Abstract
By providing new insights into the distribution of a protein’s torsion angles, recent statistical models for this data have pointed the way to more efficient methods for protein structure prediction. Most current approaches have concentrated on bivariate models at a single sequence position. There is, however, considerable value in simultaneously modeling angle pairs at multiple sequence positions in a protein. One area of application for such models is in structure prediction for the highly variable loop and turn regions. Such modeling is difficult due to the fact that the number of known protein structures available to estimate these torsion angle distributions is typically small. Furthermore, the data is “sparse” in that not all proteins have angle pairs at each sequence position. We propose a new semiparametric model for the joint distributions of angle pairs at multiple sequence positions. Our model accommodates sparse data by leveraging known information about the behavior of protein secondary structure. We demonstrate our technique by predicting the torsion angles in a loop from the globin fold family. Our results show that a template-based approach can now be successfully extended to modeling the notoriously difficult loop and turn regions.
Key words and phrases: Bayesian nonparametrics, density estimation, dihedral angles, protein structure prediction, torsion angles, von Mises distribution
1. Introduction
The field of protein structure prediction has greatly benefitted from formal statistical modeling of available data [Osguthorpe (2000); Bonneau and Baker (2001)]. More automatic methods for predicting protein structure are critical in the biological sciences as they help to overcome a major bottleneck in effectively interpreting and using the vast amount of genomic information: determining the structure, and therefore the function, of a gene’s protein product. Currently the growth of genomic data far outstrips the rate at which experimental methods can solve protein structures. To help accelerate the process, protein structure prediction methods aim to construct accurate three-dimensional models of a target protein’s native state using only the protein’s amino acid sequence.
Protein structure is typically described in terms of four categories: primary through quarternary. Primary structure consists of the linear sequence of covalently bonded amino acids that make up a protein’s polypeptide chain. Secondary structure describes the regularly repeating local motifs of α-helices, β-strands, turns and coil regions. For a single polypeptide chain, tertiary structure describes how the secondary structure elements arrange in three-dimensional space to define a protein’s fold. By allowing the polypeptide chain to come back on itself, the loops and turns effectively define the arrangement of the more regular secondary structure of α-helices and β-strands. Quarternary structure describes how multiple folded polypeptide chains interact with one another. In a typical structure prediction problem the primary structure is known, and the goal is to use this information to predict the tertiary structure.
One of the standard approaches to this problem is template-based modeling. Template-based approaches are used when the target sequence is similar to the sequence of one or more proteins with known structure, essentially forming a protein fold “family.” Typically the core of the modeled fold is well defined by regular secondary structure elements. One of the major problems is modeling the loops and turns: those regions that allow the protein’s tertiary structure to circle back on itself. Unlike the consistency of the core in a template-based prediction, the variation in the loops and turns (both in terms of length and amino acid composition) between structures with the same fold family is often quite large. For this reason current knowledge-based methods do not use fold family data. Instead of the template-based approach, they use libraries of loops which are similar in terms of length and amino acid sequence to the target. However, such library data sets do not have the same level of structural similarity as do purely within-family data sets. In this work, our approach to modeling structural data allows us to effectively extend template-based modeling to the loop and turn regions and thereby make more informed predictions of protein structure.
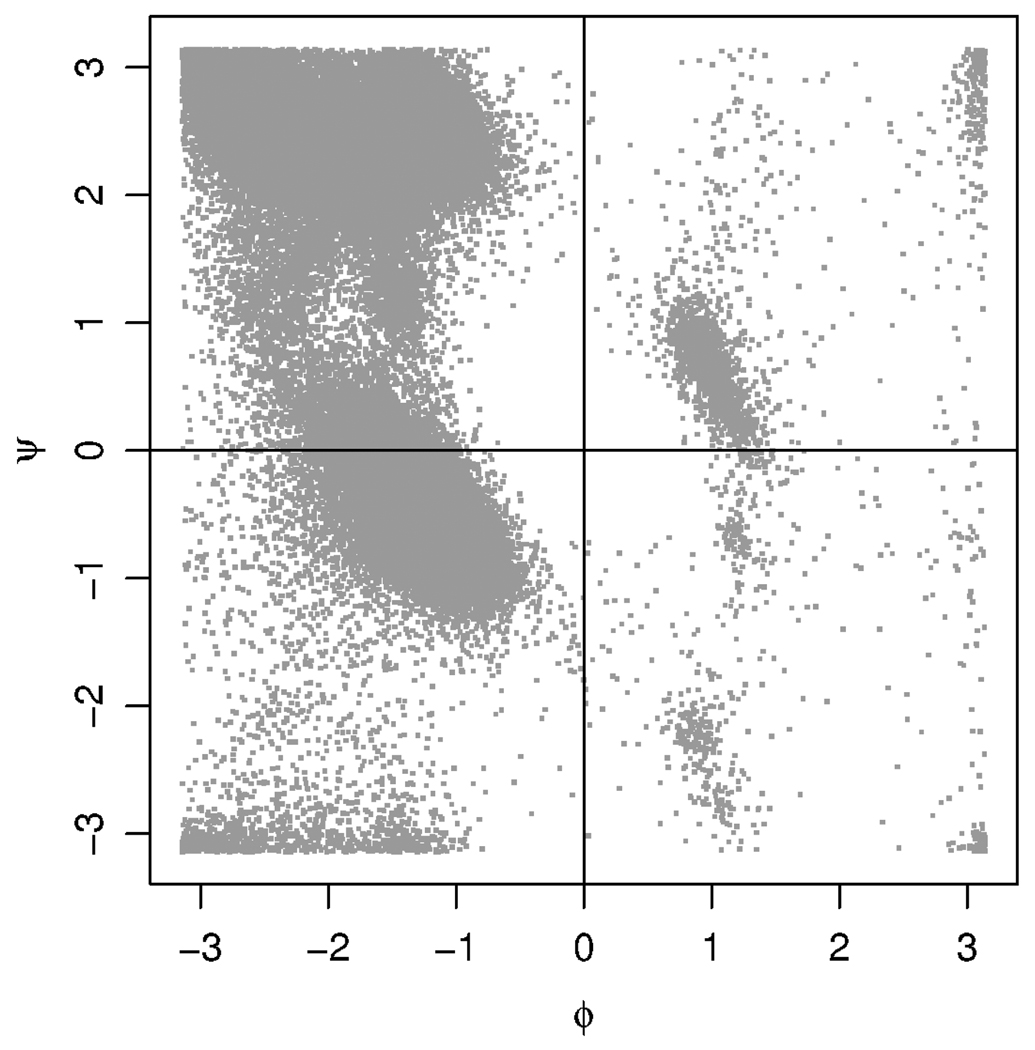
Our approach is based on the simplest representation of protein structure: the so-called backbone torsion angles. This representation consists of a (ϕ, ψ) angle pair at each sequence position in a protein, and it provides a reduction in complexity from using the 12 Cartesian coordinates for the 4 heavy backbone atoms at each position. This method for describing protein structure was originally proposed by Ramachandran, Ramakrishnan and Sasisekharan (1963), and the customary graphical representation of this type of data is the Ramachandran plot. The Ramachandran plot in Figure 1 shows the (ϕ, ψ) angles of protein positions containing the amino acid alanine. The pictured data set was obtained from the Protein Data Bank [PDB, Kouranov et al. (2006)], a repository of solved protein structures.
FIG. 1.
Ramachandran plot for the 130,965 angle pairs that make up the PDB data set for the amino acid alanine. Angles are measured in radians.
Density estimation of Ramachandran space is particularly useful for template-based structure prediction. Because a target protein with unknown tertiary structure is known to be related to several proteins with solved structures, models for bivariate angular data can be used to estimate the distribution of (ϕ, ψ) angles for a protein family, and thereby generate candidate structures for the target protein.
While there has been considerable recent work on modeling in Ramachandran space at a single sequence position [see, e.g., Ho, Thomas and Brasseur (2003); Lovell et al. (2003); Butterfoss, Richardson and Hermans (2005); Lennox et al. (2009a, 2009b)], models that accommodate multiple sequence positions remain uncommon. A notable exception is the DBN-torus method of Boomsma et al. (2008). However, this approach was developed primarily to address sampling of fragments in de novo protein structure prediction, and so specifically does not include protein family information. De novo structure prediction is used when similar proteins with known structure are unavailable and is thus inherently more difficult and less accurate than template based modeling. While template-based methods can draw on a certain amount of known information, a common complication is that protein families typically have fewer than 100 members, and often fewer than 30 members.
Not only do protein families tend to have few members, but the data within a family is “sparse,” particularly in loop regions. A template sequence for a protein structure family is generated by simultaneously aligning all of the member proteins using amino acid type at each sequence position. However, the sequences in a fold family are usually of different lengths due to different sizes of loops and turns. In such an alignment, a typical member protein is not represented at every sequence position. This leads to what we call a “sparse data” problem. Note that this is not a missing data situation, as a sequence position is not merely unobserved, but rather does not in fact exist.
A joint model for a large number of torsion angles using somewhat limited data can be enhanced by leveraging prior knowledge about the underlying structure of the data. We present a Bayesian nonparametric model incorporating a Dirichlet process (DP) with one of two possible families of centering distributions for modeling the joint distributions of multiple angle pairs in a protein backbone. Our model addresses the sparse data situation, and also accommodates a larger number of sequence positions than previously considered methods of template-based density estimation. One of our proposed centering distributions leads to a largely noninformative prior, but we also propose a family of centering distributions based on known characteristics of protein secondary structure in the form of a hidden Markov model (HMM). The inclusion of an HMM allows our model to share structural information across sequence positions. Since each secondary structure type has a distinctive footprint on the Ramachandran plot, with this process we can use an informative prior to incorporate additional information into our model.
There is precedent for the use of a hidden Markov model for protein structure prediction in the DBN-torus model of Boomsma et al. (2008). There, secondary structure information is incorporated into the state space of a dynamic Bayesian network, a generalization of an HMM, which allows the DBN-torus model to infer secondary structure when generating candidate angle pair sequences. The model generates significantly better candidates, however, when secondary structure is provided from an external secondary structure prediction method. There are other differences between the DBN-torus method and our own which result from the distinct applications of the two methods. DBN-torus is used for de novo structure prediction; it is designed to make predictions for any kind of protein, and is not customized for a particular fold family. In contrast, our method is tailored for template-based modeling. Thus, the DBN-torus model can be used even when template information is unavailable, but will miss opportunities for improvement when fold-family structure information exists.
In this paper we apply our method to the loop region between the E and F α-helices of the globin protein template, which varies between 8 and 14 sequence positions in length. By borrowing strength from neighbors containing numerous observations, our model generates informative density estimates even if relatively little data is available at a given position. This property gives our method a significant advantage in loop prediction by allowing the use of fold family data. This extension of template-based modeling to loop regions was not possible before the development of these statistical tools. We show that using our Dirichlet process mixture of hidden Markov models (DPM–HMM) in a template-based approach provides a better match to real structure data than does either a library-based method or DBN-torus.
In Section 2 we give some background on previous work in torsion angle modeling, as well as the bivariate von Mises distribution and the Dirichlet process. In Section 3 we present our model along with the informative and noninformative priors. An explanation of how to fit this model and use it for density estimation is provided in Section 4. Section 5 contains an application of our method to estimate the joint density of torsion angles in the EF loop region in the globin protein family. Finally, we discuss our conclusions in Section 6.
2. Preliminaries
We illustrate the development of our model by first exploring methods for modeling individual torsion angle pairs. Working with torsion angles requires the use of distributions specifically designed to account for the behavior of angular data. This data has the property that an angle ϕ is identical to the angle ϕ + 2kπ for all k ∈ {…, −1, 0, 1,…}. The bivariate von Mises distribution is commonly used for paired angular data.
Originally proposed as an eight parameter distribution by Mardia (1975), subclasses of the bivariate von Mises with fewer parameters are considered easier to work with and are often more interpretable. Rivest (1982) proposed a six parameter version, which has been further refined into five parameter distributions. One such subclass, known as the cosine model, was proposed by Mardia, Taylor and Subramaniam (2007), who employed it in frequentist mixture modeling of (ϕ, ψ) angles at individual sequence positions. In this paper we consider an alternative developed by Singh, Hnizdo and Demchuk (2002) known as the sine model.
The sine model density for bivariate angular observations (ϕ, ψ) is defined as
| (2.1) |
for ϕ, ψ, μ, ν ∈ (−π, π] κ1, κ2 > 0, λ ∈ (−∞, ∞), and
| (2.2) |
The parameters μ and ν determine the mean of the distribution, while κ1 and κ2 are precision parameters. The parameter λ determines the nature and strength of association between ϕ and ψ. This density is unimodal when λ2 < κ1κ2 and bimodal otherwise. One of the most attractive features of this particular parameterization of the bivariate von Mises is that, when the precision parameters are large and the density is unimodal, it can be well approximated by a bivariate normal distribution with mean (μ, ν) and precision matrix Ω, where Ω11 = κ1, Ω22 = κ2 and Ω12 = Ω21 = −λ.
Singh, Hnizdo and Demchuk (2002) fit individual sine model distributions to torsion angle data sets. Mardia et al. (2008) developed an extension of the bivariate sine model for n-dimensional angular data, but the constant of integration is unknown for n > 2, rendering it difficult to use. We instead consider a method based on a Dirichlet process mixture model.
The Dirichlet process, first described by Ferguson (1973) and Antoniak (1974), is a distribution of random measures which are discrete with probability one. The Dirichlet process is typically parameterized as having a mass parameter α0 and a centering distribution G0. Using the stick-breaking representation of Sethuraman (1994), a random measure G drawn from a Dirichlet process DP(α0G0) takes the form
where δτ is an indicator function equal to 1 if τ ∈ B and 0 otherwise, τj ~ G0, . In this form, the discreteness of G is clearly evident.
This discreteness renders the DP somewhat unattractive for directly modeling continuous data. However, it can be effectively used in hierarchical models for density estimation [Escobar and West (1995)]. Consider a data set z1,…, zn, and a family of distributions f(z|τ) with parameter τ. A Dirichlet process mixture (DPM) model takes the form
| (2.3) |
The discreteness of draws from a DP means that there is positive probability that τi = τj for some i ≠ j. For such i and j, zi and zj come from the same component distribution, and are viewed as being clustered together. The clustering induced by DPM models generates rich classes of distributions by using mixtures of simple component distributions.
While τ is generally taken to be scalar- or vector-valued, there is nothing inherent in the definition of the DP that imposes such a restriction, and more complex centering distributions have been explored [e.g., MacEachern (2000); De Iorio et al. (2004); Gelfand, Kottas and MacEachern (2005); Griffin and Steel (2006); Dunson, Pillai and Park (2007); Rodríguez, Dunson and Gelfand (2008)]. In a model for the distribution of multiple angle pairs, we propose using a hidden Markov model (HMM), a discrete stochastic process, as the centering distribution G0. In the following section we describe how to use this hidden Markov model as a component of an informative prior for protein conformation angle data.
3. Dirichlet process mixture model for multiple alignment positions
The necessary Bayesian procedures to use a DP mixture of bivariate von Mises sine distributions for modeling torsion angle data at individual sequence positions were developed by Lennox et al. (2009a, 2009b). In this section we extend this model to multiple sequence positions, and provide a noninformative prior that directly extends the single position model. In addition, we describe a method for using an HMM as a centering distribution in an informative prior for sequences of contiguous positions. We also show how to perform density estimation using our model.
Consider a protein family data set consisting of n angle pair sequences denoted x1,…, xn. Let each observation have m sequence positions, whose angle pairs are denoted xi1,…, xim for the ith sequence, with xij = (ϕij, ψij). For the moment assume that we have complete data, that is, that every xij contains an observed (ϕ, ψ) pair. Then our base model for the jth position in the ith sequence is as follows:
| (3.1) |
where θij consists of the parameters (μij, νij, Ωij), θi = (θi1, …, θim) and f(x|θ) is a bivariate von Mises sine model. The distribution G is a draw from a Dirichlet process, while H1 and H2 are the centering distributions that provide atoms of the mean and precision parameters, respectively. Note that the product H1 H2 takes the role of G0 from (2.3).
For our purposes, H2 always consists of the product of m identical Wishart distributions we call h2. This centering distribution assumes independence for the precision parameters of sequence positions given clustering information. Similarly, we do not assume a relationship between the precision parameters and the mean parameters for any sequence position, again restricting ourselves to the situation when clustering is known. The use of a Wishart prior for bivariate von Mises precision parameters is motivated by concerns about ease of sampling from the prior distribution and potential issues with identifiability. A more detailed explanation is given by Lennox et al. (2009b).
We discuss two distinct choices for H1, the centering distribution for the sequence of mean parameters (µi, νi). The first assumes a priori independence of the mean parameters across sequence positions, while the second is designed to share information across adjacent sequence positions using a hidden Markov model based on known properties of protein secondary structure.
3.1. Noninformative prior for multiple sequence positions
A straightforward extension of the existing single position DPM model takes H1 to be the product of m identical bivariate von Mises distributions we call h1. For truly noninformative priors, a diffuse von Mises distribution may be replaced by a uniform distribution on (−π, π] × (−π, π]. Both the von Mises and uniform versions of the model assume a priori independence of the centering parameters (μij, νij) across sequence positions j. However, dependence can still appear in the posterior distribution. While we refer to this as the noninformative model, and use it as such, there is no reason why informative distributions could not be used as the components of H1, nor must these components be identical. The primary distinguishing feature of this choice of model is that no assumptions are made as to the relationship between the mean parameters at the various sequence positions.
An advantage of this choice for H1 is that sequence positions j and j + 1 need not be physically adjacent in a protein. This situation could be of interest when modeling the joint distribution of amino acid residues which are not neighbors with respect to the primary structure of a protein, but which are close together when the protein is folded.
3.2. Informative DPM–HMM model for adjacent sequence positions
When considering adjacent positions, however, a model assuming independence is not making use of all available information regarding protein structure. For this situation we recommend a centering distribution H1 that consists of a hidden Markov model incorporating secondary structure information.
We call our model a Dirichlet process mixture on a hidden Markov model space, or DPM–HMM. Hidden Markov models define a versatile class of mixture distributions. An overview of Bayesian methods for hidden Markov models is given by Scott (2002). HMMs are commonly used to determine membership of protein families for template-based structure modeling, but in this case the state space relates to the amino acid sequence, also known as the primary structure [see, e.g., Karplus et al. (1997)]. We propose instead to use an HMM for which the hidden state space consists of the secondary structure type at a particular sequence position. While HMMs incorporating secondary structure have been used for de novo structure prediction methods [Boomsma et al. (2008)], they have not previously been employed for template-based strategies. We can determine both the transition probabilities between states and the distributions of (ϕ, ψ) angles for each secondary structure type based on data sets in the Protein Data Bank. Such a model provides a knowledge-driven alternative to our noninformative prior from Section 3.1 for adjacent sequence positions.
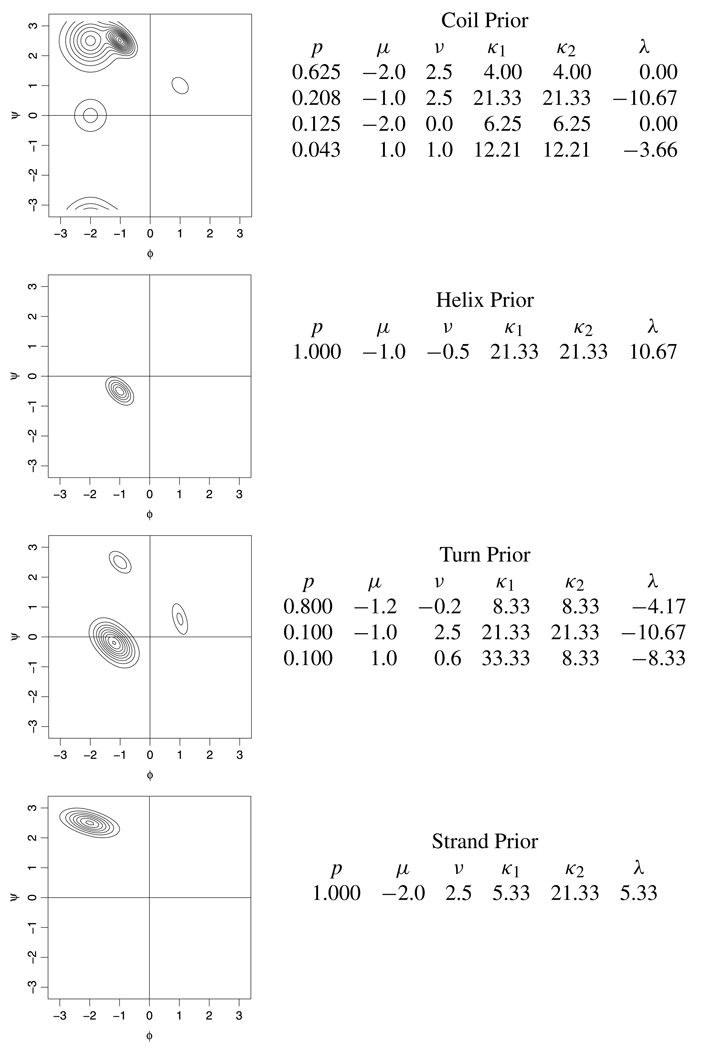
Our model has four hidden states corresponding to four secondary structure metatypes defined by the Definition of Secondary Structure for Proteins [DSSP, Kabsch and Sander (1983)] program: turn (T), helix (H), strand (E) and random coil (C). These four types are condensed from eight basic types, with all helices being characterized as (H), β-turns and G-turns combined into the class (T), and both strands and β-bulges defined as (E). The model for a realization θ from our hidden Markov model is defined as follows:
where sj defines the state of the Markov chain at position j, with sj ∈ {1, 2, 3, 4}. M(sj | sj−1) is a discrete distribution on {1, 2, 3, 4} that selects a new state type with probabilities determined by the previous state type. We set our transition probability matrix based on 1.5 million sequence position pairs from the PDB, while the initialization probabilities correspond to the stationary distribution for the chain. Note that s = (s1,…,sm) is an observation from a discrete time Markov process. We then define f(θj|sj) to be a probability distribution with parameters determined by the current secondary structure state of the chain.
Single bivariate von Mises distributions are not adequate to serve as the state distributions for the four secondary structure types. Instead, we use mixtures of between one and five bivariate von Mises sine models. The amino acids proline and glycine exhibit dramatically different secondary structure Ramachandran distributions, and so were given their own distinct sets of secondary structure distributions. Figure 2 shows the state distributions used for each secondary structure class for the eighteen standard amino acids.
FIG. 2.
Graphical and numerical representations of our von Mises mixture distributions for each of the four secondary structure states Note that this is the general set of secondary structure distributions and is not used at positions containing the amino acids proline or glycine.
Although these are distributions for the means of the bivariate von Mises distribution, we chose them to mimic the distributions of (ϕ, ψ) angles in each of these secondary structure classes, which means that they are somewhat more diffuse than necessary. The use of these secondary state distributions in conjunction with the Markov chain on the state space allows us to leverage information about secondary structure into improved density estimates, and provides a biologically sound framework for sharing information across sequence positions.
Note that our model is not to be confused with the hidden Markov Dirichlet process (HMDP) proposed by Xing and Sohn (2007). The HMDP is an implementation of a hidden Markov model with an infinite state space, originally proposed by Beal, Ghahramani and Rasmussen (2002). Their model is an instance of the Hierarchical Dirichlet Process (HDP) of Teh et al. (2006), whereas our DPM–HMM is a standard Dirichlet process with a novel centering distribution.
4. Density estimation
Recall that we are interested in estimating the joint density of x = (ϕ, ψ) angles at each sequence position for a candidate structure from some protein family. Our method, as outlined by Escobar and West (1995), involves treating our density estimate as a mixture of components f(xn+1 | θn + 1), which in our case are products of bivariate von Mises sine models, mixed with respect to the posterior predictive distribution of the parameters θn+1. This can be written as
| (4.1) |
This integral cannot be written in closed form, but can be well approximated by Monte Carlo integration. This is achieved by acquiring samples from the posterior predictive distribution for θn+1. then
| (4.2) |
While (4.2) can be evaluated for any (ϕ, ψ) sequence x, we are typically interested in graphical representations of marginal distributions at each sequence position. For this purpose we evaluate on a 360 × 360 grid at each alignment position. This general Monte Carlo approach works for joint, marginal, and conditional densities.
4.1. Markov chain Monte Carlo
All that remains is to determine how to obtain the samples from the posterior predictive distribution of θn+1, which consists of μn+1, νn+1 and Ωn+1. Fortunately, while our model is novel, the behaviors of Dirichlet process mixtures, hidden Markov models, and the bivariate von Mises distribution are well understood. The complexity of the posterior distribution prevents direct sampling, but we provide the details of a Markov chain Monte Carlo update scheme using an Auxiliary Gibbs sampler [Neal (2000)] in Appendix A.
4.2. The sparse data problem
The model as described up to this point does not fully account for the complexity of actual protein alignment data. Rather than being a simple vector xi of bivariate (ϕ, ψ) observations, the real data also includes a vector ai of length m which consists of variables indicating whether or not peptide i was observed at each sequence position. Let aij = 1 if peptide i is included at alignment position j, and 0 otherwise. This data structure is unique in several ways. Notice that ai is not only known for proteins with solved structure, but is also typically available for a target peptide sequence. Therefore, we can avoid fitting a model that includes alignment positions which are not of interest for our particular problem. This is not a true “missing data” problem as the unobserved sequence positions are not only absent from our data set, but do not exist.
Our model is able to adjust to sparse data with the following modification. Recall that the full conditional distributions could be divided up into a prior component and a data component at each sequence position. This makes it trivial to exclude an observation from the likelihood, and hence posterior distribution calculation, at sequence positions where it is not observed. For example, we can modify the full conditional distribution of the means in the DPM–HMM model, given in equation (A.3), to be
| (4.3) |
The full conditional distributions for the precision parameters and the means with a noninformative prior, equations (A.1) and (A.2), respectively, can be modified in a similar manner. The likelihood of xi|θ, is also used by the Auxiliary Gibbs sampler. Once again, adjust to absent data by removing unobserved positions from the likelihood.
This model provides a straightforward method to cope with the sparse data problem inherent in protein structure prediction. Note that the situation in which there is ample data generally but sparse data at a few sequence positions particularly highlights the value of the DPM–HMM model. Secondary structure at a sparse position can be inferred based on the surrounding positions, which can allow us to provide a better density estimate at positions with few observed data points.
5. Application: Loop modeling in the globin family
5.1. Background
A protein’s fold, or tertiary structure, consists of multiple elements of local, regular secondary structure (repeating local motifs) connected by the more variable loops and turns of various lengths. These loop and turn regions can be vital to understanding the function of the protein, as is the case in the immunoglobulin protein family where the conformation of the highly variable loops determine how an antibody binds to its target antigens to initiate the body’s immune response. These loop regions also tend to be the most structurally variable regions of the protein, and modeling their structure remains an outstanding problem in protein structure prediction [Baker and Sali (2001)]. Current knowledge-based loop modeling methods draw on generic loop libraries. Library-based methods search the Protein Data Bank for loops with entrance and exit geometries similar to those of the target loop, and use these PDB loops as templates for the target structure [e.g., Michalsky, Goede and Preissner (2003)]. Note that library-based methods differ from typical template-based modeling in that they do not confine themselves to loops within the target protein’s family. Strictly within family estimates have not previously been possible. Using the DPM–HMM model, we are able to compare a library-based approach to a purely within family template-based method for the EF loop in the globin family.
The globins are proteins involved in oxygen binding and transport. The family is well studied and has many known members. Therefore, the globin fold is suitable as a test case for template-based structure prediction methods. A globin consists of eight helices packed around the central oxygen binding site and connected by loops of varying lengths. The helices are labeled A through H, with the loops labeled according to which helices they connect. The EF loop is the longest loop in the canonical globin structure. We generated a simultaneous alignment of 94 members of the globin family with known tertiary structure using MUSCLE [Edgar (2004)]. For this alignment, positions 93–106 correspond to the EF loop.
Table 1 gives a summary of the behavior of 94 representative globins in the EF loop region. There is considerable diversity in both the length and amino acid composition of this loop. Representative loops were between 8 and 14 amino acids long, and the highly conserved regions, particularly at the tail end of the loop, exhibited considerable variability in amino acid composition.
TABLE 1.
A table giving the details on the EF loop for an alignment of 94 members of the globin family The columns are the alignment position the number of proteins represented at the position the most conserved amino acid (s) at the alignment position and the total number of distinct amino acids observed at the alignment position
| Position | # of proteins | Most conserved AA | # of AAs |
|---|---|---|---|
| 93 | 94 | LEU | 7 |
| 94 | 94 | ASP | 10 |
| 95 | 94 | ASN | 9 |
| 96 | 26 | ALA | 11 |
| 97 | 28 | GLY | 8 |
| 98 | 28 | LYS | 10 |
| 99 | 94 | LEU | 7 |
| 100 | 1 | THR | 1 |
| 101 | 2 | VAL | 1 |
| 102 | 2 | THR ARG | 2 |
| 103 | 93 | LYS | 13 |
| 104 | 94 | GLY | 15 |
| 105 | 94 | ALA | 15 |
| 106 | 94 | LEU | 10 |
We compare three different methods for loop modeling: our DPM–HMM method with globin family data, the noninformative prior model with globin family data, and a library-based approach. Library approaches generate lists of loops similar to the target and use these as templates for the target loop, generating a discrete distribution which almost surely has mass 0 at the true conformation of the unknown loop. To make this method comparable to our density-based approaches, we used our noninformative prior model on library data sets to generate a continuous density estimate. Note that all sequences in a library data set are of the same length, which means that they will never exhibit sparsity. For this reason, fitting the DPM–HMM model on the library data set would not present much improvement over the noninformative model.
5.2. Parameter settings
For each of the 94 globins in the alignment, we generated density estimates using each of the three methods in question. For the DPM–HMM and noninformative models, we excluded the target from the data set used to generate the density estimates, but used amino acid and sparse data information from the target protein. This is reasonable since primary structure based alignments are available for template modeling of an unknown protein. For the library-based estimate, we applied our noninformative prior model sequences from the coil library of Fitzkee, Fleming and Rose (2005) which have the same length as the target sequence, and have at least four sequence positions with identical amino acids. Library data sets ranged in size from 17 to 436 angle pair sequences.
For each of our models, we ran two chains: one starting with all observations in a single cluster and one with all observations starting in individual clusters. Each chain was run for 11,000 iterations with the first 1000 being discarded as burnin. Using 1 in 20 thinning, this gave us a combined 1000 draws from the posterior distribution of the parameters.
In all cases, our Wishart prior used υ = 1, and we set the scale matrix B to have diagonal elements of 0.25 and off-diagonal elements of 0. Note that we use the Bernardo and Smith (1994), pages 138–139, parameterization, with an expected value of υB−1 = B−1. Our choice of υ was motivated by the fact that this is the smallest possible value for which moments exist for the Wishart distribution, and higher values would have lead to a more informative prior. The choice of B gave an expected standard deviation of about 30 degrees and assumed a priori that there was no correlation between ϕ and ψ, which seemed to work well in practice. For our noninformative prior on the means, we took h1 to have μ0 = ν0 = 0, κ10 = κ20 = 0.1 and λ0 = 0. This provided a diffuse centering distribution.
In all cases we took the DP mass parameter α0 to be 1. However, our results were robust to departures from this value. For example, for two randomly selected proteins we gave values for α0 ranging between 0.2 and 15, giving prior expected numbers of clusters from approximately 2–30. For our first peptide the observed mean cluster number ranged from 3.96 to 4.46, while the second had values from 4.40 to 4.65. Thus, even our most extreme choices for the mass parameter changed the posterior mean number of clusters by less than 1.
5.3. Results of comparison to library
We performed pairwise comparisons for each of our models using the Bayes factor, defined as
| (5.1) |
where M1 and M2 are density estimates generated by two of our three possible models. We present the results of the analyses for our 94 leave-one-out models in Table 2.
TABLE 2.
Comparison between the DPM–HMM model on the globin family data, noninformative prior with globin data, and noninformative model with library data. The columns Model X and Model Y give the percentage of the time that the likelihood for the target conformation using Model X was greater than the likelihood of the same conformation using Model Y. This is the equivalent to a Bayes factor comparison with Model X in the numerator being greater than 1
| Loop length | Total | DPM–HMM to library (%) |
Noninf to library (%) |
DPM–HMM to noninf (%) |
|---|---|---|---|---|
| 8 | 66 | 100 | 100 | 70 |
| 10 | 3 | 67 | 67 | 67 |
| 11 | 23 | 100 | 96 | 39 |
| 13 | 1 | 100 | 100 | 100 |
| 14 | 1 | 100 | 100 | 100 |
| All | 94 | 99 | 98 | 63 |
First we will address the comparison between the DPM–HMM and noninformative models using the globin data. These models show far more similarity to each other than to the noninformative model using the library data, both in terms of the number of Bayes factors indicating superiority on each side, and the fact that those Bayes factors tended to be smaller in magnitude than those generated by comparisons to the library models. Indeed, at positions with more than 30 observations the marginal distributions generated by the two models appear to be very similar. Consider the null hypothesis that the probability that the DPM–HMM is superior to the noninformative model is less than or equal to 0.5. A binomial test of this hypothesis yields a p-value of 0.009. Of these Bayes factor results, 68 met standard criteria for substantial evidence of superiority (|log10(B) | > 1/2) [Kass and Raftery (1995)], of which 45 supported the use of the DPM–HMM model, giving a p-value of 0.005. This evidence, in addition to the fact that the combined Bayes factor, the product of all of the individual comparisons, has a value of 1038, provides overwhelming evidence in favor of using the DPM–HMM rather than the noninformative model. For this reason, in the remainder of the paper, we will only refer to the DPM–HMM model when making use of the globin data set.
Recall that the library model made use of loops of the same length as the target, and which had a certain degree of similarity in terms of amino acid sequence. Thus, the coil library does not exhibit any sparse data behavior. It is also unlikely to recapture the globin family EF loops due to the considerable variability in both length and amino acid composition. Our results indicate that the DPM– HMM model overwhelmingly outperforms the library-based method. Not only is the relevant Bayes factor greater than 1 in 93 out of 94 cases, it is greater than 100 in 92 cases. The case in which the library-based method outperformed the DPM–HMM was also significant according to the Kass and Raftery (1995) criteria, so there were no ambiguous individual cases. The combined Bayes factor was 10959, indicating that the DPM–HMM model was definitely superior to the library overall.
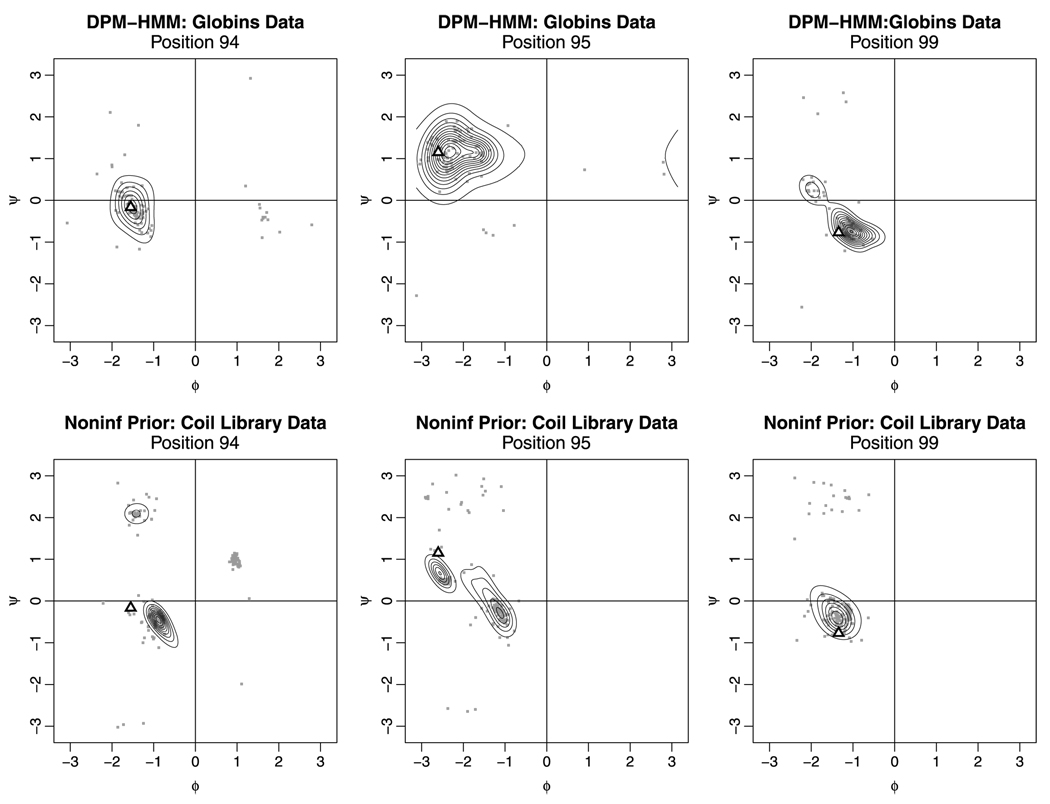
Figure 3 shows marginal density estimates generated for prototypical globin “1jebD” for both models, along with the true (ϕ, ψ) sequence for the protein for a portion of the EF loop. By searching the PDB for loops that are similar to the target in terms of length and sequence identity, the library method tends to place considerable mass in areas of conformational space that are not occupied by members of the globin family. While the members of the data set for the globin family may not match the target loop in terms of length or amino acid sequence, by virtue of being globins themselves they provide a better match to the target conformation. This pattern of improvement held true regardless of loop length. Significant improvement was found even for the length 13 and 14 loops, for which sparse data was a particular problem.
FIG. 3.
Density estimates for positions 94, 95 and 99 for protein “1jebD.” The gray dots indicate the data used to fit the model, while the triangles show the true (ϕ, ψ) conformation of the target protein.
5.4. Results of comparison to DBN-torus
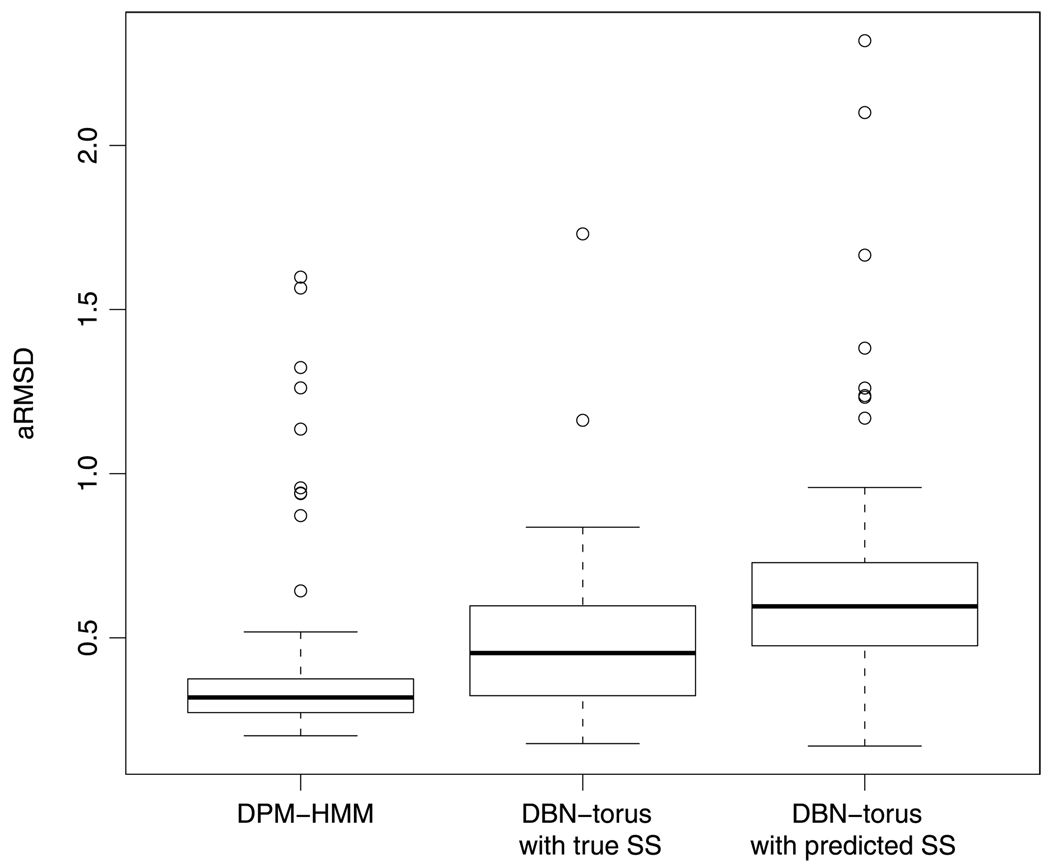
In addition to comparing the DPM–HMM to the knowledge-based library method, we have also conducted a comparison to the de novo DBN-torus sequence prediction method of Boomsma et al. (2008). Unlike the previously addressed library-based methods, DBN-torus uses continuous density estimates, but is not customized for loop regions. It can be used to generate sequences of predicted angle pairs given amino acid data, secondary structure data, or no input at all. The best results for DBN-torus are generated using amino acid data and predicted secondary structure data. For each of our 94 targets, we generated 1000 candidate draws using the DPM–HMM, DBN-torus with predicted secondary structure data from PsiPred [McGuffin, Bryson and Jones (2000)], and DBN-torus using the true secondary structure data. Although having exact knowledge of secondary structure for a target protein is unrealistic in practice, it gives an idea of how well DBN-torus can perform with optimal secondary structure prediction. We followed the strategy of Boomsma et al. (2008) of using the angular RMSD to judge the accuracy of our predictions. For each target, the best draw judged by minimum aRMSD was selected, and the results are summarized in Figure 4.
FIG. 4.
Comparison of prediction accuracy between the DPM–HMM and DBN-torus DBN-torus has been given either predicted or real secondary structure information as input Small aRMSD values here given in radians indicate predictions which are close to the target’s true tertiary structure.
The DPM–HMM provides a better minimum aRMSD estimate than DBN-torus in 75/94 cases with predicted secondary structure information and 67/94 cases with true secondary structure information. Note that even under this best case scenario, the DPM–HMM provides better predictions than does DBN-torus. This is unsurprising, as template-based methods typically outperform de novo methods where a template is available. Proteins for which DBN-torus outperforms our DPM–HMM method often contain an EF loop whose conformation is not a close match to other members of the globin family. In such cases, good conformations are more likely to be sampled from DBN-torus, which is based on the entire PDB, rather than the DPM–HMM mimicking the behavior of the other globins.
6. Discussion
We have presented a novel model for protein torsion angle data that is capable of estimating the joint distribution of around 15 angle pairs simultaneously, and applied it to extend template-based modeling to the notoriously difficult loop and turn regions. In contrast to existing methods such as library-based loop prediction and DBN-torus, our model is designed to make use of only data from highly similar proteins, which gives us an advantage when such data is available. This is a significant advance in terms of statistical models for this type of data, as well as a new approach to template-based structure prediction. In addition to providing the basic model, we proposed two possible prior formulations with interesting properties.
Our noninformative prior model, which is the direct extension of the single position model of Lennox et al. (2009a, 2009b), provides a method to jointly model sequence positions which may or may not be adjacent in terms of a protein’s primary structure. This model allows for the estimation of joint and conditional distributions for multiple sequence positions, which permits the use of innovative methods to generate candidate distributions for protein structure.
While the noninformative prior model represents a significant advance over existing methods, we also present an alternative model that incorporates prior information about protein structure. This DPM–HMM model, which uses a hidden Markov model as the centering distribution for a Dirichlet process, uses the unique characteristics of a protein’s secondary structure to generate superior density estimates for torsion angles at sequential alignment positions. We use a Bayes factor analysis to demonstrate that density estimates generated with this model are closer to the true distribution of torsion angles in proteins than our alternative ignoring secondary structure.
Regardless of our prior formulation, the model is capable of accommodating the sparse data problem inherent in protein structural data, and in the case of the DPM–HMM formulation can leverage information at adjacent sequence positions to compensate for sparse data. This allows, for the first time, the extension of template-based modeling to the loop regions in proteins. We show that within family data provides superior results to conventional library and PDB-based loop modeling methods. As loop modeling is one of the critical problems in protein structure prediction, this new model and its ability to enhance knowledge-based structure prediction represents a significant contribution to this field.
Recall that our model treats the parameters of the bivariate von Mises sine model nonparametrically through the use of the Dirichlet process prior centered on a parametric distribution. We explored the effect of this treatment relative to the parametric alternative of using the centering distribution itself as the prior for the bivariate von Mises parameters. This parametric alternative is equivalent to limiting our model to a single mixture component. Although not every sequence position gives a strong indication of multiple mixture components, there is at least one such sequence position for every loop in our data set. (See, e.g., position 94 for the coil library data set in Figure 3.) Attempts to model this data using only a single component distribution lead to poor results, particularly since our model enforces unimodality for each component via the Wishart prior. While the HMM prior does allow for a mixture of bivariate von Mises distributions, all of these components will converge to the same distribution as the number of observations increases, effectively reducing us to a single component model again. The inadequacy of such a single component model is reflected in the strong preference of the data for multiple clusters. While the prior expected number of clusters goes to 1 as the mass parameter α0 goes to 0, we found that the posterior mean number of clusters only decreased by 1 (typically from 4 to 3) when α0 decreased from 1 to 10−10.
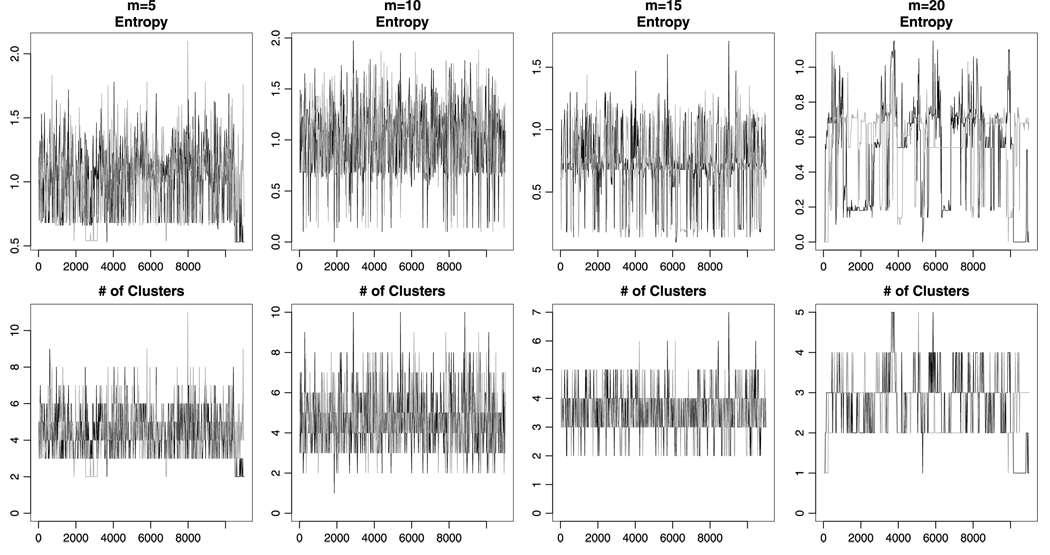
In working with our sampling schemes for both the DPM–HMM and noninformative prior models we did occasionally encounter slow mixing and convergence problems, particularly as the number of sequence positions under study increased. Figure 5 shows the effects on the total number of clusters and entropy [Green and Richardson (2001)] per iteration caused by increasing sequence length. As the number of positions under study increases, there is a greater chance of getting stuck in particular conformations, and also a subtler tendency toward having fewer observed clusters. Although in this example the effects are fairly mild, more severe issues can occur even at relatively short sequence lengths. However, even when problems appear to be evident on plots of standard convergence diagnostics, the density estimates generated by separate chains can be quite similar. For this reason we recommend comparing the density estimates generated by multiple chains in addition to the standard methods of diagnosing convergence problems.
FIG. 5.
Convergence diagnostics for density estimates using the noninformative prior model on the globin data with contiguous sequences beginning at position 93. Notice how mixing worsens as the number of sequence positions increases.
We do not recommend that our method be used for simultaneous modeling of more than about 15 sequence positions and convergence diagnostics should always be employed. The use of multiple MCMC chains with different starting configurations is also highly encouraged. Particular care should be taken with the noninformative prior model, which seems to be more prone to these sorts of problems. We did not observe any effect of sparse data on the speed of convergence or mixing.
Increases in sequence length and sample size both increase run time for our software, although sequence length is the primary practical restriction as protein families tend to have fewer than 100 members. For the analysis of the full globins data set with 5, 10, 15 or 20 sequence positions, the run times for two chains with 11,000 iterations using a 3 GHz processor were between 1 and 3.5 hours for the noninformative model and 2–8 hours for the DPM–HMM.
As the emphasis in this paper is on loop modeling, which by its very nature is limited to contiguous sequence positions, our application does not reflect the full extent of the flexibility of our model. Our general method is a good source of simultaneous continuous density estimates for large numbers of torsion angle pairs. This allows us to generate candidate models by sampling from joint distributions, or to propagate a perturbation of the torsion angle sequence at a single position up and down the chain through the use of conditional distributions. Our noninformative prior model, while less impressive than the DPM–HMM for contiguous sequence positions, can be applied to far richer classes of torsion angle sets. This allows the modeling of the behavior of tertiary structure motifs, which are composed of amino acids which are not adjacent in terms of primary structure, but which are in close contact in the natural folded state of a protein. It can even be used to investigate the structure of polypeptide complexes, as the (ϕ, ψ) positions modeled are not required to belong to the same amino acid chain. The ability to model large numbers of (ϕ, ψ) pairs simultaneously is an exciting advance which will offer new avenues of exploration for template-based modeling, even beyond the field of loop prediction.
The software used in this analysis is available for download at http://www.stat.tamu.edu/~dahl/software/cortorgles/.
Acknowledgments
The authors would like to thank J. Bradley Holmes, Jerod Parsons and Kun Wu for help with data sets, alignments, and the torsion angle calculations. We would also like to thank the editor, associate editor and referees for their helpful comments.
APPENDIX A: MARKOV CHAIN MONTE CARLO
Here we give the details of our MCMC scheme to sample from the posterior distribution. A concise description is provided in Table 3. After the state of our Markov chain has been initialized, our first step is to update the clustering associated with our Dirichlet process. We use the Auxiliary Gibbs sampler of Neal (2000) with one auxiliary component for this purpose. Having updated the clustering, we now must update the parameter values θ for each cluster by drawing values from full conditional distribution f(θ|xc), where xc = {xi : i ∈ c} and c is the set of indices for members of said cluster. Once again, this distribution is difficult to sample from directly, so we update instead using the full conditional distributions f(µ, ν|Ω, xc) and f(Ω|µ, ν, xc).
TABLE 3.
Computational procedure
|
In the case of the precision parameters Ω, the full conditional density cannot be written in closed form, but is generally well approximated by the Wishart full conditional distribution that results from the assumption that the data have a bivariate normal distribution rather than a bivariate von Mises distribution. We update Ω by implementing an independence sampler that uses this “equivalent” Wishart distribution as its proposal distribution at each sequence position. Note that under our model, the full conditional distribution of Ω does not depend on the choice of centering distribution of the mean parameters. The full conditional is proportional to
| (A.1) |
where h2 is our component Wishart prior for a single sequence position, and f is a bivariate von Mises sine model with the relevant parameters. Notice that the positions are independent given the clustering information, so it is trivial to update each Ωj separately.
After updating the precision parameters at each sequence position, we proceed to update μ and ν using an independence sampler. For our noninformative prior, with a centering distribution consisting of a single sine model, we use the update method described in Lennox et al. (2009a). In this case, with H1 = (h1)n where h1 is a bivariate von Mises distribution, the full conditional distribution is proportional to
| (A.2) |
The DPM–HMM case where H1 is defined to be a hidden Markov model is somewhat more complicated. The positions are no longer a priori, and therefore a posteriori, independent given the clustering information. In addition, the inclusion of an HMM in the model makes the nature of the full conditional distribution unclear. However, if the state chain s is known, draws from the full conditional are trivial. Therefore, we rewrite our full conditional distribution, which is proportional to
| (A.3) |
where f(μ, ν|sj) is the prior distribution determined by the state at position j. Recall that our priors are finite mixtures of bivariate von Mises sine distributions. Thus, if we can generate draws from the full conditional distribution of s, we can update μi and νi at each sequence position much as we did before. We use the forward–backward (FB) algorithm of Chib (1996) to sample the full conditional distribution of s. Note that s given μ and ν is independent of the data. Once we have the state information, generating samples from the distributions μj, νj|sj, Ωj, xcj is a straightforward process using an independence sampler, the details for which are given in Appendix B.
APPENDIX B: VON MISES MIXTURE PRIORS
We present the full conditional distribution of the mean parameters μ and ν given that the precision matrix Ω. is known and the prior is a single bivariate von Mises distribution with parameters μ0, ν0, κ10, κ20 and λ0. Using this information, we then prove that a finite mixture of bivariate von Mises distributions is a conditionally conjugate prior for this model, and present a finite mixture of sine models which serves as a good proposal distribution.
We consider now a single sequence position, and so our data set consists of the set . The full conditional distribution for a set of observations with bivariate von Mises sine model distributions and a sine model prior is an eight parameter bivariate von Mises distribution. Lennox et al. (2009a) showed that this distribution could be represented as
with parameters
| (B.1) |
where C is the appropriate constant of integration and the prior mean parameters (μ0, ν0) are treated as an additional observation (ϕ0, ψ0) from a bivariate von Mises sine model with parameters μ, ν, κ10, κ20 and λ0.
Now consider a prior distribution of the form
where Ck is the constant of integration for a von Mises sine model with parameters κ10k, κ20k and λ0k given in equation (2.2), pk ≥ 0 for k = 1,…,K and . The full conditional distribution is proportional to this distribution times the likelihood, giving
where L(μ, ν|ϕ, ψ) is the likelihood excluding the constant of integration.
Each term in the sum depends on the unknown parameters only through the product of the likelihood and a single von Mises sine distribution. This product is proportional to an eight parameter bivariate von Mises distribution with parameters given by (B.1). Call the resulting posterior parameters μ͂i, ν͂i and so on. Then the full conditional distribution is proportional to
which integrates to
where C͂k is the constant of integration for an eight parameter bivariate von Mises distribution with parameters μ͂k, ν͂k, κ͂1k, κ͂2k and λ͂k. Therefore, the full conditional distribution takes the form
where f is an eight parameter bivariate von Mises distribution and . Note that for k = 1,…,K, and .
Unfortunately computational formulas for the constant of integration of a bivariate von Mises distribution do not exist in the general case. Therefore, we do not sample directly from this full conditional distribution, but rather use an independence sampler which replaces each full conditional eight parameter distribution with a five parameter sine model, and uses the corresponding constant of integration from (2.2). This is accomplished by replacing the four parameter à with a . [This method is a direct extension of the single sine model prior case presented in Lennox et al. (2009a).] Using this sampler, we found mean and median acceptance rates around 0.52, which was comparable to the acceptance rates for the single sine model noninformative prior, which were around 0.55.
Footnotes
Supported in part by NIH/NIGMS Grant R01GM81631.
Supported in part by NIH/NHGRI Grant R01HG003319 and by NSF/DMS Grant DMS-06-05001.
Contributor Information
Kristin P. Lennox, Department of Statistics, Texas A&M University, 3143 TAMU, College Station, Texas 77843-3143, USA, lennox@stat.tamu.edu
David B. Dahl, Department of Statistics, Texas A&M University, 3143 TAMU, College Station, Texas 77843-3143, USA, dahl@stat.tamu.edu
Marina Vannucci, Department of Statistics, Rice University, MS 138, Houston, Texas 77251-1892, USA, marina@rice.edu.
Ryan Day, Department of Chemistry, University of the Pacific, 3601 Pacific Ave, Stockton, California 95211-0110, USA, rday@pacific.edu.
Jerry W. Tsai, Department of Chemistry, University of the Pacific, 3601 Pacific Ave, Stockton, California 95211-0110, USA, jtsai@pacific.edu
REFERENCES
- Antoniak CE. Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. Ann. Statist. 1974;2:1152–1174. MR0365969. [Google Scholar]
- Baker D, Sali A. Protein structure prediction and structural genomics. Science. 2001;294:93–96. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- Beal MJ, Ghahramani Z, Rasmussen CE. The infinite hidden Markov model. In: Dietterich T, Becker S, Ghahramani Z, editors. Advances in Neural Information Processing Systems 14. Cambridge, MA: MIT Press; 2002. p. 504.p. 505.p. 508. [Google Scholar]
- Bernardo JM, Smith AFM. Bayesian Theory. Chichester: Wiley; 1994. MR1274699. [Google Scholar]
- Bonneau R, Baker D. Ab initio protein structure prediction: Progress and prospects. Annu. Rev. Biophys. Biomol. Struct. 2001;30:173–189. doi: 10.1146/annurev.biophys.30.1.173. [DOI] [PubMed] [Google Scholar]
- Boomsma W, Mardia KV, Taylor CC, Ferkinghoff-Borg J, Krogh A, Hamelryck T. A generative, probabilistic model of local protein structure. Proc. Natl. Acad. Sci. USA. 2008;105:8932–8937. doi: 10.1073/pnas.0801715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfoss GL, Richardson JS, Hermans J. Protein imperfections: Separating intrinsic from extrinsic variation of torsion angles. Acta Crystallogr. D Biol. Crystallogr. 2005;61:88–98. doi: 10.1107/S0907444904027325. [DOI] [PubMed] [Google Scholar]
- Chib S. Calculating posterior distributions and modal estimates in Markov mixture models. J. Econometrics. 1996;75:79–97. MR1414504. [Google Scholar]
- De Iorio M, Müller P, Rosner GL, MacEachern SN. An ANOVA model for dependent random measures. J. Amer. Statist. Assoc. 2004;99:205–215. MR2054299. [Google Scholar]
- Dunson DB, Pillai N, Park J-H. Bayesian density regression. J. Roy. Statist. Soc. Ser. B Statist. Methodol. 2007;69:163–183. MR2325270. [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MD, West M. Bayesian density estimation and inference using mixtures. J. Amer. Statist. Assoc. 1995;90:577–588. MR1340510. [Google Scholar]
- Ferguson TS. A Bayesian analysis of some nonparametric problems. Ann. Statist. 1973;1:209–230. MR0350949. [Google Scholar]
- Fitzkee NC, Fleming PJ, Rose GD. The protein coil library: A structural database of nonhelix, nonstrand fragments derived from the PDB. Proteins. 2005;58:852–854. doi: 10.1002/prot.20394. [DOI] [PubMed] [Google Scholar]
- Gelfand AE, Kottas A, MacEachern SN. Bayesian nonparametric spatial modeling with Dirichlet process mixing. J. Amer. Statist. Assoc. 2005;100:1021–1035. MR2201028. [Google Scholar]
- Green PJ, Richardson S. Modelling heterogeneity with and without the Dirichlet process. Scand. J. Statist. 2001;28:355–375. MR1842255. [Google Scholar]
- Griffin JE, Steel MFJ. Order-based dependent Dirichlet processes. J. Amer. Statist. Assoc. 2006;101:179–194. MR2268037. [Google Scholar]
- Ho BK, Thomas A, Brasseur R. Revisiting the Ramachandran plot: Hard-sphere repulsion, electrostatics, and h-bonding in the alpha-helix. Protein Sci. 2003;12:2508–2522. doi: 10.1110/ps.03235203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Karplus K, Sjolander K, Barrett C, Cline M, Haussler D, Hughey R, Holm L, Sander C, England E. Predicting protein structure using hidden Markov models. Proteins: Structure Function and Genetics. 1997;29:134–139. doi: 10.1002/(sici)1097-0134(1997)1+<134::aid-prot18>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. J. Amer. Statist. Assoc. 1995;90:773–795. [Google Scholar]
- Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KP, Dahl DB, Vannucci M, Tsai JW. Correction to density estimation for protein conformation angles using a bivariate von Mises distribution and Bayesian nonparametrics. J. Amer. Statist. Assoc. 2009a;104:1728. doi: 10.1198/jasa.2009.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KP, Dahl DB, Vannucci M, Tsai JW. Density estimation for protein conformation angles using a bivariate von Mises distribution and Bayesian nonparametrics. J. Amer. Statist. Assoc. 2009b;104:586–596. doi: 10.1198/jasa.2009.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WBR, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: Phi, Psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- MacEachern SN. Dept. Statistics, Ohio State Univ.; Dependent Dirichlet processes. Technical report. 2000
- Mardia KV. Statistics of directional data (com: P371–392) J. Roy. Statist. Soc. Ser. B. 1975;37:349–371. MR0402998. [Google Scholar]
- Mardia KV, Hughes G, Taylor CC, Singh H. A multivariate von Mises distribution with applications to bioinformatics. Canadian J. Statist. 2008;36:99–109. MR2432195. [Google Scholar]
- Mardia KV, Taylor CC, Subramaniam GK. Protein bioinformatics and mixtures of bivariate von Mises distributions for angular data. Biometrics. 2007;63:505–512. doi: 10.1111/j.1541-0420.2006.00682.x. MR2370809. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones TD. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Michalsky E, Goede A, Preissner R. Loops in proteins (LIP)—a comprehensive loop database for homology modeling. Prot. Eng. 2003;16:979–985. doi: 10.1093/protein/gzg119. [DOI] [PubMed] [Google Scholar]
- Neal RM. Markov chain sampling methods for Dirichlet process mixture models. J. Comput. Graph. Statist. 2000;9:249–265. MR1823804. [Google Scholar]
- Osguthorpe DJ. Ab initio protein folding. Curr. Opin. Struct. Biol. 2000;10:146–152. doi: 10.1016/s0959-440x(00)00067-1. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Ramakrishnan C, Sasisekharan V. Stereochemistry of polypeptide chain configurations. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Rivest LP. Some statistical methods for bivariate circular data. J. Roy. Statist. Soc. Ser. B. 1982;44:81–90. MR0655377. [Google Scholar]
- Rodríguez A, Dunson DB, Gelfand AE. The nested Dirichlet process. J. Amer. Statist. Assoc. 2008;103:1131–1144. [Google Scholar]
- Scott SL. Bayesian methods for hidden Markov models: Recursive computing in the 21st century. J. Amer. Statist. Assoc. 2002;97:337–351. MR1963393. [Google Scholar]
- Sethuraman J. A constructive definition of Dirichlet priors. Statist. Sinica. 1994;4:639–650. MR1309433. [Google Scholar]
- Singh H, Hnizdo V, Demchuk E. Probabilistic model for two dependent circular variables. Biometrika. 2002;89:719–723. MR1929175. [Google Scholar]
- Teh YW, Jordan MI, Beal MJ, Blei DM. Hierarchical Dirichlet processes. J. Amer. Statist. Assoc. 2006;101:1566–1581. MR2279480. [Google Scholar]
- Xing EP, Sohn KA. Hidden Markov Dirichlet process: Modeling genetic inference in open ancestral space. Bayesian Anal. 2007;2:501–528. MR2342173. [Google Scholar]