis and repressing translation. Similar changes in glycolysis and in 4EBP transcription are produced by nuclear translocation of the transcription factor Foxo in response to reduced insulin/Akt signaling (Junger et al., 2003; Puig et al., 2003). To determine whether dietary sterol regulates the nuclear translocation of Foxo, we compared Foxo immunostaining in fat bodies from animals raised on YM, LDM or LDM + cholesterol. Surprisingly, Foxo was localized mainly in the nucleus in larvae fed with either LDM or with LDM + cholesterol, although it was predominantly cytoplasmic in larvae fed with YM (see Fig. S4A in the supplementary material). This suggests that cholesterol does not influence GAPDH1 or 4EBP transcription by regulating nuclear translocation of Foxo. Sterols might regulate Foxo activity within the nucleus, or act through an independent pathway. These data also suggest that an additional non-sterol lipid, which is missing from LDM, is required to maintain Foxo in the cytoplasm and allow maximal growth. Consistent with this, feeding with LDM and maximal amounts of cholesterol produced smaller flies than feeding with YM (see Fig. S4B in the supplementary material).
Sterol depletion alters abundance of specific sphingolipid variants
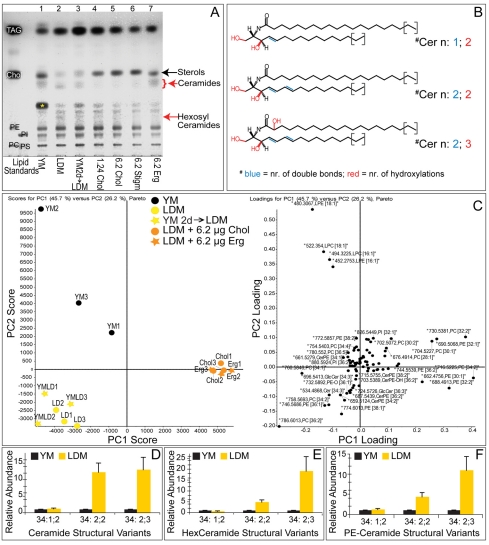
Studies in model membranes show that fluidity and ion impermeability vary greatly with sterol concentration (de Almeida et al., 2003; Haines, 2001). How do Drosophila membranes retain their biophysical properties over such a wide range of sterol concentrations? To address this, we first investigated whether specific lipids were upregulated in response to sterol depletion. TLC analysis suggested that the abundance of several non-sterol lipids changed in response to sterol depletion (Fig. 5A). To identify these lipids, we extracted them from TLC silica plates and analyzed them by MS. The lipids upregulated by sterol depletion corresponded to specific sphingolipid species.
Fig. 5.
Sterol depletion induces specific changes in the Drosophila lipidome. (A) TLC of total membrane lipids from larvae fed with different media: lane 1, larvae fed YM until 4 days AEL (third instar); lane 2, larvae fed LDM until 4 days AEL (second instar); lane 3, larvae fed YM until 2 days AEL, then LDM for 6 days (third instar); lane 4, larvae fed LDM + 1.24 mg/ml cholesterol until 5 days AEL (third instar); lane 5, larvae fed LDM + 6.2 mg/ml cholesterol until 5 days AEL (third instar); lane 6, larvae fed LDM + 6.2 mg/ml stigmasterol until 5 days AEL (third instar); lane 7, larvae fed LDM + 6.2 mg/ml ergosterol until 5 days AEL (third instar). Red arrows indicate bands that appear in response to lipid depletion and that are decreased by sterol feeding. Asterisk indicates a band that increases in larvae fed YM. Standards are as in Fig. 1. (B) Structures of ceramides detected by mass spectrometry (other sphingolipids differ only in the headgroup). These lipids contain a long chain base (LCB) with either 14 or 16 carbon atoms and N-amidated fatty acid moieties of 18 to 24 carbon atoms. Major fatty acids are 20:0 and 22:0. (C) Principal component (PC) analysis of membrane lipids. Left panel: PCA plot. Right panel: Loading plot. Top-down analysis recognized 80 lipid peaks in membrane lipid extracts (three independent experiments). Larvae were fed YM, LDM for 4 days (LDM), YM until 2 days AEL then LDM for 6 days (YM 2d→LDM), LDM + 6.2 μg/ml cholesterol or LDM + 6.2 μg/ml ergosterol. Clustering of the yellow, orange and black points reveals diet-specific changes in fly membrane lipidome. Lipid abundance was normalized to median peak ratios of sample YM1. Selected representative response profiles of various lipids are shown in Fig. S6 in the supplementary material. (D-F) Relative abundance (determined by mass spectrometry) of membrane sphingolipids from larvae fed with YM or LDM (normalized to values for larvae fed on YM). Error bars indicate standard deviations.
Sphingolipids are derived from a sphingoid base [or long chain base (LCB)], an aliphatic amino alcohol. The LCB is amide-linked to different fatty acid moieties to produce ceramide (Cer). The ceramide is then O-linked to various charged headgroups, producing sphingolipids such as hexosylceramide (HexCer) and, in Drosophila, phosphatidylethanolamine ceramide (PECer) (Lahiri and Futerman, 2007; Rietveld et al., 1999). The sphingolipids induced upon sterol depletion were unusual in that they contained an additional double bond in the LCB (Fig. 5B; see Fig. S5 in the supplementary material). Lipids with a similar sphingadiene LCB have been described in Drosophila (Fyrst et al., 2008) and Manduca (Abeytunga et al., 2004). Interestingly, a significant proportion of these sphingolipids contained N-amide-bound fatty acids that were α-hydroxylated (Fig. 5B, hydroxylation in red). Species with two double bonds in the sphingoid base and a hydroxylated fatty acid moiety are denoted n:2;3 sphingolipids and the corresponding lipids with non-hydryoxylated fatty acids are denoted n:2;2.
Independent MS profiling (Schwudke et al., 2007a) of membrane lipids from animals fed YM, LDM or LDM + sterol showed that lipid depletion caused a complex perturbation of the lipidome (Fig. 5C). Some changes were reversed by addition of ergosterol or cholesterol to LDM, suggesting that they were a specific consequence of sterol depletion (Fig. 5C; see Fig. S6 in the supplementary material). Sterol depletion caused a 10- to 20-fold increase of the major Cer, PECer and HexCer n:2;3 species, and a four- to 12-fold increase in sphingolipid n:2;2 species (Fig. 5D-F).
How are n:2;2 and n:2;3 sphingolipids produced? No enzyme that produces sphingolipid bases with two double bonds has yet been identified in any organism; however, the Drosophila genome contains a single homolog of fatty acid 2-hydroxylase (FA2H). This enzyme generates the α-hydroxy fatty acids present in some vertebrate sphingolipids (Maldonado et al., 2008; Mizutani et al., 2008; Uchida et al., 2007). To determine whether it has a similar activity in Drosophila, we altered its levels and measured the resulting changes in sphingolipid fatty acid hydroxylation. Ubiquitous overexpression of Drosophila fa2h increased the ratio of fatty acid hydroxylated to non-hydroxylated sphingolipids (see Fig. S7 in the supplementary material), but did not result in hydroxylation of phospholipid fatty acid moieties. Conversely, ubiquitous induction of RNAi against fa2h decreased the proportion of sphingolipids with hydroxylated fatty acids in sterol-depleted animals (see Fig. S7 in the supplementary material). Thus, Drosophila FA2H is a sphingolipid-specific fatty acid hydroxylase.
To test whether sterol depletion elevated sphingolipid fatty acid hydroxylation by increasing the transcription of fa2h, we performed real-time RT-PCR on larvae fed LDM or LDM + cholesterol. Sterol depletion increases fa2h mRNA levels 1.7-fold (P≤0.05) suggesting that elevated fa2h transcription contributes to increased fatty acid hydroxylation. However, additional mechanisms might be required to account for the 10 to 20-fold increase in sphingolipid fatty acid hydroxylation observed upon sterol depletion.
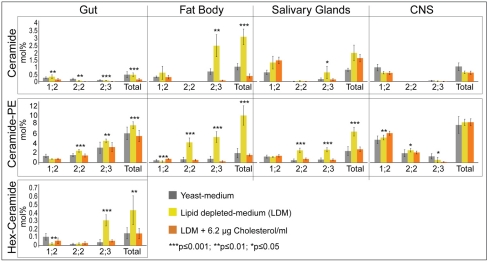
To investigate how sphingolipid composition was affected by sterol-depletion in different tissues, we quantified n:1;2, n:2;2 and n:2;3 sphingolipids in the brain, fat body, salivary gland and gut under different nutritional regimens (Fig. 6). Quantification of sphingolipids in yeast-fed larvae revealed tissue-specific differences in both total sphingolipid levels and the proportion of different sphingolipid variants. Sphingolipids in general represented a larger fraction of membrane lipids in the CNS (8.9 mol%) and gut (6.7 mol%) than in the fat body (3.0 mol%) or salivary gland (3.2 mol%). These tissues also normally contained different ratios of n:1;2, n:2;2 and n:2;3 sphingolipids. However, each tissue significantly elevated membrane levels of n:2;2 and n:2;3 sphingolipids in response to sterol depletion. These changes were most dramatic in the fat body and salivary gland, which have low membrane sphingolipid levels and few n:2;2 and n:2;3 sphingolipids under normal conditions. Interestingly, n:2;2 and n:2;3 sphingolipids increased only modestly in the CNS (Fig. 6), which retained sterol more efficiently than other tissues (Fig. 2). These data suggest that increasing n:2;2 and n:2;3 sphingolipids is a general response of many tissues to sterol depletion, and that sphingolipids respond directly to membrane sterol levels in each tissue.
Fig. 6.
Many tissues upregulate hydroxylated sphingolipids in response to sterol depletion. Mass spectrometric quantification of different sphingolipid species in tissues of larvae fed yeast medium, LDM or LDM + 6.2 μg/ml cholesterol. Asterisks indicate significant differences between larvae fed with LDM and LDM + cholesterol. Error bars indicate standard deviations.
Could increasing the amount of n:2;2 and n:2;3 sphingolipids functionally replace sterols in the membrane? On average, membrane sterol dropped from 9 to 1.5 mol% (a reduction of 7.5 mol%) upon dietary depletion (Fig. 1F), whereas total membrane sphingolipid increased by ∼ 11 mol% in the fat body and 4 mol% in the salivary gland (mainly owing to increases in n:2;2 and n:2;3 sphingolipids) (Fig. 6). Thus, the sterol lost from the membrane was replaced by a roughly similar amount of n:2;2 and n:2;3 sphingolipids, consistent with the possibility that these lipids could substitute for sterols.
To test whether increased sphingolipid levels are important for survival when membrane sterol levels drop, we examined whether reduction of sphingolipid synthesis affects viability of larvae on sterol-depleted food. Serine palmitoyl transferase catalyzes the rate-limiting step in sphingosine biosynthesis, and is encoded by lace in Drosophila. When raised on a standard diet, the hypomorphic lace mutant l(2)k05305 survives to adulthood, although in reduced numbers (Adachi-Yamada et al., 1999). However, when this lace mutant was raised on LDM + cholesterol, 90% arrested as third instar larvae (Fig. 7A). Because dietary sphingosine improves the survival of lace mutants (Adachi-Yamada et al., 1999), this arrest is likely to be caused by the lack of dietary sphingolipid in LDM. Interestingly, more than 80% of arrested lace mutant larvae remained viable for at least 10 days, by which time their heterozygote siblings had pupariated (Fig. 7A,B). When raised on LDM without cholesterol, homozygous lace mutant larvae died 3 days earlier than their heterozygous siblings (Fig. 7D). Thus, sphingolipids are particularly important for larval viability when membrane sterol levels are reduced.
Fig. 7.
Hydroxylated sphingolipids increase survival upon sterol depletion. (A) Final developmental stages reached by lace hypomorphs l(2)k05305 when fed LDM + 6.2 μg/ml cholesterol. (B) Survival of homozygous l(2)k05305 larvae over time when fed LDM + 6.2 μg/ml cholesterol. (C) RT-PCR detecting mRNA for fa2h and Actin5C in tissues from third instar larvae fed YM. (D) Effect of compromised sphingolipid production on larval survival time when on sterol-depleted food. Survival time is defined as the time at which 50% of larvae remain alive. Bars indicate the average difference in survival time of larvae with the indicated genotypes, compared with control siblings. lace l(2)k05305 larvae have reduced serine palmitoyl transferase activity. tub>fa2hi1 and tub>fa2hi2 larvae ubiquitously express distinct fa2h RNAi constructs under the control of tubulin GAL4. elavII>fa2hi1 and elavIII>fa2hi1 larvae induce fa2h1 RNAi in the nervous system using two distinct elavGAL4 drivers. adh>fa2hi1 larvae induce fa2h1 RNAi in the fat body, oenocytes and anterior midgut. lipo>fa2hi1 larvae induce fa2h1 RNAi in the fat body. npc1b>fa2hi1 larvae induce fa2h1 RNAi in the posterior and anterior midgut. Asterisks indicate significant differences from control siblings. **, P≤0.01, *, P≤0.05. (E) The percentage of tub>fa2hi1 larvae or control siblings that either die as pupae or complete adult development when fed YM. (F) The adult weight of YM-fed tub>fa2hi1 males and females and that of control siblings. (G) Mass spectrometric quantification of sphingolipid species present in the hemolymph. Error bars indicate standard deviations.
To investigate whether sphingolipid fatty acid hydroxylation increases survival when membrane sterols are low, we compared the survival of control and fa2hRNAi larvae fed LDM. Ubiquitous expression of two independent fa2hRNAi constructs caused sterol-depleted animals to die on average 20 to 44 hours earlier than sterol-depleted control animals (Fig. 7D). By contrast, fa2hRNAi has no effect on survival, developmental timing or body size of larvae fed YM (Fig. 7E,F). Thus, hydroxylated sphingolipids are important specifically under conditions of sterol depletion. This suggests that these lipids might compensate for those functions of membrane sterol required for survival.
Does one tissue act as a source for hydroxylated sphingolipids, or can each tissue produce them autonomously? RT-PCR experiments showed that the fa2h gene was transcribed in all tissues examined (Fig. 7C); thus, each tissue can synthesize its own hydroxylated sphingolipids. However, interestingly, tissue-specific RNAi-mediated knockdown of fa2h in the fat body or gut did not reduce survival on sterol-depleted food (Fig. 7D). Knockdown in the nervous system did reduce viability in some experiments; however, the effect was of marginal significance (P<0.1) on average (Fig. 7D). These data might suggest that the sensitivity to sterol depletion observed upon ubiquitous knockdown of fa2h is a cumulative effect of the reduced function of many tissues. Alternatively, it is possible that hydroxylated sphingolipids can be exchanged between tissues (with the possible exception of the CNS, which is isolated by a blood-brain barrier). The latter interpretation is suggested by MS analysis of hemolymph lipids, which identified both n:2;2 and n:2;3 sphingolipids in systemic circulation (Fig. 7G). Hemolymph density gradient fractionation showed that these lipids co-fractionate with the Drosophila lipoprotein Lipophorin (not shown). Lipophorin mobilizes lipids from both the gut and the fat body for delivery to peripheral tissues (Panáková et al., 2005). Thus, tissues unable to synthesize hydroxylated sphingolipids might acquire them from other tissues using Lipophorin.
DISCUSSION
It has been known for many years that insects do not synthesize sterols and require dietary sterol to complete development (Cooke and Sang, 1970; Hobson, 1935a; Hobson, 1935b). It was not known whether this reflected a requirement for steroid hormone biosynthesis, or whether insects might also depend on bulk membrane sterol for membrane function. If high concentrations of sterol are required in the membrane, then how do insects maintain membrane function when dietary resources are scarce or even absent? We have investigated these questions by feeding Drosophila larvae with different types and amounts of sterol. We show that Drosophila do require bulk membrane sterol, in addition to steroid hormone biosynthesis, for adult development. How do Drosophila keep membrane sterol within workable limits, even though they cannot control dietary availability? Our findings suggest that Drosophila larvae accomplish this, in part, by regulating the rate and total extent of growth. When sterols are limited in the diet, Drosophila larvae grow slowly and give rise to small adults. Providing more sterol does not increase levels in the membrane, but allows the development of larger adults. However, additional mechanisms must operate to determine tissue and membrane-specific levels of sterol accumulation. Interestingly, the gut and CNS, which accumulate the highest amounts of membrane sterol are also particularly rich in sphingolipids. As sterols pack more favorably with sphingolipids than with phospholipids (Sengupta et al., 2007), it is possible that sphingolipid-rich membranes have a higher capacity to retain sterols. Thus, the elevation of sphingolipid biosynthesis could allow some tissues to accumulate more sterols than others, even in the absence of sterol biosynthesis.
When challenged by removal of sterols from the diet, Drosophila larvae arrest growth and development. In arrested larvae, average membrane sterol levels drop sixfold to less than 2 mol% of membrane lipids without affecting larval viability. In model membranes, this amount of cholesterol is insufficient to prevent gel phase formation or maintain ion impermeability (de Almeida et al., 2003; Haines, 2001). To survive under these conditions, Drosophila increase production of sphingolipids with a doubly desaturated LCB and hydroxylated fatty acid moieties. Sphingolipid hydroxylation promotes hydrogen bonding and tighter packing (Lofgren and Pascher, 1977), and these lipids are abundant in membranes that are specialized to perform barrier functions, such as myelin and the gut apical membrane (Maldonado et al., 2008; Uchida et al., 2007). Thus, it seems plausible that these lipids might substitute for sterols in maintaining some biophysical properties of the membrane, such as impermeability. However, although changing sphingolipid composition is sufficient to allow survival when membrane sterol levels are reduced, it cannot support complete adult development. Clearly, some functions of bulk membrane sterol cannot be replaced by sphingolipids.
What are the essential functions of bulk membrane sterol in Drosophila? One clue is that sterols are more difficult to deplete in some types of cell membranes. These include axonal membranes in the brain and the apical membrane of the gut. This might suggest that the functions of these tissues require higher levels of bulk membrane sterol. Both of these membranes are probably particularly impermeable to ions. The gut apical membrane must resist the extremes of pH present in the gut lumen, and high capacitance of axonal membranes is needed for the propagation of action potentials. Another possible requirement for bulk membrane sterol is in the signaling processes that guide development. Sterol-dependent membrane microdomains regulate a wide range of signaling pathways (Pike, 2005). Growth arrest in the absence of sterol might ensure that development does not occur until sufficient membrane sterol has accumulated to support both tissue specific requirements and correct developmental signaling.
Has the loss of sterol biosynthesis forced Drosophila to evolve unique mechanisms to accommodate fluctuating membrane sterol levels, or might these mechanisms be of broader relevance? Perturbing ergosterol biosynthesis in yeast alters levels of specific sphingolipid variants (Guan et al., 2009). Thus, ancient regulatory mechanisms connect sterols and sphingolipid metabolism, even in animals that have not lost the ability to synthesize sterols. Cholesterol biosynthesis consumes 11 molecules of oxygen per sterol (Summons et al., 2006). Yeast do not synthesize ergosterol under anaerobic conditions (Rosenfeld and Beauvoit, 2003), and hypoxia downregulates cholesterol biosynthesis in vertebrate cells (Mukodani et al., 1990; Nguyen et al., 2007). It would be interesting to investigate whether hydroxylated sphingolipids might compensate for cholesterol in cells poorly served by the vascular system, and whether such cells might regulate growth in response to membrane sterol levels.
An interesting and still unanswered question is how Drosophila sense membrane sterol levels in order to regulate growth and sphingolipid biosynthesis. The sensing mechanism is likely to be independent of the SREBP pathway, which responds to phosphatidylethanolamine rather than to sterols in Drosophila (Dobrosotskaya et al., 2002; Kunte et al., 2006; Seegmiller et al., 2002). Lowering membrane sterol produces transcriptional changes in fa2h and in genes whose protein products regulate glycolysis and translation. Growth regulation by insulin/Akt signaling involves similar transcriptional changes affecting glycolysis and translation, and occurs via phosphorylation and nuclear localization of the transcription factor Foxo (Junger et al., 2003; Puig et al., 2003). However, sterols do not regulate nuclear translocation of Foxo. Might sterol levels affect Foxo activity within the nucleus? In vertebrates, Foxo physically interacts with nuclear hormone receptors, which have lipidic ligands (Dowell et al., 2003; Ganjam et al., 2009; Li et al., 2003; Ma et al., 2009). Furthermore, sirtuins modify the activity and interactions of nuclear Foxo via deacetylation (Brunet et al., 2004; Frescas et al., 2005; Li et al., 2007; Wang et al., 2007; Wang and Tong, 2009; Yang et al., 2005). Unraveling such mechanisms will provide new insights into membrane homeostasis and growth control.
Supplementary Material
Acknowledgments
We are grateful to Leonard Pallanck for npc1b mutant flies, and to Pierre Leopold for anti-Foxo. We thank Marko Brankatschk for help with filipin staining, Ali Mahmoud for constructing UAS:FA2H flies, Fanny Mende for lipid biochemistry advice and Jonathan Rodenfelds for help with real-time RT-PCR. We thank Marko Brankatschk, Elisabeth Knust, Jonathan Rodenfels and Kai Simons for critically reading the manuscript. This work was supported by the Max Planck Geselleschaft, and by grants to S.E. from the DFG and the ESF/DFG (Euromembranes). M.C. was supported by the Fundação para a Ciência e Tecnologia through the Gulbenkian Institute. D.S. was supported by Wellcome Trust/DBT India Alliance. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.044560/-/DC1
References
- Abeytunga D. T., Glick J. J., Gibson N. J., Oland L. A., Somogyi A., Wysocki V. H., Polt R. (2004). Presence of unsaturated sphingomyelins and changes in their composition during the life cycle of the moth Manduca sexta. J. Lipid Res. 45, 1221-1231 [DOI] [PubMed] [Google Scholar]
- Adachi-Yamada T., Gotoh T., Sugimura I., Tateno M., Nishida Y., Onuki T., Date H. (1999). De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Mol. Cell. Biol. 19, 7276-7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E. G., Dyer W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911-917 [DOI] [PubMed] [Google Scholar]
- Bloch K. E. (1983). Sterol structure and membrane function. CRC Crit. Rev. Biochem. 14, 47-92 [DOI] [PubMed] [Google Scholar]
- Bodenstein D. (1994). The postembryonic development of Drosophila. In Biology of Drosophila (ed. Demerec M.), pp. 283-288 New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Brankatschk M., Eaton S. (2010). Lipoprotein particles cross the BBB in Drosophila. J. Neurosci. 30, 10441-10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., London E. (2000). Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221-17224 [DOI] [PubMed] [Google Scholar]
- Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. (2004). Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011-2015 [DOI] [PubMed] [Google Scholar]
- Chang T. Y., Chang C. C., Ohgami N., Yamauchi Y. (2006). Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22, 129-157 [DOI] [PubMed] [Google Scholar]
- Chen L. L., Wang G. Z., Zhang H. Y. (2007). Sterol biosynthesis and prokaryotes-to-eukaryotes evolution. Biochem. Biophys. Res. Commun. 363, 885-888 [DOI] [PubMed] [Google Scholar]
- Clayton R. B. (1964). The utilization of sterols by insects. J. Lipid Res. 15, 3-19 [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., Carré C., Noselli S., Léopold P. (2005). Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310, 667-670 [DOI] [PubMed] [Google Scholar]
- Cooke J., Sang J. H. (1970). Utilization of sterols by larvae of Drosophila melanogaster. J. Insect Physiol. 16, 801-812 [DOI] [PubMed] [Google Scholar]
- Davidowitz G., D'Amico L. J., Nijhout H. F. (2003). Critical weight in the development of insect body size. Evol. Dev. 5, 188-197 [DOI] [PubMed] [Google Scholar]
- de Almeida R. F., Fedorov A., Prieto M. (2003). Sphingomyelin/phosphatidylcholine/cholesterol phase diagram: boundaries and composition of lipid rafts. Biophys. J. 85, 2406-2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demel R. A., De Kruyff B. (1976). The function of sterols in membranes. Biochim. Biophys. Acta 457, 109-132 [DOI] [PubMed] [Google Scholar]
- Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. (2002). Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296, 879-883 [DOI] [PubMed] [Google Scholar]
- Dowell P., Otto T. C., Adi S., Lane M. D. (2003). Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J. Biol. Chem. 278, 45485-45491 [DOI] [PubMed] [Google Scholar]
- Edgar B. A. (2006). How flies get their size: genetics meets physiology. Nat. Rev. Genet. 7, 907-916 [DOI] [PubMed] [Google Scholar]
- Espenshade P. J., Hughes A. L. (2007). Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41, 401-427 [DOI] [PubMed] [Google Scholar]
- Eugster C., Panáková D., Mahmoud A., Eaton S. (2007). Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev. Cell 13, 57-71 [DOI] [PubMed] [Google Scholar]
- Frescas D., Valenti L., Accili D. (2005). Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 280, 20589-20595 [DOI] [PubMed] [Google Scholar]
- Fyrst H., Zhang X., Herr D. R., Byun H. S., Bittman R., Phan V. H., Harris G. L., Saba J. D. (2008). Identification and characterization by electrospray mass spectrometry of endogenous Drosophila sphingadienes. J. Lipid Res. 49, 597-606 [DOI] [PubMed] [Google Scholar]
- Ganjam G. K., Dimova E. Y., Unterman T. G., Kietzmann T. (2009). FoxO1 and HNF-4 are involved in regulation of hepatic glucokinase gene expression by resveratrol. J. Biol. Chem. 284, 30783-30797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X. L., Souza C. M., Pichler H., Dewhurst G., Schaad O., Kajiwara K., Wakabayashi H., Ivanova T., Castillon G. A., Piccolis M., et al. (2009). Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol. Biol. Cell 20, 2083-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. D., Swetha M. G., Kumari S., Lakshminarayan R., Dey G., Mayor S. (2009). Analysis of endocytic pathways in Drosophila cells reveals a conserved role for GBF1 in internalization via GEECs. PLoS ONE 4, e6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines T. H. (2001). Do sterols reduce proton and sodium leaks through lipid bilayers? Prog. Lipid Res. 40, 299-324 [DOI] [PubMed] [Google Scholar]
- Hobson R. P. (1935a). On a fat-soluble growth factor required by blow-fly larvae: distribution and properties. Biochem. J. 29, 1292-1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson R. P. (1935b). On a fat-soluble growth factor required by blow-fly larvae: identity of the growth factor with cholesterol. Biochem. J. 29, 2023-2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger M. A., Rintelen F., Stocker H., Wasserman J. D., Vegh M., Radimerski T., Greenberg M. E., Hafen E. (2003). The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2, 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L., Ejsing C. S., Ekroos K., Shevchenko A., Anderson K. I., Thiele C. (2005). Polyene-lipids: a new tool to image lipids. Nat. Methods 2, 39-45 [DOI] [PubMed] [Google Scholar]
- Kunte A. S., Matthews K. A., Rawson R. B. (2006). Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3, 439-448 [DOI] [PubMed] [Google Scholar]
- Kurzchalia T. V., Ward S. (2003). Why do worms need cholesterol? Nat. Cell Biol. 5, 684-688 [DOI] [PubMed] [Google Scholar]
- Lahiri S., Futerman A. H. (2007). The metabolism and function of sphingolipids and glycosphingolipids. Cell. Mol. Life Sci. 64, 2270-2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Lee H., Guo S., Unterman T. G., Jenster G., Bai W. (2003). AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol. Cell. Biol. 23, 104-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang S., Blander G., Tse J. G., Krieger M., Guarente L. (2007). SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28, 91-106 [DOI] [PubMed] [Google Scholar]
- Lofgren H., Pascher I. (1977). Molecular arrangements of sphingolipids. The monolayer behaviour of ceramides. Chem. Phys. Lipids 20, 273-284 [DOI] [PubMed] [Google Scholar]
- Lucero H. A., Robbins P. W. (2004). Lipid rafts-protein association and the regulation of protein activity. Arch. Biochem. Biophys. 426, 208-224 [DOI] [PubMed] [Google Scholar]
- Ma Q., Fu W., Li P., Nicosia S. V., Jenster G., Zhang X., Bai W. (2009). FoxO1 mediates PTEN suppression of androgen receptor N- and C-terminal interactions and coactivator recruitment. Mol. Endocrinol. 23, 213-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E. N., Alderson N. L., Monje P. V., Wood P. M., Hama H. (2008). FA2H is responsible for the formation of 2-hydroxy galactolipids in peripheral nervous system myelin. J. Lipid Res. 49, 153-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marois E., Mahmoud A., Eaton S. (2006). The endocytic pathway and formation of the Wingless morphogen gradient. Development 133, 307-317 [DOI] [PubMed] [Google Scholar]
- Matyash V., Geier C., Henske A., Mukherjee S., Hirsh D., Thiele C., Grant B., Maxfield F. R., Kurzchalia T. V. (2001). Distribution and transport of cholesterol in Caenorhabditis elegans. Mol. Biol. Cell 12, 1725-1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Entchev E. V., Mende F., Wilsch-Brauninger M., Thiele C., Schmidt A. W., Knolker H. J., Ward S., Kurzchalia T. V. (2004). Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2, e280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., Riddiford L. M. (2007). Size assessment and growth control: how adult size is determined in insects. BioEssays 29, 344-355 [DOI] [PubMed] [Google Scholar]
- Mizutani Y., Kihara A., Chiba H., Tojo H., Igarashi Y. (2008). 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J. Lipid Res. 49, 2356-2364 [DOI] [PubMed] [Google Scholar]
- Mouritsen O. G., Zuckermann M. J. (2004). What's so special about cholesterol? Lipids 39, 1101-1113 [DOI] [PubMed] [Google Scholar]
- Mukodani J., Ishikawa Y., Fukuzaki H. (1990). Effects of hypoxia on sterol synthesis, acyl-CoA:cholesterol acyltransferase activity, and efflux of cholesterol in cultured rabbit skin fibroblasts. Arteriosclerosis 10, 106-110 [DOI] [PubMed] [Google Scholar]
- Nguyen A. D., McDonald J. G., Bruick R. K., DeBose-Boyd R. A. (2007). Hypoxia stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase through accumulation of lanosterol and hypoxia-inducible factor-mediated induction of insigs. J. Biol. Chem. 282, 27436-27446 [DOI] [PubMed] [Google Scholar]
- Panáková D., Sprong H., Marois E., Thiele C., Eaton S. (2005). Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435, 58-65 [DOI] [PubMed] [Google Scholar]
- Pike L. J. (2004). Lipid rafts: heterogeneity on the high seas. Biochem. J. 378, 281-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike L. J. (2005). Growth factor receptors, lipid rafts and caveolae: an evolving story. Biochim. Biophys. Acta 1746, 260-273 [DOI] [PubMed] [Google Scholar]
- Puig O., Marr M. T., Ruhf M. L., Tjian R. (2003). Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17, 2006-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld A., Neutz S., Simons K., Eaton S. (1999). Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J. Biol. Chem. 274, 12049-12054 [DOI] [PubMed] [Google Scholar]
- Rosenfeld E., Beauvoit B. (2003). Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 20, 1115-1144 [DOI] [PubMed] [Google Scholar]
- Rouser G., Fkeischer S., Yamamoto A. (1970). Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494-496 [DOI] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101-1108 [DOI] [PubMed] [Google Scholar]
- Schuck S., Simons K. (2004). Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J. Cell Sci. 117, 5955-5964 [DOI] [PubMed] [Google Scholar]
- Schwudke D., Hannich J. T., Surendranath V., Grimard V., Moehring T., Burton L., Kurzchalia T., Shevchenko A. (2007a). Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal. Chem. 79, 4083-4093 [DOI] [PubMed] [Google Scholar]
- Schwudke D., Liebisch G., Herzog R., Schmitz G., Shevchenko A. (2007b). Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 433, 175-191 [DOI] [PubMed] [Google Scholar]
- Seegmiller A. C., Dobrosotskaya I., Goldstein J. L., Ho Y. K., Brown M. S., Rawson R. B. (2002). The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell 2, 229-238 [DOI] [PubMed] [Google Scholar]
- Sengupta P., Baird B., Holowka D. (2007). Lipid rafts, fluid/fluid phase separation, and their relevance to plasma membrane structure and function. Semin. Cell Dev. Biol. 18, 583-590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberkang M., Havel C. M., Friend D. S., McCarthy B. J., Watson J. A. (1983). Isoprene synthesis in isolated embryonic Drosophila cells. I. Sterol-deficient eukaryotic cells. J. Biol. Chem. 258, 8503-8511 [PubMed] [Google Scholar]
- Simons K., Ikonen E. (1997). Functional rafts in cell membranes. Nature 387, 569-572 [DOI] [PubMed] [Google Scholar]
- Simons K., Vaz W. L. (2004). Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33, 269-295 [DOI] [PubMed] [Google Scholar]
- Summons R. E., Bradley A. S., Jahnke L. L., Waldbauer J. R. (2006). Steroids, tripernoids and molecular oxygen. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 361, 951-968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S. (2001). Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev. Cell 1, 453-465 [DOI] [PubMed] [Google Scholar]
- Uchida Y., Hama H., Alderson N. L., Douangpanya S., Wang Y., Crumrine D. A., Elias P. M., Holleran W. M. (2007). Fatty acid 2-hydroxylase, encoded by FA2H, accounts for differentiation-associated increase in 2-OH ceramides during keratinocyte differentiation. J. Biol. Chem. 282, 13211-13219 [DOI] [PubMed] [Google Scholar]
- Voght S. P., Fluegel M. L., Andrews L. A., Pallanck L. J. (2007). Drosophila NPC1b promotes an early step in sterol absorption from the midgut epithelium. Cell Metab. 5, 195-205 [DOI] [PubMed] [Google Scholar]
- Volkman J. K. (2003). Sterols in microorganisms. Appl. Microbiol. Biotechnol. 60, 495-506 [DOI] [PubMed] [Google Scholar]
- Wang F., Tong Q. (2009). SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol. Biol. Cell 20, 801-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Nguyen M., Qin F. X., Tong Q. (2007). SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 6, 505-514 [DOI] [PubMed] [Google Scholar]
- Yang Y., Hou H., Haller E. M., Nicosia S. V., Bai W. (2005). Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation. EMBO J. 24, 1021-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Ejsing C. S., Gaus K., de Wet B., Shevchenko A., Simons K., Harder T. (2009). Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 28, 466-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.