Abstract
Circadian clocks generate 24-h rhythms that are entrained by the day/night cycle. Clock circuits include several light inputs and interlocked feedback loops, with complex dynamics. Multiple biological components can contribute to each part of the circuit in higher organisms. Mechanistic models with morning, evening and central feedback loops have provided a heuristic framework for the clock in plants, but were based on transcriptional control. Here, we model observed, post-transcriptional and post-translational regulation and constrain many parameter values based on experimental data. The model's feedback circuit is revised and now includes PSEUDO-RESPONSE REGULATOR 7 (PRR7) and ZEITLUPE. The revised model matches data in varying environments and mutants, and gains robustness to parameter variation. Our results suggest that the activation of important morning-expressed genes follows their release from a night inhibitor (NI). Experiments inspired by the new model support the predicted NI function and show that the PRR5 gene contributes to the NI. The multiple PRR genes of Arabidopsis uncouple events in the late night from light-driven responses in the day, increasing the flexibility of rhythmic regulation.
Keywords: Arabidopsis thaliana, biological clocks, circadian rhythms, mathematical model, systems biology
Introduction
Circadian rhythms are widespread in the biology of almost all eukaryotic organisms, including plants. The self-sustained 24 h oscillations of the core clock genes drive rhythmic expression of >30% of the Arabidopsis genome (Michael and McClung, 2003; Edwards et al, 2006; Covington et al, 2008; Michael et al, 2008) and thus provide plants with an adaptive advantage to anticipate the daily changes in environmental conditions, such as light/dark cycles (Dodd et al, 2005). The current concept of the Arabidopsis circadian clock is of a genetic network consisting of morning and evening feedback loops interconnected by a central loop (Locke et al, 2006; McClung, 2006). The complexity of the mechanism, consisting of two coupled oscillators, makes mathematical modelling a useful tool for the analysis of functional properties of the system.
Our previous model (Locke et al, 2006), hereafter referred to as L2006, was based on the three-loop circuit (Figure 1). The dawn-expressed genes LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) form a morning negative feedback loop through activation of their inhibitors, PSEUDO-RESPONSE REGULATOR 9 (PRR9) and PRR7. LHY and CCA1 also control the phase of evening genes by inhibiting their expression in the morning. The important evening gene TIMING OF CAB EXPRESSION 1 (TOC1), which is expressed at dusk, regulates itself in the evening loop of the clock through inhibition of its hypothetical activator, gene Y. The evening-expressed gene GIGANTEA (GI) was previously suggested to be a component of Y (Locke et al, 2006). The evening loop feeds back to the morning loop through the hypothetical gene X, which activates LHY/CCA1 expression in the late night. Light stimulates the expression of LHY/CCA1, PRR9 and Y and thus provides daily resetting or entrainment of the clock.
Figure 1.
The main elements of the extended Arabidopsis circadian clock model. The previous circuit (Locke et al, 2006) is shown, upper right. Elements of the morning and evening oscillators are shown in yellow and grey, respectively. For clarity, proteins are shown only for ZTL, LHY modified (LHYmod) and TOC1 modified (TOC1mod). Genetic interactions, solid arrows; post-translational regulation, dashed arrows. Light inputs to gene transcription are marked by flashes. The full scheme of the model is presented in SBGN format in Supplementary Figure 1.
Although it remains a foundation for understanding the Arabidopsis circadian clock, the L2006 model did not include recent experimental findings, particularly on post-translational regulation. GI protein regulates the clock at the post-translational level by stabilization of the F-box protein ZEITLUPE (ZTL) in the presence of light (Kim et al, 2007). ZTL, in turn, is necessary for the targeting of TOC1 protein to degradation by the proteasome (Mas et al, 2003b; Kim et al, 2007). The results from gi mutants have questioned the implication that GI directly regulates TOC1 (Martin-Tryon et al, 2007), which was proposed for Y in the L2006 model. The evening-expressed gene PRR5 was not explicitly included in the model, though together with the morning genes PRR9 and PRR7, PRR5 is important for the regulation of LHY and CCA1 expression (Farre et al, 2005; Nakamichi et al, 2005, 2010). Several results (Farre and Kay, 2007; Ito et al, 2007; Kiba et al, 2007) also suggest that PRRs are regulated by light at the protein level. Finally, the model could not describe the low level of PRR9 mRNA in transgenic plants that overexpress TOC1 (TOC1-ox) (Makino et al, 2002).
Here, we have integrated the new data into the formal description of the clock gene network. First, we explicitly modelled GI and ZTL function, separating GI from Y and including the stabilization of ZTL by GI. The updated ZTL regulation retained a good match to previous data on the control of TOC1 transcription by GI, but by an unexpected mechanism. Second, we introduced a new element, the night inhibitor (NI) of LHY/CCA1 expression and suggested PRR5 as a candidate component of the NI, with an important function in controlling the phase of morning gene expression. After showing that the model could match multiple sets of molecular time-series data, we analysed the clock responses to perturbations of the light conditions combined with mutations of the clock genes. New data on prr5/prr7 double mutant plants showed a good match to the model, supporting the idea that PRR5 is an essential part of the NI. In summary, the model integrates new and existing experimental data and allows us to understand (describe, explain and predict) the complex responses of the clock to environmental and genetic perturbations.
Results
Modification of the circadian clock circuit
An extended three-loop circuit for the circadian clock in Arabidopsis is proposed. Figure 1 shows the principal scheme of the model. A more detailed description of the full reaction scheme, model assumptions and network equations are presented in Supplementary information. The proposed scheme incorporates several new features compared with the L2006 model, based on experimental data as detailed below. The LHY and CCA1 genes are represented by a single LHY/CCA1 component, as before. In contrast to the previous models (Locke et al, 2005, 2006), we identified data that constrained 35 of the 90 parameters in the model (see Supplementary information). The remaining parameters were fitted to two types of data: the quantitative profiles of molecular components of the clock and the values of free-running periods of circadian rhythms, in wild-type (wt) plants and lhy/cca1, toc1, ztl and prr7/prr9 mutants, under varying environmental conditions (see Supplementary information). The parameter values used for all calculations are shown in Supplementary Table 1. We then applied the model to analyse the mechanisms of circadian regulation under additional genetic and environmental perturbations.
Revised evening loop
We started the modification of the clock scheme from the evening loop, in which we included the post-translational regulation of TOC1 protein by GI (Kim et al, 2007). The model described the stabilization of the ZTL protein in complex with GI protein and the acceleration of TOC1 protein degradation by ZTL (Mas et al, 2003b; Kim et al, 2007). Thus, GI inhibits TOC1 function in the model at the level of TOC1 protein, through positive regulation of ZTL. Together with the inhibition of Y expression by TOC1 protein, this results in indirect activation of TOC1 mRNA expression by GI in the model, as described in a later section. Previously, GI was also considered as the main candidate for Y, the direct activator of TOC1 expression, accounting for 70% of Y activity (Locke et al, 2006). The high TOC1 mRNA levels in gi mutants (Martin-Tryon et al, 2007) suggested that this aspect of Y function was not related to GI. We, therefore, removed the direct activation of TOC1 expression by GI in the model, and discovered that the revised structure of the evening loop retained the main features of the L2006 model.
Regulation of Y and TOC1 expression under different photoperiods
The structure of the evening loop of the clock, with activation of TOC1 transcription by Y, was originally based on the data on autonomous oscillations of the evening genes in lhy/cca1 double mutant plants (Locke et al, 2005). The entrainment of rhythmic TOC1 expression by light in the lhy/cca1 mutant (Mizoguchi et al, 2002), together with the absence of direct activation of TOC1 expression by light (Matsushika et al, 2000), implied that Y was the light-responsive component of the evening loop. In addition, the evening expression of TOC1 in wt plants suggested that the expression of Y was delayed through inhibition by LHY/CCA1 protein, analogous to GI (Locke et al, 2005).
We used the revised model to predict the properties of Y and its regulation of TOC1 expression. As the structure of the evening loop is inherited from our L2006 model, its components showed qualitatively similar behaviour. The predicted profiles of Y and TOC1 mRNA expression in wt plants under light–dark cycles with various photoperiods are shown in Figure 2A. The first peak of Y after dawn is caused by the acute light activation of Y expression, whereas a second, major peak 10 h after dawn is related to the release of Y expression from its inhibition by LHY/CCA1 protein. The broad expression of Y during the light period allows a measure of ‘dusk sensing’ by the evening loop of the clock. Interestingly, the model predicts a later phase of peak TOC1 mRNA in short photoperiods compared with 9L:15D cycles, which corresponds to our experimental data (Figure 2C). The model explains this effect by the lower activation of Y expression in darkness, which slows down TOC1 accumulation in cycles with <9 h of light.
Figure 2.
Regulation of the evening components Y and TOC1 by photoperiods. Simulated expression profiles of Y (blue lines) and TOC1 (black lines) mRNA in wt (A) and lhy/cca1 mutant (B) plants are shown for 6L:18D (dotted lines); 12L:12D (solid lines) and 18L:6D (dashed lines). (C) Dependence of peak TOC1 phase on the day length in wt plants. Simulations are shown by a line and the experimental data points for the peak phase of TOC1:LUC reporter expression are taken from Edwards et al (2010). The level of each clock component in the model is normalized to its maximum level in 12L:12D (see Supplementary information).
The simulated double-peaked shape of Y mRNA profiles (Figure 2A) and the delayed phase of TOC1 in short days (Figure 2C) were also observed in the simulations of wt plants using the L2006 model (Supplementary Figures 5 and 6). However, the current model predicted a stronger response to photoperiod in the lhy/cca1 double mutant (Figure 2B and Figure 7 of Supplementary information). Figure 2B shows the simulated profiles of Y and TOC1 mRNA in lhy/cca1 plants under various photoperiods. Y mRNA is abundant in lhy/cca1 during the light period, which results in a broader profile of TOC1 mRNA in long days compared with short days. Our results on TOC1:LUC expression in lhy/cca1 mutant plants under 6L:18D and 18L:6D photoperiods confirm this prediction (Supplementary Figure 19). The characteristic profiles predicted for Y expression in lhy/cca1 suggest that candidate genes for Y could be identified by testing genome-wide expression profiles in lhy/cca1 mutant plants under various photoperiods.
Figure 7.
Regulation of LHY/CCA1 phase. Simulations are shown under entrainment of wild-type plants to various photoperiods. (A) Simulated profiles of LHY/CCA1 mRNA (black) and NI protein (blue) are shown for 6L:18D (solid lines), 12L:12D (dotted lines) and 18L:6D (dashed lines). (B) Experimental data on CCA1 mRNA for 6L:18D, 12L:12D and 18L:6D entrainment were taken from Edwards et al (2010).
Feed-forward regulation of LHY/CCA1 amplitude by TOC1 and PRR9
In addition to the revision of the structure of the evening loop, we modified the connection between morning and evening oscillators. First, the unknown activator of LHY/CCA1 expression (gene X) was replaced by a post-translational modification or complex dependent on TOC1 protein (TOC1mod), based on the data on TOC1 binding to protein complexes at the CCA1 promoter (Pruneda-Paz et al, 2009). Second, an inhibition of PRR9 expression by TOC1 was introduced, because overexpression of TOC1 was shown to reduce PRR9 mRNA to a negligible level (Makino et al, 2002; Ito et al, 2005).
Next, the model was used to analyse the regulation of LHY/CCA1 by TOC1 in more detail. The scheme of Figure 1 suggests that there are three steps in TOC1 action in the clock: (1) auto-regulation of TOC1 expression through the inhibition of Y, (2) activation of LHY/CCA1 expression and (3) inhibition of PRR9 expression. Model simulations showed that the third mechanism enhances the second, because the inhibition of PRR9 by TOC1 delays PRR9 expression relative to LHY/CCA1. LHY/CCA1 expression can, therefore, rise further under the influence of TOC1mod before the onset of inhibition by PRR9 protein. The effect of TOC1's inhibition of PRR9 on the expression profile of LHY/CCA1 is especially pronounced in constant light (LL) conditions. The expression level of PRR9 is low in LL, because the acute induction of PRR9 at lights-on does not occur in these conditions. Figure 3 shows the simulated profiles of LHY/CCA1 mRNA and PRR9 protein for wt plants and a hypothetical toc1 mutant that is unable to inhibit PRR9 expression. The release of PRR9 from inhibition by TOC1 results in a higher level of PRR9 protein, which in turn reduces the amplitude of LHY/CCA1 expression and shortens the period in this partial toc1 mutant.
Figure 3.
Importance of PRR9 inhibition by TOC1 for high-amplitude oscillations of LHY/CCA1 in constant light. Simulated profiles of LHY/CCA1 mRNA (black lines) and PRR9 protein (magenta lines) are shown for wild-type plants (solid lines) and for a simulated partial toc1 mutant that specifically lacks PRR9 inhibition by TOC1 (dashed lines).
The model predicts that the phenotype of toc1 null mutants is similar to the partial toc1 mutant described above (Supplementary Figure 9). Thus, the experimentally observed 4 h shortening of the period and decrease of LHY/CCA1 amplitude in the toc1 mutant compared with wt (Mas et al, 2003a) can be mostly ascribed to the above effect of the absence of PRR9 inhibition by TOC1, rather than direct activation of LHY/CCA1. The proposed mechanism of LHY/CCA1 regulation by TOC1 is known in the biology of transcriptional networks as a coherent feed-forward motif, when one transcription factor affects another and both of them jointly regulate a target gene (Mangan and Alon, 2003). In this case, TOC1 regulates LHY/CCA1 through a double negative connection through PRR9, in parallel with activation through TOC1mod.
Updating of ZTL effects on the clock
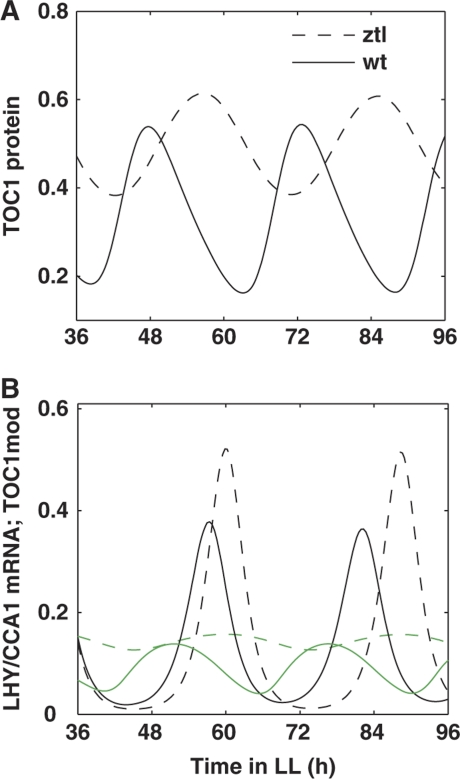
The L2006 model included ZTL-dependent degradation of TOC1 protein, but it did not include the regulation of ZTL by the clock and light (Locke et al, 2006). Updating the mechanism of ZTL regulation by including its stabilization by GI in the presence of light allowed us to explore ZTL action in the clock. We found that in the absence of ZTL, the level of TOC1 protein is higher and its rhythm has lower amplitude in ztl mutants compared with wt plants, which corresponds to experimental observations (Mas et al, 2003b). Figure 4 shows TOC1 protein (panel A) and TOC1mod protein (panel B) profiles in wt and ztl mutant plants in LL conditions. The change of TOC1 profile, in turn, results in the delay of the LHY/CCA1 mRNA peak (Figure 4B) and is in agreement with the experimentally observed, long period of the ztl mutant (Mas et al, 2003b).
Figure 4.
Mechanism of ZTL function under LL conditions. Simulated profiles for wild-type and ztl mutant plants are shown by solid and dashed lines, respectively. (A) TOC1 protein; (B) LHY/CCA1 mRNA (black) and TOC1mod protein (green).
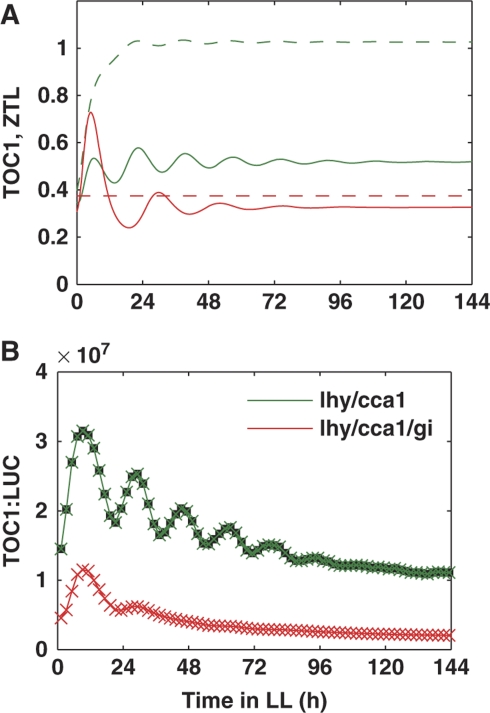
To understand the influence of ZTL on the evening loop, we revisited our previous data on the kinetics of TOC1 expression in the lhy/cca1 double and lhy/cca1/gi triple mutant plants in LL conditions (Locke et al, 2006). Our simulations showed that the ZTL level would be low in lhy/cca1/gi (Figure 5A), because of the rapid degradation of ZTL in the absence of GI protein (Kim et al, 2007). The lower ZTL increased TOC1 protein levels in lhy/cca1/gi compared with lhy/cca1. Next, the negative feedback from TOC1 to Y resulted in decreased Y mRNA in lhy/cca1/gi. As Y activates TOC1 expression, this resulted in a lower level of TOC1 mRNA and reduction of oscillation amplitude in the lhy/cca1/gi mutant plants (Figure 5A and B). This prediction of the model suggested that GI stabilization of ZTL causes an unexpected, indirect activation of TOC1 expression, consistent with the 2.5-fold increase of mean TOC1:LUC expression observed in lhy/cca1 compared with lhy/cca1/gi (Locke et al, 2006). In summary, the above simulations of the wt, ztl and lhy/cca1/gi mutants showed good agreement of the updated ZTL mechanism with existing experimental data.
Figure 5.
Mechanism of TOC1 regulation by GI. The predicted (A) and experimental (B) profiles of TOC1 expression in lhy/cca1 double and lhy/cca1/gi triple mutants. Green lines correspond to lhy/cca1 and red lines to lhy/cca1/gi. (A) TOC1 mRNA expression is shown by solid lines and the total content of ZTL protein is shown by dashed lines. (B) Bioluminescence of transgenic mutant plants carrying the TOC1:LUC reporter, in the lhy/cca1 double and lhy/cca1/gi triple mutant backgrounds (taken from our previous data (Locke et al, 2006)).
Regulation of LHY/CCA1 expression by a wave of inhibitors allows dawn and dusk sensitivity
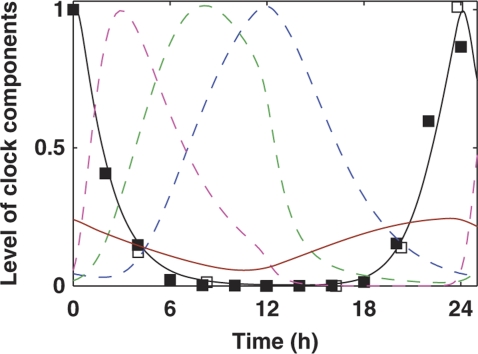
The morning loop was extended by introducing a new element, the NI of LHY/CCA1 (NI). Introducing NI was originally motivated by the levels of LHY and CCA1 mRNA, 100- to 1000-fold lower than their dawn peak, that were observed in the middle of the night (Figure 6; Supplementary Figure 1). The identity of NI is discussed in the next section. Its effect, together with the day-phased inhibitors PRR9 and PRR7 (Farre et al, 2005), guarantees that LHY/CCA1 mRNA quickly falls after dawn and stays at a very low level until the middle of the night. Figure 6 shows the simulated profile of LHY/CCA1 mRNA together with experimental data and the wave of inhibitor proteins. The high level of activator TOC1mod at night (Supplementary Figure 8) ensures a steep rise of LHY/CCA1 mRNA before dawn, when the inhibitor proteins degrade (Figure 6).
Figure 6.
Regulation of LHY/CCA1 expression by the wave of inhibitors and the activator TOC1mod. Simulations are shown during 12L:12D entrainment of wild-type plants. LHY/CCA1 mRNA is shown by the black line. Inhibitor proteins PRR9, PRR7 and NI are shown by dashed magenta, green and blue lines, respectively. TOC1mod is shown by the brown line. Experimental data on LHY/CCA1 expression are shown by filled squares (Edwards et al, 2010) and open squares (Farre et al, 2005).
The model provides sequential expression of PRR9, PRR7 and NI using a simple, effective formulation, because the molecular details underlying the striking ‘PRR wave’ of mRNA profiles (Matsushika et al, 2000) are unclear. PRR9 expression in the model is activated by light at dawn and by LHY/CCA1 protein, consistent with data (Ito et al, 2005). PRR9 protein activates PRR7, and PRR7 activates NI. A modified form of LHY/CCA1 protein, LHYmod, is introduced to activate PRR7 and NI. The PRR9/PRR7/NI activation cascade and LHYmod have no other function in the model. Phosphorylation of CCA1 protein, which is known to affect timing in vivo (Daniel et al, 2004), provides one example of the type of post-translational modification envisaged for LHYmod. Alternative formulations are discussed in Supplementary information.
Modelling the PRR/NI wave allowed us to explore the functional impact of multiple genes with divergent timing and light regulation in the morning loop. In addition to light-activated PRR9 transcription, the model includes the light-dependent stabilization observed for PRR9 and PRR7 proteins and assumed for NI (Farre and Kay, 2007; Ito et al, 2007; Kiba et al, 2007). The PRR/NI wave has the potential to respond both to dawn, when PRR9 is activated, and to dusk, when the PRR/NI proteins are degraded, and thus to alter circadian timing in response to changing photoperiods. Figure 7A shows simulated LHY/CCA1 mRNA profiles under short 6L:18D photoperiods, standard 12L:12D photoperiods and long 18L:6D photoperiods. PRR9 was activated strongly after dawn in all photoperiods, matching published data (Matsushika et al, 2000; Supplementary Figure 3). The model predicts that the PRR7 and NI proteins will fall to low levels during the long dark interval in 6L:18D, owing to their higher degradation rate in darkness (Figure 7A; Supplementary Figure 3). Consequently, LHY/CCA1 expression is expected to be de-repressed earlier in short photoperiods compared with long photoperiods. This corresponds closely to our experimental data, in which CCA1 mRNA levels rise 3 h earlier in short photoperiods compared with long photoperiods (Figure 7B).
Thus, the inclusion of the PRR/NI wave improved the entrainment of the model to various photoperiods compared with L2006 (Supplementary Figure 4). In addition, the PRR/NI wave allowed the model to entrain in a broad range of T cycles (light/dark cycles of total duration T), again in contrast with L2006 (Supplementary Figure 6).
Validation of the inhibitor wave and the identity of NI
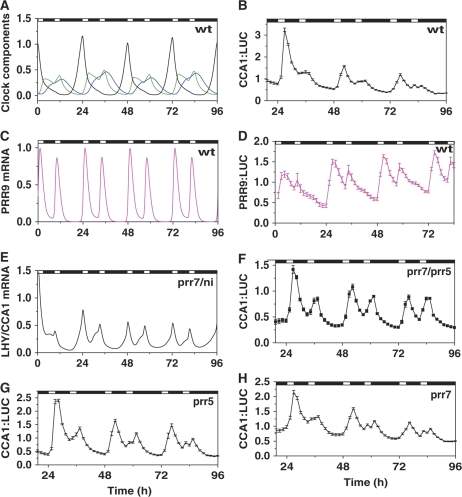
We used a classical protocol of circadian biology, the skeleton photoperiod (Pittendrigh and Daan, 1976), to test the predicted regulation of LHY/CCA1 by the inhibitor wave. The skeleton photoperiod uses two short light treatments per daily cycle to entrain the clock, separating the contributions of light at dawn and dusk. Previous work had shown that the Arabidopsis clock could be entrained by skeleton photoperiods as expected (Millar, 2003), but that >3 h light per cycle was required to maintain normal plant development (Millar, unpublished results). Simulations were, therefore, conducted to match the experiments: initial entrainment in 12L:12D was followed by simulated skeleton photoperiods with 3 h light treatments starting at dawn and ending at dusk (3L:6D:3L:12D). The LHY/CCA1 mRNA profile showed correct entrainment, with a strong peak at the first (‘dawn’) pulse and only a small shoulder of expression at the second pulse (Figure 8A). The L2006 model failed to match this entrained pattern (see Supplementary information). PRR9, in contrast, was predicted to show strong expression in response to both light pulses (Figure 8C).
Figure 8.
Theoretical and experimental profiles of clock components during skeleton entrainment. Plants were transferred from 12L:12D to skeleton entrainment with ‘dawn’ and ‘dusk’ pulses of light, each of 3 h duration (3L:6D:3L:12D), at time 0 h in simulations (A, C, E) and experiments (B, D, F–H). Black lines correspond to simulated LHY/CCA1 mRNA in wild-type (A) or prr7/ni double mutants (E), or normalized luminescence from transgenic CCA1:LUC plants in wild-type (B) or prr7/prr5 (F), prr7 (H) or prr5 (G) mutant backgrounds. (A) Simulated levels of inhibitor proteins PRR7 and NI in wild type are shown by green and blue lines. (C) Simulated levels of PRR9 mRNA are shown by the magenta line. Normalized luminescence from wild-type PRR9:LUC plants is shown in panel D. Luminescence data represent the mean of data from 9 to 18 individual plants, in which each plant's trace was normalized to the mean luminescence level for the plant. Data shown are representative of two independent experiments.
The responses of wt plants to skeleton photoperiods were tested experimentally by measuring the bioluminescence of the CCA1:LUC and PRR9:LUC reporter genes. Our data confirmed the prevalence of the ‘dawn’ response in CCA1 expression (Figure 8B), contrasting with major light induction of PRR9:LUC expression after both light pulses (Figure 8D). The slower decrease and delay in the peak of CCA1:LUC and PRR9:LUC activity (Figure 8B and D) compared with the simulated mRNA in the model (Figure 8A and C) may result from a slower degradation rate of LUC mRNA and protein (Finkenstadt et al, 2008) compared with rapid degradation of CCA1 and PRR9 mRNA (Ito et al, 2007; Yakir et al, 2007, 2009). Supplementary Figure 17 shows the faster decrease of CCA1 and PRR9 mRNA in qPCR measurements compared with CCA1:LUC and PRR9:LUC, supporting this interpretation. We conclude that light input pathways are functional during both light pulses in the skeleton photoperiod, yet LHY/CCA1 was only weakly induced at dusk.
The model explains the mechanism of LHY/CCA1 expression at ‘dawn’, because the inhibitory PRR7 and NI proteins are degraded during the longer period of darkness that represents the night phase (Figure 8A). In contrast, the PRR/NI inhibitors are strongly expressed during the day phase, which results in a low LHY/CCA1 response to the ‘dusk’ pulse of light.
The model predicted that the absence of both PRR7 and NI would result in the loss of the ‘dawn preference’ of LHY/CCA1 expression in the skeleton protocol (Figure 8E). The existing data suggested that PRR5 is a good candidate for NI, because PRR5 together with PRR7 inhibits LHY and CCA1 RNA accumulation in darkness (Nakamichi et al, 2005) and data published during revision of this manuscript revealed physical association of PRR5 to LHY and CCA1 promoters (Nakamichi et al, 2010). CCA1:LUC expression in the prr7/prr5 double mutant responded at nearly equal levels to both light pulses (Figure 8F). This result indicates that PRR5 is a good candidate for NI, because it participates in the inhibition of LHY/CCA1 expression at night together with PRR7.
To test the relative importance of PRR7 and PRR5 in vivo, we analysed the behaviour of the prr7 and prr5 single mutant plants under skeleton conditions. Our experiments showed some impairment of CCA1 inhibition in the ‘dusk’ light pulse in both single mutants (Figure 8G and H) compared with wt plants (Figure 8B). The difference between the prr7/prr5 double mutant and the single mutants showed that both PRR7 and PRR5 normally contribute to repress CCA1 expression in the late day.
Discussion
The main aim of this study was to understand better the structure of the circadian clock circuit based on existing and new experimental data. To achieve this, we improved our previous modelling approach (Locke et al, 2006) by including more kinetic data to build and constrain the model. Quantitative time-series data from dynamic perturbation experiments are common in circadian biology and are essential to this approach. Experiments to measure specific parameter values are also helpful, because the sensitive parameters in complex biological models cannot generally be located a priori. Each measured parameter value, even with associated experimental uncertainty, progressively constrains the high-dimensional search space of biologically realistic values. The final model's behaviour may be robust (insensitive) to variation in many parameter values, but this does not justify a post hoc deprecation of laborious parameter measurements (Gutenkunst et al, 2007). Measuring the degradation rates of the LHY/CCA1 and TOC1 mRNA, for example, was a relatively tractable experiment (Supplementary Figure 18), and these were unexpectedly among the most sensitive parameters in our model (Supplementary Figures 15 and 16).
We were able to introduce several new components and interactions into the clock circuit (Figure 1) using these constraints, starting from the revision of the evening loop. The effects of the GI gene on the clock are not completely understood. The L2006 model suggested that GI was part of the hypothetical component Y—an activator of TOC1 transcription (Locke et al, 2005). However, recent data (Kim et al, 2007; Martin-Tryon et al, 2007) show that GI influences TOC1 not on the transcriptional, but on the protein level. We, therefore, modified the structure of the evening loop by separating GI and Y functions in the model. Y increases TOC1 mRNA level, whereas GI decreases TOC1 protein level through stabilization of ZTL protein. The decreased TOC1 protein tends to relieve repression of Y by TOC1, acting in the evening negative feedback loop. Increased Y expression in turn activates TOC1 expression. Both Y (directly) and GI (indirectly) activate TOC1 expression in the model. This allows us to give a new explanation of the previous experimental data from the gi/lhy/cca1 triple mutant (Locke et al, 2006; Figure 5), which show that the gi mutation reduces TOC1 expression.
However, we included only one function of GI in the model. In addition to the interaction with ZTL, GI can function as a scaffold cofactor in the formation of multi-protein activator complexes at gene promoters (Sawa et al, 2007). This raises the possibility that GI can also accelerate the expression of some clock genes, similarly to the hypothetical activation of TOC1 by Y. Experimental data show a variation of the periods of gi mutant alleles from short to long compared with wt plants (Park et al, 1999; Gould et al, 2006). The shortening of the period of some gi mutants was less pronounced in blue light (Martin-Tryon et al, 2007), which is necessary for GI–ZTL interaction, suggesting that a perturbation of the GI–ZTL interaction could be responsible for the longer period of gi alleles that cause amino-acid substitutions, such as gi-596 (Gould et al, 2006). This observation is consistent with the 2 h period lengthening of the simulated gi mutant relative to wt in our model (data not shown), which incorporated only the effect of the GI–ZTL interaction upon the degradation of TOC1 protein. The short periods observed in other gi mutant alleles are possibly related with the loss of other functions of GI as a scaffold, which might affect protein targets other than TOC1. We do not yet have data to include this function of GI explicitly in the model, so only Y retained the function in transcriptional activation, and simulated y mutants had short periods in our model (data not shown), as in Locke et al (2005, 2006).
We also used the model to study the ZTL effects on the clock. We showed that the long period of ztl mutant is related to changes in TOC1 protein levels. This is consistent with experimental data, which show that the main effect of ZTL is related with TOC1: the ztl/toc1 double mutant has a short period close to the toc1 single mutant (Mas et al, 2003b). In addition to its effect on TOC1, ZTL is known to participate in the degradation of PRR5 (Kiba et al, 2007), which is probably responsible for some part of NI activity. The potential for ZTL-like control of NI protein degradation was taken into consideration by increasing the rate of NI protein degradation in darkness, analogous to PRR9 and PRR7.
Our current understanding of the Arabidopsis clock leaves some open questions about the structure of the evening loop and its connection to the morning loop. Mostly, they are related with Y, the unknown TOC1 transcriptional activator. The model predicts that the characteristic property of Y is its broad expression in lhy/cca1 double mutants in the daytime, which may allow identification of this gene in future. This profile is most obvious in long days, in which Y mRNA is predicted to rise to a second peak before dusk (Figure 2B). Another question is related with the multiple functions ascribed to TOC1. The data on TOC1 co-expression with the transcription factor LUX in constant light, and short- and long-photoperiod conditions, as well as in multiple mutant backgrounds (Hazen et al, 2004, 2005), suggest that some TOC1 functions in the model might be realized through LUX in vivo. New experimental data on LUX expression and LUX functions will be required to understand how LUX contributes to the evening loop and its connection to the morning loop of the clock (Hazen et al, 2005).
An important aspect of this study is related to regulation of the important morning components LHY and CCA1, which in turn control the expression of multiple genes with evening and morning elements in their promoters (Harmer and Kay, 2005). The central function of LHY and CCA1 in the clock may explain the precise regulation of their expression by multiple inhibitors (Locke et al, 2006; McClung, 2006). When the LHY and CCA1 profiles are altered, the phase of all other clock components is changed. Here, we introduced NI, an inhibitor of LHY/CCA1 expression in the model, based on existing data (Nakamichi et al, 2005, 2010; Pruneda-Paz et al, 2009).
Our model predicts that regulation of the wave of inhibitor proteins PRR9, PRR7 and NI by light is important for correct phasing of the clock. The simulated profiles of LHY/CCA1 expression match our experimental data under multiple light conditions including various photoperiods, constant light conditions and in skeleton photoperiods, in which the clock is reset by ‘dawn’ and ‘dusk’ pulses of light. The model shows that the wave of inhibitors may contribute to the observed response of morning genes to the time of dusk (Figure 7B), whereas dusk sensitivity was previously ascribed only to the evening loop (Locke et al, 2006). The combined, PRR9/7 gene of the L2006 model did not match either this dusk-responsive behaviour or the strong induction of PRR9 observed at dawn, because the regulation of that single component represented a difficult compromise between the two patterns. Consequently, the morning loop in L2006 showed no response to changing photoperiods (Edwards et al, 2010; Supplementary Figure 4), and parameters of the PRR7/9 component were the most sensitive in the model (Locke et al, 2006; Treenut Saithong and Millar, unpublished results). The L2006 model was also relatively slow to reach stable entrainment, because it lacked strong light activation of PRR9 (see Supplementary information). The introduction of three inhibitor components (PRR9, PRR7 and NI) instead of one (combined PRR7/9) greatly increased the flexibility of entrainment and the robustness of the present model to parameter changes, compared with the L2006 model. These advantages arise generically from the duplication and divergent regulation of clock components. Organisms with large genomes show this effect in clock gene families, such as the plant PRR genes and the mammalian Period genes, but other biochemical mechanisms could evolve to provide the same properties. The differential regulation of a single clock gene in distinct, though coupled, cells offers one alternative, as in Drosophila.
The model predicted that the initial phase of LHY/CCA1 transcription in the late night depends on PRR7 and NI proteins, which are present at night. Such regulation of LHY/CCA1 would not result simply from the PRR9 protein profile, which falls at the start of the night (Nakamichi et al, 2010; Supplementary Figure 3). Considering PRR5 as a candidate for NI based on previous data (Nakamichi et al, 2005), we investigated in more detail the effects of PRR7 and PRR5 on LHY/CCA1 expression. Our experimental data on the prr7 and prr5 single and prr7/prr5 double mutant plants showed that both PRR7 and PRR5 are important for the higher induction of LHY/CCA1 at dawn compared with dusk under skeleton photoperiods, in agreement with the model prediction (Figure 8). The higher level of LHY and CCA1 expression at dusk in the prr7 and prr7/prr5 mutants was expected from published data (Farre et al, 2005; Nakamichi et al, 2005). The increase of CCA1 mRNA levels at dusk in prr5 was not expected from previous data under normal photoperiods (Eriksson et al, 2003), emphasizing the usefulness of the skeleton photoperiod protocol in understanding the clock mechanisms. Thus, our data on prr7/prr5 and prr5 mutants confirm that PRR5 is a good candidate for NI. During revision of this manuscript, experimental results showing that PRR5 associates with and inhibits expression of LHY and CCA1 further supported this hypothesis (Nakamichi et al, 2010).
Previous data showed that the prr7/prr5 double mutant alters the entrained phase in normal photoperiods (Yamashino et al, 2008). Our model also shows a phase shift (data not shown), but the extent of the shift is dependent on the quantitative contributions and mutual regulation of the PRR genes, which cannot yet be defined from data. In addition, recent experimental data (Pruneda-Paz et al, 2009) show the inhibition of CCA1 by another dusk-expressed gene, CHE (TCP21), the product of which interacts with TOC1 at the CCA1 promoter. On the basis of these data we expect that PRR7 and PRR5 may work together with CHE protein at night in the inhibition specifically of CCA1. Thus, the NI component of our model may represent a protein complex with elements additional to PRR5.
The model simulations showed that, in addition to inhibitors, the activator TOC1mod has an impact on LHY/CCA1 expression by determining the amplitude of the LHY/CCA1 mRNA rhythm. The model predicts that TOC1 has two, coherent effects on LHY/CCA1 expression. As well as direct activation of LHY/CCA1, TOC1 inhibits activation of PRR9 expression by LHY/CCA1, which further increases the amplitude of LHY/CCA1 expression (Figure 3). Thus, inhibition of PRR9 by TOC1 at night increases robustness of oscillations. In the morning, however, PRR9 is quickly induced by light. The molecular mechanism of the switch from inhibition to activation at the PRR9 promoter is unknown. In our model, we assumed that the acute induction of PRR9 expression by light through protein P does not depend on TOC1, allowing a robust acute induction of PRR9 by both pulses of light in the skeleton photoperiods (Figure 8C). However, the observed PRR9 response to the ‘dusk’ pulse of light is smaller than the response to the ‘dawn’ pulse (Figure 8D). This would be consistent with some minor inhibition of PRR9 light induction by TOC1. Future studies of the protein complexes at the PRR9 promoter will allow a more detailed description of PRR9 inhibition by TOC1 and the competition between TOC1 and light activation for the PRR9 promoter.
The model emphasises that LHY/CCA1 regulation must switch from inhibition by PRR9, PRR7 and PRR5 in the early night to activation by TOC1mod in the late night, but the molecular mechanism of this switch is unknown. One possible scenario is that the function of CHE, and perhaps other similar DNA-binding proteins, is modulated through binding of different effectors, such as PRR proteins other than TOC1 (PRR1). In that case, competition between PRR proteins that inhibit or activate CCA1 might also be important in the modulation of the morning loop by evening components. Thus, our approach both highlights general principles of circadian circuits and also focuses experimental work, from introducing an inhibitory function for PRR5 to considering how this function must end in the night.
Materials and methods
Computational and experimental methods are described in detail in Supplementary information.
Supplementary Material
Supplementary information, Supplementary figures S2–19
The version of this supplementary SBML file originally posted online on 21 September 2010 contained minor errors due to the use of the name "time" in the light function and the parameter "default". These errors have been corrected by substitution of "time" with "t" and "default" with "def" and the file replaced on 22 November 2010.
Acknowledgments
This work was supported by BBSRC awards E015263 and F005466. The Centre for Systems Biology at Edinburgh is a Centre for Integrative and Systems Biology supported by BBSRC and EPSRC award D019621. This work has made use of the resources provided by the Edinburgh Compute and Data Facility (ECDF). The ECDF is partially supported by the eDIKT initiative. We are grateful to the Edinburgh Parallel Computing Centre (EPCC; http://www.epcc.ed.ac.uk/) for computing time on their BlueGene/L service.
Author contributions: AP, SKH, KK, KDE and AJM designed experiments; SKH, KK, KDE and AWT performed experiments; TM supplied biological materials; AP, KS and AJM designed computational analysis; AP and KS performed computational analysis; AP and AJM wrote the paper with comments from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM (2004) CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc Natl Acad Sci USA 101: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Akman OE, Knox K, Lumsden PJ, Thomson AW, Brown PE, Pokhilko A, Kozma-Bognar L, Nagy F, Rand DA, Millar AJ (2010) Quantiatiave analysis of regulatory flexibility under changing environmental conditions. Mol Syst Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson ME, Hanano S, Southern MM, Hall A, Millar AJ (2003) Response regulator homologues have complementary, light-dependent functions in the Arabidopsis circadian clock. Planta 218: 159–162 [DOI] [PubMed] [Google Scholar]
- Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Farre EM, Kay SA (2007) PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J 52: 548–560 [DOI] [PubMed] [Google Scholar]
- Finkenstadt B, Heron EA, Komorowski M, Edwards K, Tang S, Harper CV, Davis JR, White MR, Millar AJ, Rand DA (2008) Reconstruction of transcriptional dynamics from gene reporter data using differential equations. Bioinformatics (Oxford, England) 24: 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP (2007) Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol 3: 1871–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Kay SA (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Borevitz JO, Schultz TF, Harmon FG, Pruneda-Paz JL, Ecker JR, Kay SA (2004) Mapping LUX ARRHYTHMO, a novel myb transcription factor essential for circadian rhythms, and other circadian clock mutants by oligonucleotide array genotyping. In 15th International Conference on Arabidopsis Research. Berlin, Germany
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nakamichi N, Kiba T, Yamashino T, Mizuno T (2007) Rhythmic and light-inducible appearance of clock-associated pseudo-response regulator protein PRR9 through programmed degradation in the dark in Arabidopsis thaliana. Plant Cell Physiol 48: 1644–1651 [DOI] [PubMed] [Google Scholar]
- Ito S, Nakamichi N, Matsushika A, Fujimori T, Yamashino T, Mizuno T (2005) Molecular dissection of the promoter of the light-induced and circadian-controlled APRR9 gene encoding a clock-associated component of Arabidopsis thaliana. Biosci Biotechnol Biochem 69: 382–390 [DOI] [PubMed] [Google Scholar]
- Kiba T, Henriques R, Sakakibara H, Chua NH (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19: 2516–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, Li L, Sauro H, Schilstra M, Shapiro B, Snoep JL, Hucka M (2006) BioModels database: a free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Res 34: D689–D691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognar L, Gould PD, Feher B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ (2006) Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 2005 0013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon EL, Kreps JA, Harmer SL (2007) GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA (2003a) Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA (2003b) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426: 567–570 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McClung CR (2006) Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR (2003) Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol 132: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, Givan SA, Yanovsky M, Hong F, Kay SA, Chory J (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ (2003) A suite of photoreceptors entrains the plant circadian clock. J Biol Rhythms 18: 217–226 [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G (2002) LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T (2005) The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol 46: 609–619 [DOI] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, Kim HJ, Kay SA, Nam HG (1999) Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol 106: 291–331 [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Hassidim M, Green RM (2007) CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiol 145: 925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM (2009) Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol 150: 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Ito S, Niwa Y, Kunihiro A, Nakamichi N, Mizuno T (2008) Involvement of Arabidopsis clock-associated pseudo-response regulators in diurnal oscillations of gene expression in the presence of environmental time cues. Plant Cell Physiol 49: 1839–1850 [DOI] [PubMed] [Google Scholar]
Synopsis only
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ (2004) The neurospora circadian system. J Biol Rhythms 19: 414–424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Supplementary figures S2–19
The version of this supplementary SBML file originally posted online on 21 September 2010 contained minor errors due to the use of the name "time" in the light function and the parameter "default". These errors have been corrected by substitution of "time" with "t" and "default" with "def" and the file replaced on 22 November 2010.