Abstract
With the accumulation of data on complex molecular machineries coordinating cell-cycle dynamics, coupled with its central function in disease patho-physiologies, it is becoming increasingly important to collate the disparate knowledge sources into a comprehensive molecular network amenable to systems-level analyses. In this work, we present a comprehensive map of the budding yeast cell-cycle, curating reactions from ∼600 original papers. Toward leveraging the map as a framework to explore the underlying network architecture, we abstract the molecular components into three planes—signaling, cell-cycle core and structural planes. The planar view together with topological analyses facilitates network-centric identification of functions and control mechanisms. Further, we perform a comparative motif analysis to identify around 194 motifs including feed-forward, mutual inhibitory and feedback mechanisms contributing to cell-cycle robustness. We envisage the open access, comprehensive cell-cycle map to open roads toward community-based deeper understanding of cell-cycle dynamics.
Keywords: comprehensive map, large-scale network, yeast cell cycle
Introduction
The eukaryotic cell replication division cycle process involves a sequence of biological events by which one cell grows and divides into two daughter cells, replicating all its components and dividing them evenly among the daughters (Mitchison, 1971; Murray and Hunt, 1993; Morgan, 2006). It is an important process underlying biological growth, reproduction and development. The eukaryotic cell cycle is characterized by precise spatio-temporal coordination between hundreds of proteins (∼800 genes in budding yeast) and its deregulation has been implicated in a wide variety of human diseases—most importantly, cancer.
The eukaryotic cell cycle is viewed as a cyclical progression through four phases: G1, S, G2 and M, orchestrated primarily by cyclins proteins, which interact with cyclin-dependent protein kinases (Cdks) controlling the activities of other executor proteins (EPs) (Csikasz-Nagy et al, 2009). In budding yeast, the process starts in G1, when the cell grows and commits to division under appropriate conditions. Subsequent activation of Clb5 drives the cell into S phase, in which DNA is synthesized and chromosome replication occurs. Followed by a ‘gap' phase (G2), the cell enters M phase for chromosome separation and cell division, the entry and exit into which is controlled by activation and degradation of Clb2. After the M phase, the cell moves back into G1.
The complex molecular machinery governing crucial events of cell cycle (DNA replication, mitosis) are highly conserved among eukaryotes, particularly human beings and yeast. Thus, insights into the molecular mechanisms governing cell proliferation have been successfully obtained by genetic studies on fission yeast (Nurse, 1997; Moser and Russell, 2000) and budding yeast (Nasmyth, 1996; Mendenhall and Hodge, 1998). The availability of the complete genome (Goffeau et al, 1996), together with ease in genetic tractability, large-scale experimental omics data and efficient growth under laboratory conditions make the budding yeast, Saccharomyces cerevisiae, an ideal model organism. A vast corpus of knowledge on yeast cell-cycle complexes, interactions and pathways are scattered across different databases, such as the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/), Yeast Protein Complex Database (http://yeast.cellzome.com/) to name a few, or in the text of biological literature.
Concurrent strides have been made in applying mathematical analyses to investigate cell division cycle, with particular emphasis on budding yeast. Although the early models based on ordinary differential equations (ODEs) were proposed in the mid-1970s (cf. Chapter 10 of Goldbeter, 1996), subsequent generation of models have evolved to incorporate the growing knowledge of cell-cycle biology (Hyver and Le Guyader, 1990; Goldbeter, 1991; Norel and Agur, 1991; Tyson, 1991; Chen et al, 2004).
With a wealth of high precision experimental data available on the budding yeast, construction of a large-scale molecular interaction map of cell-cycle process is pertinent for systems-level understanding of the fine-grained control orchestrated by the molecular components. In this work, we endeavor to provide a comprehensive model of the molecular entities involved in various stages of the budding yeast cell cycle, derived from careful study of high-level molecular biology literature. Before embarking on a navigational tour of the comprehensive map, we enumerate the important challenges and considerations in the construction of the cell-cycle map:
Focus: The focus of the current version of the map is to capture the biological facts related to molecular interactions involved in different stages of the cell-cycle process in budding yeast. Specifically, the interactions involved in G1, S and G1/S transitions are captured in the map together with current consensus on M phase mechanisms. Structural modifications, such as histone modification, septin ring formation, DNA replication machinery and spindle pole body pose a challenge in graphical representation, and the current map only represents the component molecules involved in these processes. In order to facilitate ease of representation and comprehension, compartment information of the molecules has been largely ignored in the map, whereas spatial information has been partially represented through cellular transport reactions in which such transports have a critical function in the cellular process.
Representation: An important consideration in the construction of a molecular interaction map is the representation format, which should be standardized across various computational tools while being expressive enough to readily incorporate available biological facts. In this paper, a choice was made to use Systems Biology Graphical Notation (SBGN) standard (Le Novere et al, 2009) partially implemented in CellDesigner™ software (Kitano et al, 2005) while using Systems Biology Markup Language (SBML) (Hucka et al, 2003) as the model storage and exchange format.
Curation: One of the issues in constructing maps of molecular interactions is the accuracy of representation—particularly in case of conflicting evidences or ambiguity of representation from different references. In this work, we have endeavored to include interactions that have been experimentally verified in multiple reports. Possible alternative interpretations and conflicting reports have been documented through annotations (layered texts and reaction notes) as explained later.
Analysis: The comprehensive molecular interaction map provides a framework to analyze the architecture of the molecular pathways regulating cell cycle. In this direction, we first analyze the topological properties of the map to identify important hub molecules. Next, we abstract the map to a tri-planar view, assigning molecules to signaling, core or structural planes. This view facilities identification of function of molecular components, while revealing regulatory mechanisms between the planes. On the basis of the planar structure, we develop a comparative motif analysis technique to identify core, recurring motifs, which contribute to cell-cycle robustness—feed-forward control, mutual regulations and feedback control mechanisms.
The overarching aim of this work is to present a common ground for the dissemination of knowledge related to the yeast cell-cycle interactions. We endeavor to aggregate the consensus molecular interactions scattered across the current yeast literature, in a comprehensive interaction map, which can be accessed through computational means for the development of mathematical models together with community-wide collaborative enhancements to the interaction network.
Consensus reconstruction of comprehensive cell-cycle network
The principal focus of this work is the consensus reconstruction of the cell-cycle molecular interaction map for the budding yeast. The current map is developed using CellDesigner™ 4.1, which is widely used network editor developed by The Systems Biology Institute and complies with well-accepted standard such as SBML (Hucka et al, 2003) and SBGN (Le Novere et al, 2009). The diagram uses the process diagram notation schema (Kitano et al, 2005) to represent proteins and their specific modifications, protein complexes and genes, as well as various protein transformations (binding, unbinding, phosphorylation and so on.) and their effects on activation or inhibition of chemical reactions, including transcriptional activation or inhibition (a complete list of symbols used in CellDesigner™ is available on the CellDesigner™ website; http://celldesigner.org/).
It is pertinent to note here that most existing maps, such as protein–protein interaction (PPI) networks, aim to capture the interaction of the proteins without delving into biochemical details. However, the cell-cycle map focused in this paper endeavors to provide a mechanism-oriented view, capturing details such as phosphorylation sites, complex formations, cellular transports and other bioprocesses as obtained from the literature. This provides a high-granular dimensionality to the knowledge level contained in the map than obtained from traditional PPI networks (Ito et al, 2000, 2001; Schwikowski et al, 2000; Uetz et al, 2000).
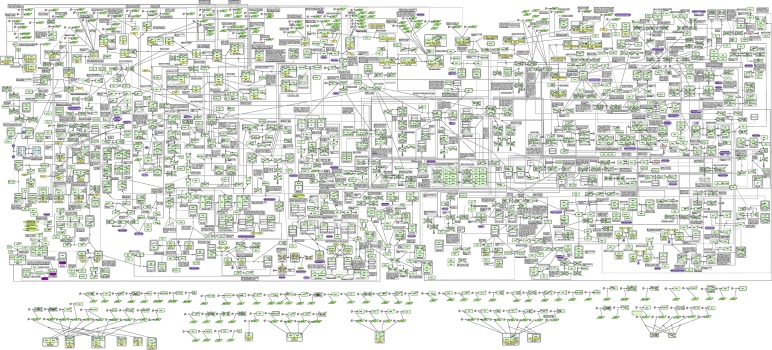
The bird's eye view of the comprehensive yeast cell-cycle molecular interaction map is depicted in Figure 1 (the source xml file is in Supplementary information S1). The molecular interactions captured in this map focus not only on core cell-cycle mechanisms, but also on various checkpoints (DNA damage, morphogenetic and spindle assembly checkpoints and so on.) and parts of signaling systems such as pheromone and heat-shock response pathways. The map layout reflects each phases of cell cycle (G1-S-G2-M) aligned from left to right. Transcriptional regulations are displayed on the upper part of the map. Changes in localization that are related to regulation are described using transport reactions. The current comprehensive map has a total of 880 species, which are represented in 475 proteins and 107 genes and RNAs. The species are involved in 732 reactions and regulations (among them, 147 protein associations and dissociations, 360 state transitions, 180 transcriptions and 42 transport reactions). The corpus of experimental literature covered includes ∼600 papers published (Supplementary information S2). Table I provides the overall statistics of the map.
Figure 1.
A comprehensive molecular interaction map for budding yeast cell cycle. This map was created using CellDesigner™ 4.1. The graphical representation is compliant with systems biology graphical notation (SBGN) (Le Novere et al, 2009). A total of 880 species and 732 reactions are included (for the further properties, see Table I). This image is also available as SBML (see Supplementary information S1). This image is also available as high resolution PDF.
Table 1. The statistical properties of yeast cell-cycle controlling map in this study.
| aA full list of publications referred in the map is available in Supplementary information S2. | |
|---|---|
| bA full ORF list is available in Supplementary information S3. | |
| Number of species | 880 |
| Proteins | 475 (/880) |
| Genes and RNAs | 107 (/880) |
| Number of reactions | 732 |
| Associations and dissociations | 147 (/732) |
| State transitions | 360 (/732) |
| Transcriptions and translations | 180 (/732) |
| Transports | 42 (/732) |
| Number of referencesa | 586 |
| Number of ORFsb | 373 |
In the rest of the section, we endeavor to provide an overview of the map reconstruction process to facilitate the readers in navigating through the map.
Collection and representation
We followed a top-down approach in the construction of the map, focusing first on review articles, which provide an overview of each pathway (e.g. pheromone response, DNA damage checkpoint, mitotic exit) involved in yeast cell cycle. The reviews identify the major components involved in different phases of cell cycle together with canonical interactions between them. Although these reviews are important building blocks for the map, they provide only a base of the molecular interactions as no experiments were carried out in reviews to substantiate the interactions.
Therefore, we focus next on detailed examination of all references involving direct evidences for the interactions described in reviews, identifying specific reaction mechanisms, residue sites for modifications and so on. For some cases, in which only the occurrence of the modification was confirmed, but the sites were not detected, additional cross-referencing was conducted to indicate specific sites on the map. In case of specific proteins in which a large number of modification sites were reported, the sites were represented as a single residue with annotations defining the actual position of the sites.
Each gene encoding the major components was enriched through their functional annotations in different databases (e.g. SGD) and related papers curated from PubMed using the corresponding MeSH terms. The open reading frame (ORF) list of the genes in the map is in Supplementary information S3. At this stage, the map revealed interactions between cell-cycle pathways, modifications with functional annotations as well as molecular species, which have reported interactions with some components of the core pathways. This approach enables the network focus to be fixed on cell-cycle mechanisms, while capturing lateral pathways, particularly those related to cell signaling, which interact and cross-talk with them.
Transcriptional regulation has been separately treated in the current map to provide a comprehensive view of the transcriptional regulatory network related to cell cycle. First, transcription factors (TFs) regulate multiple genes as a whole. Therefore, though a TF binds to the binding site upstream of each gene, one binding site for each TF was represented in the map to simplify the visualization. The consensus sequence of a binding site was described in the layered text. Second, there are multiple experimental methods to substantiate a transcriptional regulation and at times, it is difficult to decide the ‘direct' evidence. To avoid this problem, we followed the procedure below.
First, we focused on references in which detailed experiments were specifically carried out in some genes (e.g. the first paper, which revealed the function of the transcriptional factor or a sequence of the binding site).
Next, we used a data-driven approach, curating transcriptional information from databases, such as the Yeast Search for Transcriptional Regulators And Consensus Tracking (http://www.yeastract.com/), a curated repository of transcriptional regulations for S. cerevisiae. The database is mainly based on three kinds of evidences (ChIP-on-chip, microarrays, existence of a potential-binding site). Therefore, we searched and added regulations for each gene if they were verified with multiple methods (at least two different experiments). These regulations were notated in the layered text.
Annotations
As mentioned in the introduction, systematic annotation and curation are fundamental to the construction of a large-scale map. In this work, we annotate the specific molecular reactions with specific information on the genes and proteins involved (providing ORF IDs where available), references of relevant papers along with comments on them (PubMed IDs). In this map, we trace down to the original paper that reports specific interactions using biochemical experiments and often provides direct support of such interactions. However, it is controversial what can be viewed as direct experimental support for transcriptional regulations. Thus, multiple papers are referred to make our best effort to give maximum accuracy to the map. ‘Unknown catalysis' and ‘unknown inhibition' are used when such interactions can be assumed from genetics, but without knowing detailed mechanisms. For such cases, detailed notes are added using layered text function of CellDesigner™ 4.1 (Supplementary information S4). Papers referred are noted in layered text format and can be viewed in a printable poster format (refer to link to poster version of the map in http://www.systems-biology.org/001/yeast/YeastCellCyclePosterEdition.pdf). PubMed ID is also stored in the SBML notes field so that users can directly access papers through PubMed from the CellDesigner™ model file (Supplementary information S1). The layered text functionality provides the user with a ready reckoner for the scientific evidence associated with a molecular reaction.
Community-based annotation and quality improvement
Given the size of the map and its potential usage, it is critically important that the map is maintained to be high quality. Although we developed this map with extreme care, we must admit that the map cannot be error free. Unfortunately, there is no established process of creating error-free molecular interaction maps or databases of such kind. In fact, recent report indicates that error rate turned out to be very high for protein databases ranges from 5 to 63% (Schnoes et al, 2009). It was also reported that our approach provides better quality in terms of accuracy and coverage over interaction database (Bauer-Mehren et al, 2009). Thus, we are hoping that error rate is relatively low. However, it is also true that only limited efficacy exists for self- or intra-group cross-checking on accuracy of the map. Thus, we decide to take ‘thousands eyes approach' in which errors shall be spotted by anyone in the community and immediately fed back to the community through the community sharing of information.
In addition, new discoveries are constantly reported that is not readily included in the map. Therefore, it is essential that a platform should be provided to facilitate a continuous error-correction process and incorporation of new discoveries.
In order to implement this process, Payao has been developed and launched to provide a community-based annotation and curation of molecular interaction maps. It is a web-based system for sharing and curation of pathways (http://www.payaologue.org/) (Matsuoka et al, 2010). In other words, the aim of Payao is to provide a Google Map (http://maps.google.com) equivalent for biological pathways, wherein researchers can share large-scale, curated and annotated (minimum information requested in the annotation of biochemical models, MIRIAM compliant; Le Novere et al, 2005) network maps that are SBGN and SBML compliant. Such maps can be created using software such as CellDesigner™ and publish it to the online community. With the built-in tagging and collaborative system, the community can participate in enhancing the biological entities in the map or navigating their specific areas of interest. We envision that the availability of the comprehensive map through Payao (Supplementary information S13) will provide a community knowledge base for up-to-date discussions and exchange of information, thereby accelerating knowledge enhancement on eukaryotic cell cycle.
Global analysis of the cell-cycle network
In order to gain insights into the structure and control mechanisms governing spatio-temporal signaling of cell-cycle components, we conducted systematic analysis on the comprehensive map—identifying topological properties, a control structural view of the components and finally a comparative motif analysis to elucidate important regulatory motifs contributing to the inherent robustness of the cell-cycle mechanism. The detailed definitions of the following analyses can be found in Supplementary information S12.
One of the important properties of a molecular network is the identification of the hub nodes (i.e. the nodes, which have high connectivity with other nodes in the network) in the model. The degree or connectivity of a molecule (represented as a node) is defined as the total number of molecules with which it interacts (i.e. is involved in some biochemical reaction). As the elemental property of the map, Table II lists the top 15 nodes of high degree from 1822 nodes (among them, 880 nodes are species) and are considered to be the hubs of cell-cycle network. Among these genes, six are kinase, seven are TF, one ubiquitin ligase and the phosphastase Cdc14, which are well known to have central function in yeast cell-cycle dynamics. On the other hand, >90% of nodes are under degree 5 (Supplementary information S5). One may argue that degree statistics from literature-based map may have a sociological bias in which well-known molecules are studied intensively than less well-known molecules, thus their interactions are better revealed. Although this is a valid point to make, such bias can be checked by using further analysis on the comprehensive map based on available PPI data elsewhere. However, the focus of the current work has been to curate the latest knowledge on yeast cell-cycle reactions and provide a standard compliant, community-based platform on the basis of which further analysis can be performed. In addition, well-known molecules are studied more than others because of their importance in cellular function that is often correlated with numbers of interactions.
Table 2. Top 15 hub nodes of high degree.
| Rank | Specie name | Specie classification | Closeness centrality | Degree |
|---|---|---|---|---|
| aThese nodes involve a protein expressed from an essential gene. | ||||
| 1 | Cdc14a | Phosphatase | 0.175 | 45 |
| 2 | Clb2 Cdc28a | Kinase | 0.176 | 29 |
| 3 | Cln2 Cdc28a | Kinase | 0.164 | 22 |
| 4 | Swi4 Swi6 | Transcription factor | 0.158 | 20 |
| 5 | Mbp1 Swi6 Stb1 | Transcription factor | 0.141 | 19 |
| 6 | Cdc5a | Kinase | 0.154 | 16 |
| 6 | Swi5 | Transcription factor | 0.147 | 16 |
| 8 | Clb5 Cdc28a | Kinase | 0.153 | 15 |
| 8 | Ace2 | Transcription factor | 0.147 | 15 |
| 10 | Cdc28a | Kinase | 0.147 | 14 |
| 10 | Hog1 | Kinase | 0.131 | 14 |
| 12 | Tec1 Ste12 | Transcription factor | 0.134 | 13 |
| 12 | Mcm1 Fkh2 Ndd1a | Transcription factor | 0.136 | 13 |
| 12 | Mcm1 ECBa | Transcription factor | 0.136 | 13 |
| 15 | Cdh1 APC | Ubiquitin ligase | 0.141 | 12 |
Control structure
Broad-level understanding of the basic mechanism governing synchronization and checkpoint-based control of cell-cycle events has been studied by Tyson (1991) and Novak and Tyson (1993) and more recently by Li et al (2004); Csikasz-Nagy et al (2006, 2009) and Novak et al (2007). In order to present an overview of the control structure implicit in the cell-cycle map, we visualize the map as a tri-planar control structure consisting of the following:
Signaling and checkpoint plane: This plane consists of molecules involved in controlling the progression of cell cycle by transducing external environmental signals (e.g. Mck1, Slt2/Mpk1, Fus3) or checkpoint molecules (e.g. Mad2, Rad53, Bub1-3).
Cell-cycle core plane: This plane consists of the molecular entities, which form the heart of the cell-cycle process, namely cyclins.
Structural plane: This plane represents the molecules, which are associated with controlling the structure of the cell during cell-cycle events. These molecules can be viewed as EPs (Csikasz-Nagy et al, 2009), which capture the inputs from the cell-cycle core plane and transform them into feedback signals, pre-dominantly as physiological signals. Molecules belonging to this plane include DNA replication complexes such as Sld2, Mcm3, and Cdc6 or mitotic spindle regulators such as Ase1, Cin8 and Fin1.
Figure 2A shows a schematic of the planar view with the flow of information and control signals between three planes. As seen from the figure, the signaling and checkpoint plane sends signals to control the progression of cell cycle, either in response to external signals such as stress or in the form of physiological feedbacks coming from structural complexes. The core molecules involved in the cell-cycle core plane interact with the structural as well checkpoint and signaling planes in controlling the different phases of the cell cycle.
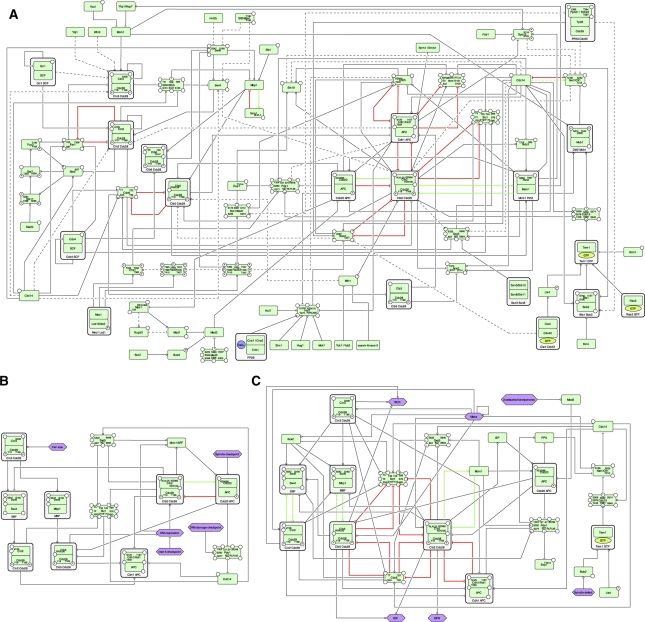
Figure 2.
An overview of the control structure of budding yeast cell cycle. (A) Tri-planar view of cell-cycle regulation in yeast (signaling, cell-cycle core and structural planes). (B) Illustrative regulatory interactions mapped to the different planes. Green and red arrows indicate activation and inhibition, respectively.
Figure 2B provides illustrative examples of the regulatory motifs involved between the different planes, located according to their position relative to the tri-planar view in Figure 2A. Represented in the diagram are the inhibitory effects from the Hog1, Mck1, Slt2 and Rad53 kinases to the cell-cycle-related molecules of Hsl1 and Swi6 (Madden et al, 1997; Mizunuma et al, 2001; Sidorova and Breeden, 2003; Clotet et al, 2006). In addition, the negative regulatory interactions between the Fus3 molecule and the Cln2–Cdc28 complex through the Far1 molecule illustrate the exchange of control information between the signaling/checkpoint plane and the cell-cycle core plane (Peter et al, 1993; Peter and Herskowitz, 1994; Henchoz et al, 1997; Gartner et al, 1998). The control mechanisms from the core to the structural plane are illustrated through the well-studied regulations of the DNA replication complex and mitotic spindle by the B-type cyclin (Clb5–Cdc28) and the anaphase-promoting complex (APC) ubiquitin ligase (Cdh1-APC), respectively (Juang et al, 1997; Elsasser et al, 1999; Nguyen et al, 2000; Hildebrandt and Hoyt, 2001; Masumoto et al, 2002; Woodbury and Morgan, 2007).
As reported by Csikasz-Nagy et al (2009), various feed-forward regulatory motifs exist between the cell cycle and structural planes. In order to facilitate the study of the planes, we created planar maps, which contain a subset of the comprehensive molecular interactions focusing only on the reactions associated with the molecules in a plane. The source XML files for core plane and signaling plane maps are provided in Supplementary information S6-7. However, as mentioned earlier, the representation of the structural plane and associated modifications are only captured in the current map through the list of participating molecules and their complexes (Supplementary information S8 enumerates the list of structural plane molecules).
One of the important issues of the planar view on the molecular interaction map is that the abstraction does not provide clear boundaries for delineating the planes and many molecules can be arguably captured in multiple planes. For instance, Cdc6 is a structural component of the pre-replicative complex, which also inhibits the activity of B-type cyclins by its direct binding (Elsasser et al, 1996). Moreover, the physiological feedbacks provided from the structural plane to the signaling and checkpoint planes (e.g. morphological changes) are difficult to capture in the current graphical notation schema. However, the tri-planar view of the cell-cycle interactions captures the important regulatory controls implicated in the progression of the yeast cell cycle and provides a framework to identify control motifs, which we study next.
Comparative motif analysis
Robustness of budding yeast cell cycle has been discussed in various studies (Nasmyth, 1996; Cross, 2003; Chen et al, 2004; Li et al, 2004; Moriya et al, 2006; Braunewell and Bornholdt, 2007) and regulatory motifs, such as feed-forward, feedback and mutual regulations, have been identified as fundamental structures conferring dynamic behavior and control to biochemical networks (Tyson et al, 2003; Novak and Tyson, 2008). Li et al identified inherent robustness in the cell-cycle network, identifying the G1 state and biological pathways associated with it as a global attractor and a globally attracting trajectory of the dynamics, respectively. Csikasz-Nagy et al (2009) identified the feed-forward regulation of cell cycle by transcription and phosphorylation of EPs regulated by cell-cycle core elements (namely Cdk1). In order to identify regulatory motifs in our comprehensive map and compare them to published models in an equitable manner, we elucidate a systematic motif comparison framework developed as part of this study.
Various network abstractions have been used in studying the properties of large-scale molecular networks. Boolean networks are a well-suited abstraction for analyzing regulatory architectures of biochemical networks, as highlighted by the study of Li et al (2004) (referred hereafter as the Li model) (Faure and Thieffry, 2009; Faure et al, 2009). On the other hand, protein–protein interaction networks (PINs) are another representation format for regulatory networks based on experimentally determined and verified data sets. In budding yeast, a large amount of PPI data have been accumulated, for example, using yeast two-hybrid analysis (Ito et al, 2000, 2001; Uetz et al, 2000; Maslov and Sneppen, 2002). However, in general, PINs can only suggest existence of interactions between components, but cannot determine either their direction or type (activation or inhibition). An integrated analysis of cell-cycle control was presented by Chen et al (2004) (referred hereafter as the Chen model). The model, based on biochemical rate equations, captures the important molecular components of yeast cell cycle, building a consensus picture of the phenotypic properties for over 100 genetically engineered strains. In this study, we focus on the dynamic Chen model and the Boolean Li model for comparative motif analysis, with our network map.
As mentioned above, the Chen model was originally developed to reproduce various phenotypic behaviors in over 100 mutants, and is represented as ODEs. Thus, in order to identify motif structures in the model, it needs to be represented as an interaction graph. In this study, the ODE model was converted into Boolean model based on the Jacobian matrix, which described sensitivities between components. Regulatory types of interactions in the ODE model were determined by signs of the Jacobian elements: activation if it is positive, inhibition if it is negative or none if it is constitutively equal to zero (see Supplementary information S12 for details). The Chen model as well as our comprehensive map involves multiple states (e.g. phosphorylated or not, cytoplasmic or intra-nuclear) for each protein. Therefore, we selected a unique active state for each protein defining it as a node corresponding to the protein. Finally, the Li model was manually imported from Supporting Table 3 in Li et al (2004) and the three networks were represented in a common format, the Cytoscape SIF (Supplementary information S9). The network interaction view of the three cell-cycle models is shown in Figure 3. In this manner, we implemented totally 18 nodes and 31 edges for the Li model, 31 nodes and 76 edges for the Chen model, and 78 nodes and 175 edges for our model (Table III).
Figure 3.
Directed graph views of abstracted interaction models used for comparative motif analyses. Each node represents the active state of species and edges between nodes indicate regulations (e.g. phosphorylation, degradation, transcriptional regulation). The bold colored lines indicate mutual regulations (green for activation and red for inhibition). (A) Interaction of core cycle entities in this study. The edge with dotted line represents the regulation with only genetic, but no direct evidence for the interaction. See annotations in the original network (Supplementary information S1) for details of each interaction. (B) Interaction of core cycle entities in Li et al (2004). (C) Interaction of core cycle entities in Chen et al (2004). See Supplementary information S12 for the further explanation.This image is also available as high resolution PDF.
Table 3. The statistical properties of yeast cell-cycle regulatory map in this study.
| Li et al (2004) | Chen et al (2004) | This study | |
|---|---|---|---|
| aWe ignored the types of regulations, activation or inhibition when counting. | |||
| bThe number in brackets indicates the total number of mutual regulations including mutual inhibitions and activations. That also means the number of feedback loops with 2 hops. | |||
| cThe number of all feedbacks with 3, 4 or 5 hops is shown in brackets. Therefore, the difference between the numbers inside and outside brackets indicates the number of positive feedback loops. | |||
| Number of nodes | 18 | 31 | 78 |
| Number of edges | 31 | 76 | 175 |
| Number of motifs | |||
| Feed-forward regulationsa | 5 | 9 | 68 |
| Mutual inhibitions or activationsb | 0 (/1) | 8 (/8) | 8 (/11) |
| Number of negative feedbacksc | |||
| 3 hops | 1 (/1) | 2 (/5) | 7(/15) |
| 4 hops | 2 (/4) | 8 (/15) | 23 (/40) |
| 5 hops | 1 (/3) | 7 (/11) | 35 (/60) |
Next, we enumerated three significant motifs in these models—feed-forward, feedback and mutual regulations. The details of the algorithm for counting these motifs are explained in Supplementary informations S10 and S12. The algorithm for counting all feedback loops, which are also called closed paths, with less than 6 hops was implemented based on Johnson's method (Johnson, 1975). Mutual inhibitions and activations can be considered as positive feedback loops with 2 hops. The total number of motifs identified in the comparative study is listed in Table III. As seen from the data, the number of motifs identified increases with the size of the network (largest for our comprehensive map), indicating a global pattern of control throughout the cell-cycle network.
Mutual inhibition and activation are one of the elemental motifs for making an irreversible switch-like behavior in biochemical circuit (Tyson et al, 2003). Although eight mutual regulations were detected in the Chen and our work, no mutual regulations were detected in the Li model. Surprisingly, although the number of feedback regulations in our model was much larger than the Chen model, most of the mutual regulations in our model were also detected in the Chen model except for mutual regulations involving newly added components such as Acm1 and Cdc5 (Supplementary information S11 gives the complete list of all identified motif components). Thus, the Chen model has captured mutual regulations, such as Clb2 and Sic1, Clb2 and Cdh1, and Clb2 and Mcm1, for the basis of bi-stable switches that enables robust oscillatory behavior of the cell-cycle process (Chen et al, 2000; Ciliberto et al, 2003; Cross and Siggia, 2005; Ingolia, 2005).
Negative feedback regulations with more than 3 hops have significant function for creating driving force of oscillations (Elowitz and Leibler, 2000; Tyson et al, 2003). Accordingly, feedback regulations essential to the cell-cycle system were commonly observed in the Chen and our network (e.g. Cdc20, Clb2 and Mcm1, Cdh1, Clb2, SBF and Cln2). On the other hand, positive feedback stabilizes the oscillation by creating bistability (Tyson et al, 2003; Novak and Tyson, 2008). In addition, positive feedbacks also contribute to make various steady states, which correspond to cell-cycle arrest points. Thus, the combination of positive and negative feedback regulations provides a fundamental mechanism of robust oscillatory response. As shown in Table III, the number of both positive and negative feedback regulations identified in our model was significantly larger compared with the other models. This result highlights the predominance of such complex control structures across the network, although all of them might not apparently affect phenotypic behaviors. The effect of these potential regulatory motifs on robustness would need to be investigated further.
Finally, the number of feed-forward regulations in our network (68) was much larger than in the Li and Chen model (5 and 9, respectively). The large part of them consists of a target protein and the characteristic regulator pair such as a kinase and an ubiquitin ligase (Clb2 and Cdc20), a kinase and a TF (Clb2 and Mcm1), and a phosphtase and a kinase (Cdc14 and Cdc5) (Supplementary information S11). Recently, Csikasz-Nagy et al (2009) have reported that a large number of ‘executor' proteins in cell-cycle system were regulated by a feed-forward loop (FFL) consisting of cyclin and a TF. In addition, the motif analysis revealed a large amount of feed-forward regulations even in ‘cell-cycle core' and ‘check points', which were only partially captured in the other two models.
The function of FFLs generally depends on the type and relative activity of each regulation, and, thus is less evident than feedback loops. However, several types of FFLs have been theoretically studied (Mangan and Alon, 2003; Tyson et al, 2003; Csikasz-Nagy et al, 2009). Coherent FFLs can confer robust signal transduction, and incoherent loops also can assure transient activation of an EP against signal input.
Questions may be raised that how does the map contribute to biological discoveries, in particular how does such control motifs actually contribute to robustness of the cellular system. In this study, we have shown that there are numbers of feedback and feed-forward control loops that was identified in our study, but not captured in Chen's study. In our previous paper (Moriya et al, 2006), we have shown that the Chen model behave less robustly than the real yeast against dose change of 30 cell-cycle-related genes. We hypothesized that some of such discrepancies are due to missing regulatory motifs in the Chen model. One of such regulations missing in the Chen model, but present in our map, is a feed-forward regulation involving Clb2, Cdc20 and Pds1. In our recent study (Kaizu et al, 2010), we have shown that, in fact, a feed-forward regulation (Clb2, Cdc20 and Pds1) that was not captured in the Chen model contributed to the robustness of cell cycle in vivo.
In summary, the results from the motif analysis highlight the recurring theme of structural control governing functional robustness in a global perspective across the entire yeast cell-cycle network.
Discussion
The yeast cell-cycle map elucidated in this work represents the initial step for capturing molecular-level information in a standardized format. It opens up avenues for integrating this information into realistic computational models, which can be shared across the community. As examples of such focused studies, we can cite reconstruction of mammalian RB/E2F pathway (Calzone et al, 2008), human cell-cycle events (Kohn, 1999), comprehensive maps of EGFR pathway (Oda et al, 2005) or Toll-like receptor signaling pathway (Oda and Kitano, 2006) to name a few. The yeast cell-cycle map endeavors to enlarge this collection of mechanistic pathway networks.
One of the challenges encountered in the construction of the consensus molecular network is the representation of structural modifications such as conformational changes in signaling proteins, arrangements of large scaffolding proteins in a particular three-dimensional orientations and so on. For example, the specific orientation of molecular components in forming the septin ring structure is only captured in terms of a complex of constituent molecules and does not represent their ring-like arrangement. Future versions of visualization standards would need to provide suitable graphical symbols to represent them more realistically. It is pertinent to note here that large-scale global analysis results on yeast, for example, identification of the substrates Cdk1 by Ubersax et al (2003), have been curated during the map reconstruction phase. However, as a lot of the information from these studies are unconfirmed by subsequent experiments and/or does not provide precise mechanisms and effects of interactions, the current cell-cycle map only incorporates cross-validated reactions from such global data sets.
Finally, usage of such maps for advancing science and their application needs to be clarified. One might argue that simply representing interactions in form of the map does not provide scientific insights, thus has only a limited value. For such an argument, it is important to remember what insights an atlas of the world provides us. Insights are in the eye of the observer, not on the map. The other argument may point to the complexity of the map, and claim that simplified diagrams are better or sufficient. Although usage of simplified small-scale diagrams are well-accepted, solely depending on such informal cartoons hide the reality of the biological system that are essentially highly complex. It is far more productive to focus our efforts on how to represent and make best use of comprehensive maps that is far more faithful to the reality than simplified diagrams.
Supplementary Material
This is the SBML-compliant CellDesigner™ 4.1 xml file of the original cell cycle comprehensive map represented in Figure. 1. The poster edition of the comprehensive map is available at: http://www.systems-biology.org/001/yeast/YeastCellCyclePosterEdition.pdf (This is a large file over 15MB)
This is the list of references used for the curation of the cell cycle comprehensive map in this paper.
This is the comprehensive list of Open Reading Frames (ORFs) for the genes in the comprehensive map.
This is the snapshot view of the comprehensive map representing the layered text annotation scheme employed to capture relevant information for a reaction in the map.
This is the network properties of the comprehensive yeast cell cycle map; degree distribution (a; Left vertical dotted line indicates the average of degrees, and right line indicates the boundary with top 15 species.), path length distribution (b; Red arrow indicates the average of path lengths.), closeness centrality distribution (c), and average neighborhood connectivity distribution (d).
This is the original XML file for the core plane of the comprehensive map.
This is the original XML file for the signaling plane of the comprehensive map.
This is the list of the structural plane components.
This is the archive file which contains three SIF files for motif analysis: The Li model (LiF_2004.sif), the Chen model (ChenKC_2004.sif), and this study (ThisStudy.sif).
Materials and methods employed for the analysis in the paper.
A list of regulatory motifs identified from the comprehensive map.
This is the Python implementation of the motif computation algorithm used in this study. The original extension is '.py'.
Navigation of the yeast cell cycle map on Payao, the community-curation platform for biological pathways.
Acknowledgments
ERATO-SORST Program of Japan Science and Technology Agency (JST), Genome Network Project of Ministry of Education, Culture, Sports, Science, and Technology, NEDO Fund for International Standard Formation from New Energy Development Organization and Okinawa Institute of Science and Technology in part support this research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bauer-Mehren A, Furlong LI, Sanz F (2009) Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol Syst Biol 5: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunewell S, Bornholdt S (2007) Superstability of the yeast cell-cycle dynamics: ensuring causality in the presence of biochemical stochasticity. J Theor Biol 245: 638–643 [DOI] [PubMed] [Google Scholar]
- Calzone L, Gelay A, Zinovyev A, Radvanyi F, Barillot E (2008) A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol Syst Biol 4: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Calzone L, Csikasz-Nagy A, Cross FR, Novak B, Tyson JJ (2004) Integrative analysis of cell cycle control in budding yeast. Mol Biol Cell 15: 3841–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KC, Csikasz-Nagy A, Gyorffy B, Val J, Novak B, Tyson JJ (2000) Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol Biol Cell 11: 369–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto A, Novak B, Tyson JJ (2003) Mathematical model of the morphogenesis checkpoint in budding yeast. J Cell Biol 163: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F (2006) Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J 25: 2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR (2003) Two redundant oscillatory mechanisms in the yeast cell cycle. Dev Cell 4: 741–752 [DOI] [PubMed] [Google Scholar]
- Cross FR, Siggia ED (2005) Shake it, don't break it: positive feedback and the evolution of oscillator design. Dev Cell 9: 309–310 [DOI] [PubMed] [Google Scholar]
- Csikasz-Nagy A, Battogtokh D, Chen KC, Novak B, Tyson JJ (2006) Analysis of a generic model of eukaryotic cell-cycle regulation. Biophys J 90: 4361–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csikasz-Nagy A, Kapuy O, Toth A, Pal C, Jensen LJ, Uhlmann F, Tyson JJ, Novak B (2009) Cell cycle regulation by feed-forward loops coupling transcription and phosphorylation. Mol Syst Biol 5: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL (1999) Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol Biol Cell 10: 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Lou F, Wang B, Campbell JL, Jong A (1996) Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell 7: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Naldi A, Lopez F, Chaouiya C, Ciliberto A, Thieffry D (2009) Modular logical modelling of the budding yeast cell cycle. Mol Biosyst 5: 1787–1796 [DOI] [PubMed] [Google Scholar]
- Faure A, Thieffry D (2009) Logical modelling of cell cycle control in eukaryotes: a comparative study. Mol Biosyst 5: 1569–1581 [DOI] [PubMed] [Google Scholar]
- Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G (1998) Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol 18: 3681–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (1996) Life with 6000 genes. Science 274: 546, 563–547 [DOI] [PubMed] [Google Scholar]
- Goldbeter A (1991) A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc Natl Acad Sci USA 88: 9107–9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbeter A (1996) Biochemical Oscillations and Cellular Rhythms: The Molecular Bases of Periodic and Chaotic Behaviour. Cambridge, UK: Cambridge University Press [Google Scholar]
- Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies RJ, Peter M (1997) Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev 11: 3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA (2001) Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol Biol Cell 12: 3402–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Hyver C, Le Guyader H (1990) MPF and cyclin: modelling of the cell cycle minimum oscillator. Biosystems 24: 85–90 [DOI] [PubMed] [Google Scholar]
- Ingolia N (2005) Cell cycle: bistability is needed for robust cycling. Curr Biol 15: R961–R963 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y (2000) Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB (1975) Finding all the elementary circuits of a directed graph. SIAM J Sci Comput 4: 77–84 [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275: 1311–1314 [DOI] [PubMed] [Google Scholar]
- Kaizu K, Moriya H, Kitano H (2010) Fragilities caused by dosage imbalance in regulation of the budding yeast cell cycle. PLoS Genet 6: e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H, Funahashi A, Matsuoka Y, Oda K (2005) Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 23: 961–966 [DOI] [PubMed] [Google Scholar]
- Kohn KW (1999) Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell 10: 2703–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Novere N, Finney A, Hucka M, Bhalla US, Campagne F, Collado-Vides J, Crampin EJ, Halstead M, Klipp E, Mendes P, Nielsen P, Sauro H, Shapiro B, Snoep JL, Spence HD, Wanner BL (2005) Minimum information requested in the annotation of biochemical models (MIRIAM). Nat Biotechnol 23: 1509–1515 [DOI] [PubMed] [Google Scholar]
- Le Novere N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, Bergman FT, Gauges R, Ghazal P, Kawaji H, Li L, Matsuoka Y, Villeger A, Boyd SE, Calzone L, Courtot M et al. (2009) The systems biology graphical notation. Nat Biotechnol 27: 735–741 [DOI] [PubMed] [Google Scholar]
- Li F, Long T, Lu Y, Ouyang Q, Tang C (2004) The yeast cell-cycle network is robustly designed. Proc Natl Acad Sci USA 101: 4781–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Sheu YJ, Baetz K, Andrews B, Snyder M (1997) SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275: 1781–1784 [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002) Specificity and stability in topology of protein networks. Science 296: 910–913 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Ghosh S, Kikuchi N, Kitano H (2010) Payao: a community platform for SBML pathway model curation. Bioinformatics 26: 1381–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62: 1191–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM (1971) The Biology of Cell Cycle. London: Cambridge University Press [Google Scholar]
- Mizunuma M, Hirata D, Miyaoka R, Miyakawa T (2001) GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J 20: 1074–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (2006) The Cell Cylcle: Principles of Control. London: New Science Press, Ltd [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Kitano H (2006) In vivo robustness analysis of cell division cycle genes in Saccharomyces cerevisiae. PLoS Genet 2: e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Russell P (2000) Cell cycle regulation in Schizosaccharomyces pombe. Curr Opin Microbiol 3: 631–636 [DOI] [PubMed] [Google Scholar]
- Murray A, Hunt T (1993) The Cell Cycle: An Introduction. USA: Oxford University Press [Google Scholar]
- Nasmyth K (1996) Viewpoint: putting the cell cycle in order. Science 274: 1643–1645 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ (2000) Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol 10: 195–205 [DOI] [PubMed] [Google Scholar]
- Norel R, Agur Z (1991) A model for the adjustment of the mitotic clock by cyclin and MPF levels. Science 251: 1076–1078 [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ (1993) Numerical analysis of a comprehensive model of M-phase control in Xenopus oocyte extracts and intact embryos. J Cell Sci 106 (Pt 4): 1153–1168 [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ (2008) Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9: 981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak B, Tyson JJ, Gyorffy B, Csikasz-Nagy A (2007) Irreversible cell-cycle transitions are due to systems-level feedback. Nat Cell Biol 9: 724–728 [DOI] [PubMed] [Google Scholar]
- Nurse P (1997) The Josef Steiner Lecture: CDKs and cell-cycle control in fission yeast: relevance to other eukaryotes and cancer. Int J Cancer 71: 707–708 [DOI] [PubMed] [Google Scholar]
- Oda K, Kitano H (2006) A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2: 2006 0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H (2005) A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol 1: 2005 0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I (1993) FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73: 747–760 [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I (1994) Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science 265: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Schnoes AM, Brown SD, Dodevski I, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S (2000) A network of protein-protein interactions in yeast. Nat Biotechnol 18: 1257–1261 [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Breeden LL (2003) Rad53 checkpoint kinase phosphorylation site preference identified in the Swi6 protein of Saccharomyces cerevisiae. Mol Cell Biol 23: 3405–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ (1991) Modeling the cell division cycle: cdc2 and cyclin interactions. Proc Natl Acad Sci USA 88: 7328–7332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Chen KC, Novak B (2003) Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol 15: 221–231 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO (2007) Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol 9: 106–112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This is the SBML-compliant CellDesigner™ 4.1 xml file of the original cell cycle comprehensive map represented in Figure. 1. The poster edition of the comprehensive map is available at: http://www.systems-biology.org/001/yeast/YeastCellCyclePosterEdition.pdf (This is a large file over 15MB)
This is the list of references used for the curation of the cell cycle comprehensive map in this paper.
This is the comprehensive list of Open Reading Frames (ORFs) for the genes in the comprehensive map.
This is the snapshot view of the comprehensive map representing the layered text annotation scheme employed to capture relevant information for a reaction in the map.
This is the network properties of the comprehensive yeast cell cycle map; degree distribution (a; Left vertical dotted line indicates the average of degrees, and right line indicates the boundary with top 15 species.), path length distribution (b; Red arrow indicates the average of path lengths.), closeness centrality distribution (c), and average neighborhood connectivity distribution (d).
This is the original XML file for the core plane of the comprehensive map.
This is the original XML file for the signaling plane of the comprehensive map.
This is the list of the structural plane components.
This is the archive file which contains three SIF files for motif analysis: The Li model (LiF_2004.sif), the Chen model (ChenKC_2004.sif), and this study (ThisStudy.sif).
Materials and methods employed for the analysis in the paper.
A list of regulatory motifs identified from the comprehensive map.
This is the Python implementation of the motif computation algorithm used in this study. The original extension is '.py'.
Navigation of the yeast cell cycle map on Payao, the community-curation platform for biological pathways.