Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis
Chemotaxis in bacteria is transduced by ligands that either directly or indirectly bind transmembrane receptors. This work reveals that the nature of ligand binding, rather than overall receptor abundance is the physiological determinant of chemotaxis.
Keywords: bacterial chemotaxis, FRET, signal integration, signal transduction, transport
Abstract
In chemotaxis of Escherichia coli and other bacteria, extracellular stimuli are perceived by transmembrane receptors that bind their ligands either directly, or indirectly through periplasmic-binding proteins (BPs). As BPs are also involved in ligand uptake, they provide a link between chemotaxis and nutrient utilization by cells. However, signalling by indirectly binding ligands remains much less understood than signalling by directly binding ligands. Here, we compared intracellular responses mediated by both types of ligands and developed a new mathematical model for signalling by indirectly binding ligands. We show that indirect binding allows cells to better control sensitivity to specific ligands in response to their nutrient environment and to coordinate chemotaxis with ligand transport, but at the cost of the dynamic range being much narrower than for directly binding ligands. We further demonstrate that signal integration by the chemosensory complexes does not depend on the type of ligand. Overall, our data suggest that the distinction between signalling by directly and indirectly binding ligands is more physiologically important than the traditional distinction between high- and low-abundance receptors.
Introduction
Chemotactic bacteria follow chemical gradients in their environment by performing temporal comparisons of chemoeffector concentrations (Berg and Brown, 1972; Macnab and Koshland, 1972). In Escherichia coli, sensing and processing of stimuli are performed by complexes that consist of four types of attractant-specific chemoreceptors, a histidine kinase CheA, and an adaptor protein CheW (Gegner et al, 1992). Attractant binding to the periplasmic part of a receptor homodimer, the functional chemoreceptor unit—hereafter simply referred to as a ‘receptor'—inhibits CheA autophosphorylation, thus reducing phosphotransfer to the motor regulator CheY. The signal transduction pathway also includes a phosphatase CheZ, and an adaptation system that consists of a methyltransferase, CheR, and a methylesterase, CheB. Adaptation to a persistent attractant stimulus works through receptor methylation on four specific glutamate residues, which increases CheA activity and decreases sensitivity to the attractant (Borkovich et al, 1992; Li and Weis, 2000; Levit and Stock, 2002).
Although all E. coli chemoreceptors are homologous, two classes of ligands can be distinguished based on their binding properties. Amino acids bind directly to the periplasmic domains of the so-called major receptors Tar and Tsr, and constitute one class. A second class is represented by sugars and dipeptides, which bind to minor receptors Trg and Tap, respectively, through periplasmic substrate-binding proteins (BPs) of ATP-binding cassette (ABC) transporters. An interesting exception is the sugar maltose, which binds indirectly to the major receptor Tar (Hazelbauer, 1975; Manson and Kossmann, 1986). Early studies of chemotaxis that relied on accumulation of bacteria into attractant-filled microcapillaries already indicated differences in the magnitude and concentration range of responses mediated by the two types of ligands (Mesibov and Adler, 1972; Adler et al, 1973; Mesibov et al, 1973), but the origin of those differences remained largely unclear.
Apart from the mode of ligand binding, major and minor receptors differ in their copy numbers, with the total number of minor receptors previously estimated to be <1000 copies per cell compared with ∼10 000 for major receptors (Li and Hazelbauer, 2004), and this difference was frequently assumed to explain the apparently weaker responses mediated by minor receptors. Yet another difference lies in the truncation for the minor receptors of the last 20 amino acids of the C-terminus, which includes the binding site for the adaptation enzymes, making minor receptors dependent on the proximity of neighbouring major receptors for efficient adaptation (Li and Hazelbauer, 2005).
The receptor–kinase sensory complexes are organized in large clusters that localize at the cell poles and in smaller clusters along the cell body (Maddock and Shapiro, 1993; Sourjik and Berg, 2000), where receptors of different ligand specificities are intermixed (Ames et al, 2002). Activities of neighbouring receptors are allosterically coupled, resulting in amplification of receptor-mediated stimuli (Li and Weis, 2000; Gestwicki and Kiessling, 2002; Sourjik and Berg, 2002, 2004; Lai et al, 2005), and thus allowing E. coli to generate highly sensitive responses that approach physical limits of sensitivity (Berg and Purcell, 1977; Bialek and Setayeshgar, 2005). The allosteric interactions between receptors have been described using a number of mathematical models (Bray et al, 1998; Duke et al, 2001; Sourjik and Berg, 2004; Mello and Tu, 2005; Keymer et al, 2006). The most commonly applied are Monod–Wyman–Changeux type (MWC) models, which assume that receptors within clusters exist in tightly coupled allosteric signalling complexes, or teams, of 10–20 receptors, with all receptors in one team switching synchronously between inactive (off) and active (on) states (Sourjik and Berg, 2004; Mello and Tu, 2005; Keymer et al, 2006). As receptors of different types are intermixed within clusters (Ames et al, 2002), allosteric interactions between receptors also provide a means of signal integration (Sourjik and Berg, 2004).
So far, most of the effort to investigate signal processing by allosteric chemoreceptor teams has concentrated on the directly binding ligands of the major receptors Tar and Tsr, L-aspartate (or its non-metabolizable analogue α-methyl-DL-aspartate, MeAsp) and L-serine (or its non-metabolizable analogue α-aminoisobutyrate, AiBu), respectively. The goal of this work was to systematically compare intracellular signalling by the directly binding ligands of major receptors with that mediated by the indirectly binding sugars D-maltose (specific for Tar), D-ribose and D-galactose (specific for Trg), and the dipeptide Pro-Leu (specific for Tap). To quantify signalling parameters of individual ligands, we determined relative expression levels of the corresponding receptors and extended the MWC model of allosteric teams to describe indirect ligand binding through BPs. Our results show that the signalling strength and dynamic range of a specific ligand are determined both by its binding properties and by the expression levels of its receptor and the respective BP. We conclude that the indirect mode of ligand binding allows greater flexibility in the modulation of response sensitivity, but at a cost of a narrower dynamic range. Chemotactic response to mixtures of effectors showed that simultaneous stimulation by ligands of different receptors is additive, and that adaptation to ligands of one receptor does not interfere with signalling by other receptors. On the basis of these and other data, we discuss possible evolutionary origins of the two modes of ligand binding in relation to strategies of coupling between chemotaxis and transport.
Results
Responses towards directly and indirectly binding ligands
To measure intracellular responses, we used a FRET reporter based on the CheY-YFP/CheZ-CFP pair (Sourjik and Berg, 2002; Sourjik et al, 2007). The FRET assay determines the relative intracellular level of the complex formed by phosphorylated CheY-YFP and CheZ-CFP. As the amount of this complex is proportional to the rate of CheY phosphorylation, it provides a direct readout of the intracellular kinase activity (Supplementary Figure S1). It has been shown before that CheY-YFP/CheZ-CFP FRET changes linearly with the kinase activity within the physiological activity range (Sourjik and Berg, 2002; Sourjik et al, 2007; Endres et al, 2008). As wild type for our experiments, we used a cheY cheZ derivative of E. coli K-12 strain LJ110 (Zeppenfeld et al, 2000) whose parent W3110 was used for early studies on sugar taxis (Hazelbauer et al, 1969; Adler et al, 1973). This strain shows saturating expression of galactose BP (GBP) (see below), which makes it convenient for comparative study of amino acid and sugar taxis.
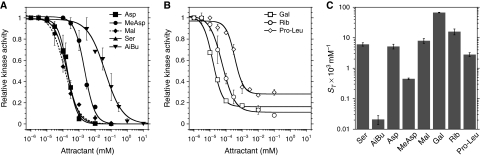
When pre-adapted in buffer before stimulation, wild-type cells responded to the natural ligands of major and minor receptors in a similar concentration range, with EC50 values—the ligand concentrations at the half-maximal response—being 193 and 126 nM for the Tar ligands aspartate and maltose, respectively, 165 nM for the Tsr ligand serine, 15 and 63 nM for the Trg ligands galactose and ribose, respectively, and 356 nM for the dipeptide Pro-Leu, a ligand of Tap (Figure 1A and B). Lower sensitivity was observed for the response to the non-metabolizable ligands of Tar (MeAsp) and of Tsr (AiBu), with EC50 values of 2.2 and 48 μM, respectively. Response amplitudes to saturating stimuli of all ligands were similar, although a residual activity of 10–30% of the pre-stimulus level was observed in cells saturated with minor receptor ligands (Figure 1B). The resulting threshold sensitivity ST, defined as EC50−1, which reflects the lower limit of the concentration range that can be sensed by chemotactic cells, was highest for sugars and only slightly lower for dipeptides and natural amino acids (Figure 1C; Supplementary Table SI).
Figure 1.
Response of major and minor receptors to attractants. Dose responses of wild-type cells to ligands of major (A) and minor receptors (B). Cells were stimulated by step-like addition of increasing amounts of attractant. The following abbreviations are used here and throughout: Asp, L-aspartate; MeAsp, α-methyl-DL-aspartate; Ser, L-serine; AiBu, α-aminoisobutyrate; Mal, maltose; Gal, galactose; Rib, ribose; Pro-Leu, proline-leucine dipeptide. Resulting initial changes in kinase activity were measured using a FRET activity reporter (see main text and Materials and methods). After each stimulation, the cells were re-adapted to buffer. The response for each step was normalized to the response of buffer-adapted cells towards a saturating stimulus of 100 μM MeAsp. (C) Threshold sensitivity ST, determined as EC50−1 from Hill fits to the dose-response curves of individual experiments. Error bars indicate standard errors.
To minimize the effect of ligand depletion by cells at low-ligand concentrations, these experiments were performed at the highest flow rate possible in our setup, whereby the content of the entire flow chamber was constantly exchanged nearly every second (see Materials and methods). Although such a high flow rate generally increased measurement errors, it resulted in a significant increase in response amplitudes to the lowest concentrations of some ligands when compared with a five-fold lower flow rate (Supplementary Figure S2), and led to modestly higher estimates of the effective threshold sensitivity (Supplementary Table SI). Such reduction of the response threshold by ligand uptake is consistent with previous observations made in capillary assays (Ordal and Adler, 1974; Hazelbauer, 1975; Zhang et al, 1999), and its extent was ligand-dependent as expected from differences in the rates of ligand uptake. Only minor effects were observed for aspartate or galactose, whereas depletion effects for serine or ribose were substantially larger. However, in all cases, the effective reduction in ligand concentration was on the order of EC50, meaning that ligand depletion should not affect the response behaviour at higher ligand concentrations.
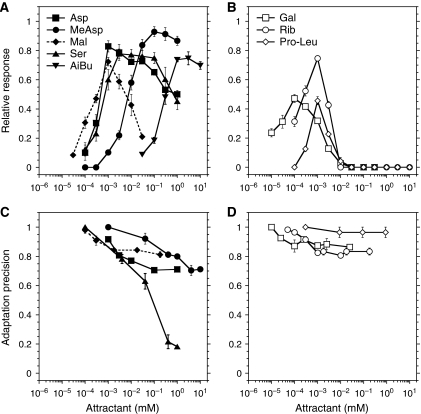
Dynamic range of response
The dynamic range over which a sensor can discriminate stimuli of varying strengths is an important property of any sensory system. We determined the dynamic range for a particular ligand by increasing its concentration in approximately three-fold steps (Figure 2A and B; Supplementary Figure S3A and B). In contrast to dose-response measurements, cells in these experiments were allowed to adapt to the current ambient concentration before being stimulated with a next step of increased concentration. Responses to directly binding ligands showed a wide dynamic range and large response amplitudes across this range (Figure 2A). In most of the range, each three-fold increase in aspartate, MeAsp, serine, or AiBu concentration elicited a saturating response, bringing the kinase activity initially close to zero. In contrast, the Tar-mediated response to the indirectly binding ligand maltose was limited to a relatively narrow concentration range, covering only 2–3 orders of magnitude of attractant concentration (Figure 2A), and the dynamic ranges of the responses mediated by indirectly binding ligands of minor receptors were even narrower (Figure 2B). The measured dynamic range may be modestly narrowed for some directly and indirectly binding ligands by their depletion, to the same extent as in the dose-response measurements (Supplementary Table SI; Supplementary Figure S2). However, the effects of depletion are limited to very low ligand concentrations and should neither affect the height at the peaks nor the overall shapes of the dynamic range curves.
Figure 2.
Dynamic range and adaptation precision. Dynamic range (A, B) and adaptation precision (C, D) for major (A, C) and minor (B, D) receptors. For dynamic range measurements, concentrations were raised in three-fold steps, and the cells were allowed to adapt prior to each subsequent stimulation. The response for each step was calculated relative to the response towards a saturating stimulus (100 μM MeAsp for buffer-adapted cells) and is plotted against the final concentration of ligand. Precision of adaptation was defined as the adapted FRET value in the presence of a given concentration of ambient ligand, normalized to the adapted value of FRET in buffer. In these experiments, duration of adaptation was limited to 45 min. Error bars indicate standard errors.
In the high-concentration regime, the dynamic range of the directly binding ligands was apparently limited by the gradually decreasing value of the adapted kinase activity, that is by the imprecision of adaptation (Figure 2A and C; Supplementary Figure S3A and E). The imprecision of adaptation was particularly pronounced for serine, consistent with previous observations (Berg and Brown, 1972). In contrast, adaptation to the indirectly binding ligand maltose (Figure 2C) and minor-receptor ligands (Figure 2D; Supplementary Figure S3B) was relatively precise, indicating that the dynamic range of these ligands was limited by saturation of the sensory system rather than by failure of adaptation, as illustrated in Supplementary Figure S3C–F.
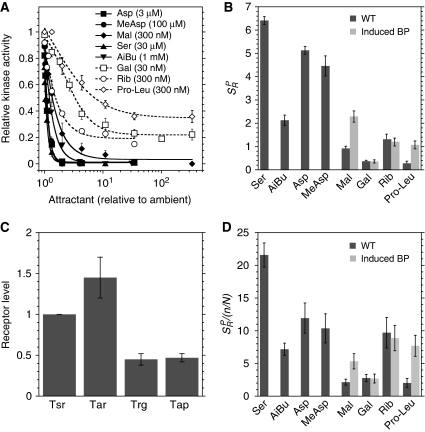
Response sensitivity
Another apparent difference between ligands was in the response amplitude at the peak of the dynamic range, which reflects the response sensitivity SR to relative changes in concentration of a particular ligand, defined as the ratio between the fractional change in activity (ΔA/A) and the fractional change in ligand concentration (ΔL/L). The value of response sensitivity at the peak of the dynamic range, SRP, could in principle be estimated from our dynamic range measurements (Figure 2A and B), but such direct estimation of SRP was rather imprecise, and not possible for the directly binding ligands because the three-fold concentration steps were already saturating for these ligands. To determine SRP more precisely, dose-response curves were acquired for cells that were pre-adapted to ambient concentrations around the peak of the dynamic range. Plotting these dose-response curves against the relative ligand concentration suggests that indirectly binding ligands are on average less potent in inactivating allosteric receptor teams in wild-type cells (Figure 3A and B). However, this difference was reduced by the overexpression of BPs from salicylate-inducible expression constructs in the wild-type background (Figure 3B).
Figure 3.
Response sensitivity of major and minor receptors. (A) Dose-response curves for cells that were pre-adapted to an ambient concentration of the respective ligand around the peak of the dynamic range (shown in brackets). Kinase activity is plotted as a function of final ligand concentration, normalized to the respective ambient concentration. (B) Response sensitivity at the peak of the dynamic range, SRP, for WT and otherwise wild-type cells each expressing a plasmid-encoded periplasmic binding protein (BP) at maximum induction of 10 μM salicylate. See text for the definition of SRP. (C) Native expression levels of receptors in wild-type cells normalized to Tsr, determined by immunoblot as described in Materials and methods and in Supplementary Figure S4. (D) SRP values from (B) normalized to the receptor fraction. Error bars indicate standard errors.
Quantification of receptor levels
Allosteric models of signal processing by the receptor complexes predict that the response mediated by a particular ligand depends on the number n of specific receptors in a typical signalling team of overall size N. The observed differences in SRP and dynamic range might thus be partly due to differences in expression of specific receptors. We therefore quantified the relative cellular levels of all four chemoreceptors by immunoblot analyses, using antibodies raised against signalling domains of Tar and Trg. Because of the high-sequence homology of the signalling domains, these antibodies recognize all chemoreceptors, but with varying specificity. To calibrate the relative antibody specificity for individual receptors, we compared signal intensities in immunoblots with defined amounts of receptor fusions to YFP, adjusted using both the YFP fluorescence and immunoblotting with GFP-specific antibody that equally well recognizes YFP (see Supplementary Figure S4, Materials and methods, and Supplementary data). The αTar antibody was found to recognize Tar and Tsr with comparable specificity (Tar/TsrαTar: 1.39±0.23), and was therefore used to determine the relative levels of these receptors. The αTrg antibody has similar specificity for minor receptors and Tsr (Trg/TsrαTrg: 1.64±0.13; Tap/TsrαTrg: 1.23±0.08) and was used for the relative quantification of the minor receptors.
Our results showed that Tap and Trg are less abundant than Tar and Tsr, but in contrast to previous estimates (Li and Hazelbauer, 2004), we only observed a two- to three-fold difference in their levels (Figure 3C; Supplementary Figure S4D). The absolute expression level of Tsr was estimated to be around 6000 copies per cell (Supplementary Figure S5). Comparable receptor ratios were observed for the more conventionally used chemotaxis strain RP437 (Supplementary Figure S4E). It should be noted that the ratio between the levels of major receptors is known to depend on the optical density of the culture (Salman and Libchaber, 2007; Kalinin et al, 2010), and we demonstrated a similarly steep dependence for the relative levels of Trg and Tap to Tsr (Supplementary Figure S6). Nevertheless, as the same growth conditions were used to evaluate receptor levels and response parameters in our experiments, we could use the obtained relative receptor amounts to normalize the response sensitivity for respective ligands. Assuming that the relative expression levels reflect the receptor fraction in a signalling team (n/N), we can normalize SRP by that fraction to obtain values that are independent of n and thus directly reflect the signalling properties of individual ligands (Figure 3D). Such normalization confirmed that minor and major receptor ligands can signal with similar strengths, although some indirectly binding ligands showed weaker responses even after normalization. Surprisingly, sensitivity of the response to serine was markedly higher than for other directly binding ligands, including the Tsr ligand AiBu. This could not be explained solely by binding properties of serine to Tsr, but was apparently due to a Tsr-independent response to serine in the range of concentrations used to determine SRP (Supplementary Figure S7). This Tsr-independent response was also observed in a Tar-only strain (data not shown), indicating that the response may be mediated by Tar, but its exact nature remains to be investigated.
Dependence of response parameters on expression levels of periplasmic BPs and receptors
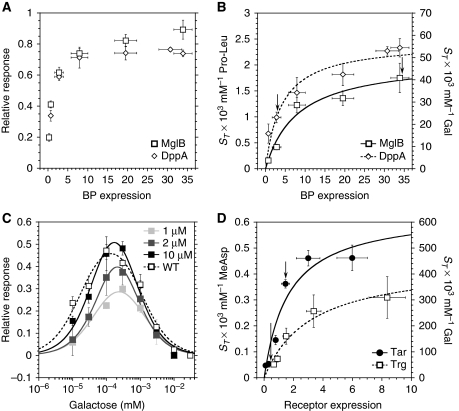
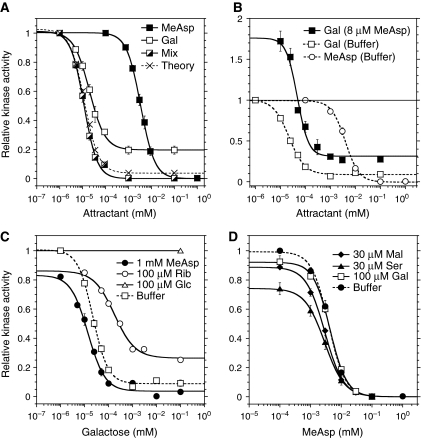
We observed that the response to indirectly binding ligands strongly depends on expression levels of BPs. Raising levels of BPs specific for galactose (MglB) and Pro-Leu (DppA) through expression from a salicylate-inducible plasmid construct in mutants deleted for the respective endogenous BPs gradually increased response amplitude (Figure 4A) and ST (Figure 4B) for the corresponding ligands. Consistent with that, an increase in ST for all indirectly binding ligands except galactose was also observed upon overexpression of the respective BPs in the wild-type background (Supplementary Table SI). Expression of the BPs also affected the values of SRP (Figures 3B, 3D, and 4C; Supplementary Figure S8A–C; and Supplementary Table SI), whereby the height of the peak of the dynamic range increased with [BP] and the position of the peak shifted slightly towards lower ligand concentrations, as predicted by our mathematical model of signalling by indirectly binding ligands (see below). The value of ST (Figure 4B) and the dynamic range (Figure 4C) for galactose in wild-type cells, as well as the results of BP overexpression in WT cells (Supplementary Table SI), confirm that the GBP (or MglB) is fully induced under our growth conditions, whereas BPs for dipeptides, ribose, and maltose are not.
Figure 4.
Dependence of chemotactic response on binding protein (BP) and receptor expression. (A) Response amplitudes to saturating stimuli of dipeptide Pro-Leu at varying levels of DppA or to galactose at varying levels of MglB. (B) Threshold sensitivity ST of the response to the dipeptide Pro-Leu at varying levels of DppA or to galactose at varying levels of MglB. Arrows mark the native BP expression level estimated from ST of wild-type cells. (A, B) Dipeptide BP DppA was expressed at different levels of salicylate induction in a ΔdppA strain, and the galactose BP MglB was expressed in a ΔmglB strain. Expression of BPs was assumed to be proportional to the expression of yellow fluorescent protein (YFP) from the same promoter, measured by FACS and used for the X axis. The data were fitted using the function ST=STmax[BP]/(C+[BP]), where C is a constant (see equation (3)). (C) Dynamic range of the response to galactose at varying expression of MglB. The three highest expression levels in the ΔmglB background from (A) are shown, with darker grey levels corresponding to higher expression. The wild-type dynamic range is shown for comparison. Data were fitted using the function SR=C1C2[L]/((C2+[L])([L]+C3)), where C1, C2, and C3 are constants (see equation S18 in Supplementary data). (D) Threshold sensitivity of the response to MeAsp at varying levels of Tar or to galactose at varying levels of Trg. Tar was expressed at different levels of salicylate induction in a Δtar strain; Trg was expressed in the wild-type strain. Receptor expression levels were determined as in Figure 3C and normalized to the native expression of Tsr. Data were fitted using the function ST=STmax[R]/([R]+C), where C is a constant describing the expression level of all other receptors. Response amplitudes were nearly saturating at all measured expression levels. Arrows mark the native expression levels of Trg and Tar. Error bars indicate standard errors.
Similarly, increased expression of Tar and Trg resulted in a gradual increase in ST for MeAsp and galactose, respectively (Figure 4D). Receptor overexpression further led to an expansion of the dynamic range and an increased SRP, as illustrated for Trg (Supplementary Figure S8D). The dependence of ST on the level of receptor expression can be fitted well by assuming simple scaling with the fraction of that particular receptor in the total receptor pool (Figure 4D). Best-fit values for the total receptor level relative to Tsr yield estimates of 3.1±0.8 and 3.6±1.2 for Tar and Trg titrations, respectively, consistent with the estimate obtained by immunoblotting (3.4±0.3; Supplementary Table SI). The similarity of titration curves for Tar and Trg confirms that major and minor receptors behave essentially identically in allosteric complexes.
Mathematical model of minor receptor signalling
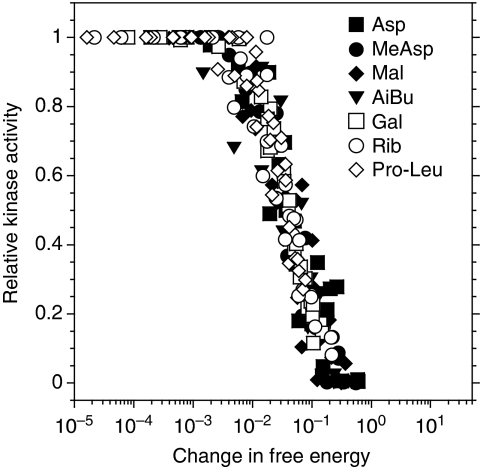
The allosteric MWC model for receptors predicts that a tightly coupled team of receptors turns on or off as a whole, with the activity A, that is the probability of being on, determined solely by the free-energy difference between the on and off states summed over all receptors in the team, 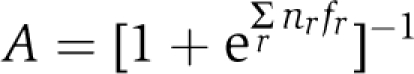 , where fr is the free-energy difference for receptors of type r (with energies expressed in units of the thermal energy kBT), and nr is the number of such receptors in the team (Mello and Tu, 2005; Endres and Wingreen, 2006; Keymer et al, 2006; Hansen et al, 2008). For receptors that directly bind ligand, with [L] as the ligand concentration,
, where fr is the free-energy difference for receptors of type r (with energies expressed in units of the thermal energy kBT), and nr is the number of such receptors in the team (Mello and Tu, 2005; Endres and Wingreen, 2006; Keymer et al, 2006; Hansen et al, 2008). For receptors that directly bind ligand, with [L] as the ligand concentration,
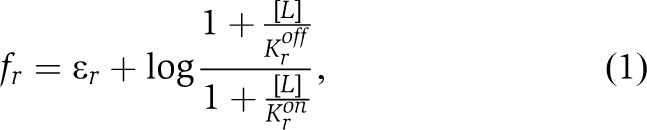 |
where Kron and Kroff are the binding constants in the on and off states for a specific type of receptor r and ɛr is the offset energy in the absence of ligand. For receptors that indirectly bind ligand, with [BP] the BP concentration,
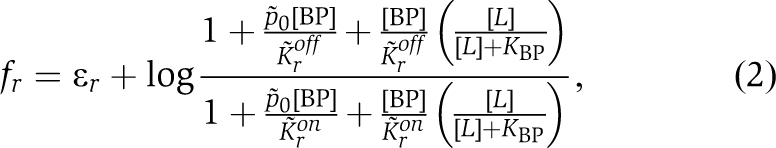 |
where K̃ron and K̃roff are binding constants of an on and off receptor to the closed BP, p̃0 reflects the proportion of closed BP in the absence of ligand, and KBP is a binding constant of BP to ligand (see Supplementary data for full derivations). As precise adaptation returns the total free-energy difference to a fixed value, the response of activity to a change in ligand concentration depends only on the resulting free-energy change. Therefore, experimental dose-response curves should collapse when plotted as a function of this free-energy change. For different receptor types, the free-energy change must be weighted by the receptor proportion to yield an average, 〈δf〉=(nr/N)δfr, where nr is the number of receptors of type r in a team of N receptors. Using the receptor ratios from Figure 3C, we calculated the change in free energy upon addition of more ligand to cells adapted to ambient ligand concentration, and from the best-data collapse obtained the binding parameters given in Supplementary data. The results for WT cells (Figure 5) prove the ability of the model to account for responses of both kinds of ligands. From the free-energy expressions, both the threshold sensitivity ST and the response sensitivity SR can be calculated. For both direct and indirect ligand binding, ST and SR are proportional to the number nr of ligand-specific receptors in a signalling team, as observed experimentally (Figure 4D). The indirect ligand-binding model predicts that significant on receptor binding to BP would cause a decrease in both ST and SR at high [BP]. However, this decrease of sensitivity at high [BP] is not observed (Figure 4A and B). Instead, both ST and SR saturate at the highest levels of [BP] measured. From the model, we conclude that ST and SR saturate with increasing [BP] as off receptors begin to significantly bind BP in the absence of ligand. In the regime of parameters inferred from the data collapse (K̃roff≪[BP]≪K̃ron, and p̃0≪1), ST and SRP have the simple forms,
Figure 5.
Collapse of receptor activity when plotted as a function of free-energy change. Results are shown for wild-type cells responding to indicated ligands. Binding parameters were obtained through the collapse of normalized dose-response and dynamic-range curves of both wild-type cells and cells at different BP induction levels. The free-energy model for BP-binding ligands (Approximation 1) and binding parameters are described in Supplementary data. The observed ratios Tar: Tsr: Tap: Trg of 1.5:1:0.5:0.5 were used for the collapse.
In the same experimental regime, the peak in response sensitivity also shifts with [BP] as
 |
From the estimated parameters, the response sensitivity SRP for receptors that indirectly bind ligand is much lower than the maximum possible value of nr (1−A0). For receptors that bind ligand directly, as long as adaptation remains precise or nearly so, loss of response sensitivity is due to binding of ligand by on receptors, but for BP-binding receptors, in addition to binding of closed BP by on receptors, additional losses in sensitivity occur because: (i) receptors bind to open BPs, (ii) there are some closed BPs even in the absence of ligand, and (iii) the BP concentration may be non-optimal. These additional factors help explain the lower response sensitivities observed for receptors that bind BPs compared with receptors that bind ligand directly (Figure 3A and B). Our mathematical model and the obtained data collapse thus not only allow us to evaluate response parameters for the indirectly binding ligands, but also yield a better understanding of the advantages and disadvantages of the two distinct modes of ligand binding.
Signal integration
In its natural environment, E. coli typically encounters complex mixtures of chemoeffectors, but signal integration by cells stimulated with combinations of different ligands has not been well studied until now. To investigate signal integration, we first compared responses to simultaneous excitation with two different ligands with responses elicited by these ligands individually. As illustrated for the example of galactose and MeAsp (Figure 6A), we first determined dose-response curves for each attractant individually, and then stimulated the same cells with a mixture of approximately equipotent concentrations of the two ligands, for example 10 nM galactose and 1 μM MeAsp. For sub-saturating stimuli, such stimulation produced response amplitudes that were approximately twice the size of the response amplitudes elicited by corresponding concentrations of galactose or MeAsp individually, indicating simple summation of the chemotactic responses. Further computational analysis confirmed that this summation could be accounted for by additivity of changes in free energy because of binding of galactose and MeAsp to the mixed signalling teams (Figure 6A, dashed line). Consistent with this summation of signal, when cells were pre-adapted to 8 μM MeAsp and then stimulated with different concentrations of galactose while removing MeAsp, the sign of the response depended solely on the relative strengths of both stimuli (Figure 6B). Nearly no response was observed for simultaneous addition of one and removal of the other ligand at approximately equipotent concentrations, 50 nM galactose and 8 μM MeAsp in this example. Similar results were obtained for other ligands (data not shown). In all cases, signal integration did not depend on the type of ligand binding.
Figure 6.
Responses to simultaneous stimulation with multiple ligands. (A, B) Signal integration. (A) Integration of stimuli of the same sign. Dose responses were acquired for buffer-adapted cells to MeAsp and galactose separately, and then to simultaneous addition of the both MeAsp and galactose at concentrations found to be equipotent from the single-attractant dose-response curves. Responses to the galactose–MeAsp mixture are plotted against galactose concentration. Crosses and dashed line indicate model prediction for the response to the galactose–MeAsp mixture assuming summation of the free energies of ligand binding to the mixed signalling teams. Lines here and throughout are Hill fits to the data. (B) Integration of stimuli of opposing signs. Cells were pre-adapted to 8 μM MeAsp and then stimulated by indicated concentrations of galactose in the buffer. The solid horizontal line indicates the adapted kinase activity at 8 μM MeAsp. No response was observed upon stimulation with the galactose concentration equipotent to 8 μM MeAsp. Below this concentration, the response was negative (increase in kinase activity), whereas above this concentration, the response was positive (decrease in kinase activity). Responses of buffer-adapted cells to galactose and to MeAsp are shown for a reference. A slight difference in the amplitude of the saturated response for galactose in (A, B) is presumably due to variance in the native expression level of GBP. (C, D) Effects of adaptation to other ligands on dose responses. (C) Responses to galactose of cells adapted to buffer, 1 mM MeAsp, 100 μM ribose, or 100 μM glucose. (D) Responses to MeAsp of cells adapted to buffer, 100 μM galactose, 30 μM serine, or 30 μM maltose.
We further addressed the question of how adaptation to a saturating concentration of one ligand affects responses to other ligands. For two ligands that are sensed by different receptors, the response to a second ligand was nearly unaffected by the presence of the first (Figure 6C and D; Table I), although a slight increase in the threshold sensitivity was observed in most cases. Such an increase may be explained by the imprecise adaptation to the ambient ligand, as lower adapted activity of the receptor team A0 is expected to increase ST (see equation 30 in Supplementary data). The increase in sensitivity may also be due in part to an increased receptor team size, following the increase in the receptor methylation level upon adaptation (Endres et al, 2008). Interestingly, no significant interference with the threshold sensitivity ST could be observed between MeAsp and maltose, the directly and indirectly binding ligands of Tar (Figure 6D; Table I), which confirms previous observations that these ligands can be sensed independently even though they share a receptor (Mowbray and Koshland, 1987; Gardina et al, 1998). In contrast, the indirectly binding ligands sensed by Trg clearly affected each other's responses, with sensitivity and response amplitude towards galactose being markedly reduced upon adaptation to ribose, and vice versa (Figure 6C; Table I). This clearly indicated competition of the ligand-loaded periplasmic BPs for Trg, consistent with previous observations (Adler et al, 1973; Strange and Koshland, 1976). The observed asymmetry in the mutual effects of galactose and ribose on each other's responses (Table I) may indicate a higher affinity of the ligand-loaded ribose BP to Trg (Yaghmai and Hazelbauer, 1993). However, in none of the cases was the response completely blocked by the presence of another ligand, suggesting that even at saturating levels of stimulation receptors are not fully occupied by either BP at our wild-type BP concentrations.
Table 1. Effects of adaptation to a saturating level of an ambient ligand on responses to other ligands.
| Condition | Attractant | Ambient | EC50 (without ambient) (μM)a,b | EC50 (with ambient) (μM) |
|---|---|---|---|---|
| Directly binding ligands sensed by different receptors | MeAsp | 30 μM Ser | 4.0±0.4 | 3.0±0.2 |
| Ser | 1 mM MeAsp | 0.47±0.06 | 0.40±0.03 | |
| Indirectly and directly binding ligand sensed by different receptors | Gal | 1 mM MeAsp 30 μM Ser | 0.023±0.002 | 0.014±0.002 |
| Gal | 1 mM MeAsp | 0.015±0.002 | ||
| MeAsp | 100 μM Gal | 4.0±0.4 | 4.4±0.15 | |
| Indirectly and directly binding ligand sensed by the same receptor | MeAsp | 30 μM Mal | 4.0±0.4 | 3.0±0.2 |
| Mal | 1 mM MeAsp | 0.18±0.01 | 0.23±0.01 | |
| Indirectly binding ligands recognized by the same receptor via different BPs | Gal | 100 μM Rib | 0.023±0.002 | 0.18±0.03 |
| Rib | 100 μM Gal | 0.16±0.02 | 0.34±0.02 | |
| Indirectly binding ligands recognized via the same BP | Gal | 100 μM Glc | 0.023±0.002 | No response |
| Glc | 100 μM Gal | 0.026±0.004 | 0.19±0.02 | |
| aExperiments were performed at a flow rate of 500 μl/min. | ||||
| bError bars indicate standard errors. | ||||
Even stronger competition was observed between galactose and glucose, which signal to Trg through the same BP (GBP). As the sensitivity of response of buffer-adapted cells to either ligand is essentially the same (Table I), the adaptation to a saturating concentration of glucose completely inhibited the response to galactose, consistent with the competitive binding of both ligands to GBP (Anraku, 1968; Adler et al, 1973). In contrast, a saturating amount of galactose only shifted the response to glucose towards higher concentrations, where the response to glucose is most likely mediated by the phosphotransferase system (PTS; Adler et al, 1973; Adler and Epstein, 1974). Response to glucose with comparable EC50 (0.18±0.03 μM) and response amplitude was also observed in the trg strain. The PTS thus apparently provides a backup system that allows cells to sense a high enough concentration of glucose, a preferred carbon source, even when the periplasmic BP is saturated by another ligand.
Discussion
In this work, we have quantitatively compared signalling mediated by ligands for all four E. coli chemoreceptors. In contrast to the common classification in chemotactic sensing, which primarily distinguishes between major (high abundance) and minor (low abundance) receptors, our data suggest that minor receptors are significantly more abundant than previously believed, constituting more than a quarter of the total receptor pool, and that the main distinction thus must be made between directly and indirectly binding ligands. Our analysis of response features that are expected to be important for cells to navigate in chemical gradients—threshold sensitivity ST, dynamic range, and response sensitivity to fractional changes in ligand concentration SR—showed that responses mediated by these two types of ligands are distinctly different.
The discrepancy of our results with the previously reported values of receptor abundance (Hazelbauer et al, 1981; Li and Hazelbauer, 2004) can be partly attributed to differences in the culture growth conditions, as the relative levels of receptors strongly depend on the optical density of the culture (Salman and Libchaber, 2007; Kalinin et al, 2010; Supplementary Figure S6). Moreover, the total relative levels of minor receptors, including those of Tap, were previously estimated only indirectly (Hazelbauer et al, 1981; Li and Hazelbauer, 2004 and references therein), which apparently led to a significant underestimation. Most importantly, our study uniquely enabled a direct comparison between the levels and response of individual receptors, because both were measured under identical growth conditions.
Similarity of the normalized response sensitivity values, SRP, for different ligands suggest that all receptors in an allosteric signalling team can mediate responses of comparable magnitude and sensitivity, irrespective of their type and the mode of ligand binding. This is consistent with the existing mathematical description of chemosensory complexes, which predicts that the response sensitivity primarily depends on the fraction of ligand-specific receptors in a signalling team, and further validates our estimates of the relative expression levels of individual receptors. In addition, response sensitivity for both modes of binding is influenced by the ratio of binding affinities of the ligand towards on and off states of receptors (equation S16 in Supplementary data), explaining the residual variation of sensitivity towards directly binding ligands after normalization for the level of receptor expression. An exception was the response towards serine, which showed distinctly higher sensitivity, apparently because of a second, Tsr independent, sensory system.
Stimulation with different ligand combinations demonstrated that responses mediated by different receptors in the sub-saturating stimulus range are additive. This confirms predictions of allosteric models of signalling, which suggest that the sign and the magnitude of the response are determined by the net change in the stimulus strength, that is by the free-energy change due to ligand binding. Receptors in clusters thus integrate signals using ‘majority voting' to navigate in mixed gradients, consistent with early analyses of chemotaxis using capillary assays (Adler and Tso, 1974). Moreover, we observed that the response to a given ligand is not substantially affected by the adaptation to saturating levels of other ligands, which apparently allows the chemotaxis system to maintain high sensitivity at high levels of background stimulation. An exception is signalling by ligands that compete for the same receptor or for the same BP, in which case the response is reduced or even entirely abolished by high levels of competing ligands.
How can cells regulate their ligand preferences? Our experiments confirmed that the response sensitivity for both modes of binding grows in proportion to the receptor fraction in a signalling team. For the indirectly binding ligands, however, the response further depends on the binding characteristics and availability of the associated periplasmic BP, and a response with maximal sensitivity is only observed when BPs are expressed in excess of receptors, which is consistent with previous studies (Manson et al, 1985). The effects of BP expression on response are unlikely to be related to the depletion of free ligand-bound BPs from the periplasm by their association with membrane transporters, because the expression levels of BPs are believed to be much higher than the levels of their transporters (Manson et al, 1985; Higgins et al, 1990). Rather, our analysis suggests that the uninduced expression of most BPs under our growth conditions is not sufficient to saturate receptors upon ligand binding. Furthermore, we observed that, even when BPs are overexpressed, the response sensitivity for some indirectly binding ligands can be significantly lower. This is illustrated by the comparison of two Trg ligands, galactose and ribose: whereas SRP for ribose is essentially the same as for the directly binding ligands, the value for galactose is nearly three-fold lower. Our mathematical model of signalling by the indirectly binding ligands suggests that such loss of sensitivity can be explained by the residual BP binding to receptor even in the absence of ligand, as this would reduce the signalling effect of ligand binding.
The dependence of response towards a particular ligand on the relative expression levels of receptors and of BPs means that sensitivity can be dynamically controlled by cells, dependent on growth or environmental conditions. From this perspective, indirect ligand binding provides an additional flexibility in regulation. This flexibility comes, however, at the cost of a narrower dynamic range. Although the extension of the dynamic range towards low concentrations is likely to have been modestly underestimated for some ligands because of their depletion, the dynamic range of the indirectly binding ligands is clearly much narrower than that of the directly binding ligands. For direct ligand binding, the major extension of the dynamic range is achieved by the adaptation system (Supplementary Figure S3C and E). Adaptive receptor methylation maintains the sensitivity of the system at high-ligand concentrations and returns the kinase activity to a preset level by means of effectively tuning the ligand affinity of receptors between Kroff and Kron (Endres and Wingreen, 2006; Mello and Tu, 2007; Hansen et al, 2008). Thus, the response range for major receptors extends until either (i) receptors become fully methylated and can no longer adapt to added ligand, or (ii) receptors become fully saturated with ligand (i.e. [L]>Kron) and can no longer respond to added ligand. Consistent with this picture, the dynamic range for the directly binding ligands spans 4–5 orders of magnitude (Mesibov and Adler, 1972; Sourjik and Berg, 2002). Our data suggest that for serine, the dynamic range is indeed limited by the range of receptor adaptation (case i). In contrast, the dynamic range for aspartate appears to be restricted instead by receptor saturation (case ii), given the estimated value for Kron of 20 μM of Tar receptors for aspartate and the observed relatively precise adaptation up to 1 mM aspartate.
In contrast, the dynamic range for indirect binding cannot be extended by adaptation, because the range is limited by the occupancy of the BP, and the affinity of the latter for its ligand cannot be adjusted by the adaptation system working on the receptor (Supplementary Figure S3D and F). As a result, cells respond to galactose, ribose, dipeptides, or maltose over only about two orders of magnitude of ligand concentration, consistent with the results of previous capillary assays (Mesibov et al, 1973). As a consequence, adaptation to saturating concentrations of indirectly binding ligands results in much smaller changes in receptor methylation than adaptation to directly binding ligands (Supplementary Figure S9).
Given the advantages and disadvantages of the two modes of ligand sensing, what could have led to their evolutionary fixation? For indirectly binding ligands, it can be speculated that coupling of the ABC transport and chemotaxis systems allows better coordination of the rate of ligand-specific nutrient uptake and the chemotactic response. This prevents cell accumulation at excessively high concentrations of attractants and allows a concerted regulation of chemotaxis and transport according to environmental conditions. The expression of periplasmic BPs is typically inducible (Koman et al, 1979), so that cells can simultaneously upregulate both uptake and chemotaxis in the presence of the respective ligand. In contrast to response regulation through the level of receptor expression, such BP-dependent regulation allows cells to specifically increase chemotaxis towards one particular ligand without upregulating responses to all other ligands sensed by the same receptor. Moreover, cells would chemotactically follow gradients of the indirectly binding ligand only as long as the uptake system remained unsaturated, and moving up the gradient increased the efficiency of nutrient uptake. At concentrations above transporter saturation, cells will be indifferent to the gradient of that ligand and can instead follow gradients of other attractants. Taxis to carbohydrates such as glucose is further connected to their uptake through the PTS transporters in a receptor-independent manner (Postma et al, 1993), which allows cells to selectively extend the dynamic range of the chemotaxis system, again in a tight coupling with the dynamic range of the uptake. Taken together, the indirect sensing of sugars and dipeptides appears to present an evolutionarily optimal strategy to maximize the overall nutrient uptake by specific systems.
This contrasts with the direct sensing of amino acids, which allows cells to follow gradients over a much larger span of concentrations. The physiological reason for this difference remains unclear. In contrast to sugars, the uptake of amino acids such as serine and aspartate in E. coli is primarily performed by multiple symporters (Schellenberg and Furlong, 1977; McFall and Newman, 1996; Kim et al, 2002), which are not known to mediate any chemotactic responses. The broad dynamic range for these amino acids may therefore enable cells to accumulate at concentrations corresponding to the functional range of symporters without a direct coupling to transport. In addition, the broader dynamic range may be required for the nutrition-unrelated function of chemotaxis towards amino acids, such as signal exchange between bacteria (Budrene and Berg, 1991; Park et al, 2003).
Materials and methods
Attractants
L-aspartic acid (>99% purity), α-methyl-DL-aspartic acid (MeAsp), L-serine, 2-aminoisobutyric acid (98% purity), D-(+)-maltose monohydrate (min. 99% purity), D-(+)-galactose (min. 99% purity), D-(+)-glucose, D-(−)-ribose (min. 99% purity), and Pro-Leu were purchased from Sigma. At this purity grade, maltose and galactose are declared to contain <0.3 and <0.1% glucose, respectively. This glucose contamination may lead to an unspecific Trg-mediated response at around 1 mM of these ligands, which set the limits of the concentration range of maltose and galactose used in this study.
Strains and plasmids
All strains and respective genotypes, plasmids and primers used in this study are listed in Supplementary Table SII. SN1 and VS275 were generated by the pAMPts homologous recombination system of allele exchange (Sourjik and Berg, 2000). SN11 was made by P1 transduction with donor VS139 and recipient SN1.
SN27, SN28, and SN31 were made by P1 transduction using respective donor strains from the Keio collection (Baba et al, 2006) and SN1 as recipient. The λ-Red system (Datsenko and Wanner, 2000) was used to generate both SN25 and SN23. Here, the KanR cassette was amplified from Keio strain JW1875 using primers Vic113 and Vic146 and transformed into recipient strains SN1 and VS275 expressing the Red-System from the helper plasmid pKD46 by electroporation. After selecting strains on kanamycin (50 μg/ml), the resistance cassette in all strains was flipped out using the curable temperature-sensitive plasmid pCP20 that encodes FLP recombinase.
Preparation of cells
E. coli K-12 strain LJ110 Δ(cheY cheZ) and its derivatives were used for all FRET measurements. Cells were grown in tryptone broth supplemented with antibiotics (100 μg/ml ampicillin; 34 μg/ml chloramphenicol) depending on the plasmids present. IPTG was added to a final concentration of 50 μM to induce expression of CheY-YFP and CheZ-CFP from pVS88 (pTrc99a; AmpR; Sourjik and Berg, 2004). BP and receptor expression from pKG110 derivatives was induced by salicylate in a range of 0–10 μM as indicated. After growth to OD600 of 0.45 at 34°C and 275 r.p.m., cells were harvested by centrifugation (10 min at 5000 g) and resuspended in the original culture volume of tethering buffer (10 mM KPO4, 0.1 mM EDTA, 1 μM methionine, 10 mM lactic acid, 67 mM NaCl, pH 7). Protein expression was shut down for at least 30 min at 8°C, before cells were attached to a polylysine-coated coverslip and placed into a flow chamber of 50 μl volume (Berg and Block, 1984; Sourjik et al, 2007). The chamber was kept under constant flow of tethering buffer (300 μl/min for dynamic range and adaptation measurements, 500 μl/min for sensitivity measurements, and 2500 or 500 μl/min for dose-response measurements) by a syringe pump (Harvard Apparatus 22) that was stopped briefly to add and remove attractants.
Data acquisition and analysis
Measurements were done as described before (Sourjik and Berg, 2002) on a Zeiss Axio Imager.Z1 microscope equipped with a 40x/0.75 EC Plan-Neofluar objective and controlled by Axiovision software. CFP fluorescence of a dense monolayer was excited at 436/20 nm through a 455 nm dichroic mirror by a 75 W Xenon lamp attenuated 500-fold with neutral density filters. CFP and YFP emissions were detected through 480/40 nm band pass and 520 nm long-pass emission filters, respectively, and signals were collected with an integration time of 1 s by Peltier-cooled photon counters (Hamamatsu) equipped with a PCI-6034 counting board connected to a computer with custom written LabView7 software (both from National Instruments). FRET values were calculated as described previously (Sourjik and Berg, 2002; Sourjik et al, 2007; see Supplementary methods for details).
Receptor quantification
Receptor quantification was performed using immunoblot and FACS analyses as described in Supplementary data and in Supplementary Figures S4 and S5. For YFP quantification using FACScan (Becton Dickinson), cells were prepared as for FRET and diluted 1:20. For immunoblots, cells were harvested by centrifugation and resuspended in 1 × Laemmli buffer to give the same YFP concentration, determined by the FACS value, in all samples. Samples were boiled and separated by 10% SDS–PAGE, with several consecutive dilutions of each sample being applied to each gel (Supplementary Figure S4). Proteins were then transferred to a 0.2-μm pore-size Hybond ECL nitrocellulose membrane using tank blotting. Receptor detection was performed using primary polyclonal αTrg (kindly provided by G Hazelbauer) or αTar antibody at 1:5000 dilution and IRDye® 800 conjugated secondary antibody (Rockland) at 1:10 000 dilution. Membranes were scanned with an Odyssey® Imager (LI-COR) and protein bands were quantified using ImageJ software (http://rsbweb.nih.gov/ij). The background signal from empty areas of the membrane was subtracted. Only bands in the linear intensity range were used for subsequent analyses. Specificity of αTrg and αTar antibodies for individual receptors was determined by using YFP fusions to receptors as a reference. To check the consistency of YFP loading in the reference samples, control immunoblotting was performed using αGFP monoclonal antibody (JL-8; Clontech) and IRDye 700 conjugated secondary antibody (Rockland).
Estimation of relative BP expression
Expression of BPs was assumed to be proportional to the expression of YFP controlled by the same promoter (pnahG), determined by the FACScan analysis. For FACS, cells expressing eYFP from pVS118 and carrying an empty pTrc99a vector were induced over a range of 0–10 μM salicylate, and grown and prepared as for FRET experiments.
Supplementary Material
Acknowledgments
We thank GL Hazelbauer for the gift of αTrg antibody. This work was supported by grants RGP66/2005 from the Human Frontier Science Program, SO 421/7-1 and SO 421/3-3 from the Deutsche Forschungsgemeinschaft and GM082938 from the National Institutes of Health. VS and NSW acknowledge the hospitality of the Aspen Center for Physics.
Author contributions: VS, SN, and NSW designed the research; SN performed the experimental research; CH and NSW performed the mathematical modelling; VS, SN, CH, and NSW wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adler J, Epstein W (1974) Phosphotransferase-system enzymes as chemoreceptors for certain sugars in Escherichia coli chemotaxis. Proc Natl Acad Sci USA 71: 2895–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J, Hazelbauer GL, Dahl MM (1973) Chemotaxis toward sugars in Escherichia coli. J Bacteriol 115: 824–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J, Tso WW (1974) ‘Decision'-making in bacteria: chemotactic response of Escherichia coli to conflicting stimuli. Science 184: 1292–1294 [DOI] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS (2002) Collaborative signalling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA 99: 7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku Y (1968) Transport of sugars and amino acids in bacteria. I. Purification and specificity of the galactose- and leucine-binding proteins. J Biol Chem 243: 3116–3122 [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC, Block SM (1984) A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J Gen Microbiol 130: 2915–2920 [DOI] [PubMed] [Google Scholar]
- Berg HC, Brown DA (1972) Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239: 500–504 [DOI] [PubMed] [Google Scholar]
- Berg HC, Purcell EM (1977) Physics of chemoreception. Biophys J 20: 193–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialek W, Setayeshgar S (2005) Physical limits to biochemical signaling. Proc Natl Acad Sci USA 102: 10040–10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Simon MI (1992) Attenuation of sensory receptor signalling by covalent modification. Proc Natl Acad Sci USA 89: 6756–6760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D, Levin MD, Morton-Firth CJ (1998) Receptor clustering as a cellular mechanism to control sensitivity. Nature 393: 85–88 [DOI] [PubMed] [Google Scholar]
- Budrene EO, Berg HC (1991) Complex patterns formed by motile cells of Escherichia coli. Nature 349: 630–633 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke TA, Le Novere N, Bray D (2001) Conformational spread in a ring of proteins: a stochastic approach to allostery. J Mol Biol 308: 541–553 [DOI] [PubMed] [Google Scholar]
- Endres RG, Oleksiuk O, Hansen CH, Meir Y, Sourjik V, Wingreen NS (2008) Variable sizes of Escherichia coli chemoreceptor signaling teams. Mol Syst Biol 4: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres RG, Wingreen NS (2006) Precise adaptation in bacterial chemotaxis through ‘assistance neighborhoods'. Proc Natl Acad Sci USA 103: 13040–13044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardina PJ, Bormans AF, Manson MD (1998) A mechanism for simultaneous sensing of aspartate and maltose by the Tar chemoreceptor of Escherichia coli. Mol Microbiol 29: 1147–1154 [DOI] [PubMed] [Google Scholar]
- Gegner JA, Graham DR, Roth AF, Dahlquist FW (1992) Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70: 975–982 [DOI] [PubMed] [Google Scholar]
- Gestwicki JE, Kiessling LL (2002) Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415: 81–84 [DOI] [PubMed] [Google Scholar]
- Hansen CH, Endres RG, Wingreen NS (2008) Chemotaxis in Escherichia coli: a molecular model for robust precise adaptation. PLoS Comput Biol 4: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL (1975) Maltose chemoreceptor of Escherichia coli. J Bacteriol 122: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Engstrom P, Harayama S (1981) Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol 145: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Mesibov RE, Adler J (1969) Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc Natl Acad Sci USA 64: 1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Gallagher MP, Hyde SC, Mimmack ML, Pearce SR (1990) Periplasmic binding protein-dependent transport systems: the membrane-associated components. Philos Trans R Soc Lond B Biol Sci 326: 353–364 [DOI] [PubMed] [Google Scholar]
- Kalinin Y, Neumann S, Sourjik V, Wu M (2010) Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J Bacteriol 192: 1796–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS (2006) Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci USA 103: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Ogawa W, Tamai E, Kuroda T, Mizushima T, Tsuchiya T (2002) Purification, reconstitution, and characterization of Na(+)/serine symporter, SstT, of Escherichia coli. J Biochem 132: 71–76 [DOI] [PubMed] [Google Scholar]
- Koman A, Harayama S, Hazelbauer GL (1979) Relation of chemotactic response to the amount of receptor: evidence for different efficiencies of signal transduction. J Bacteriol 138: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai RZ, Manson JM, Bormans AF, Draheim RR, Nguyen NT, Manson MD (2005) Cooperative signalling among bacterial chemoreceptors. Biochemistry 44: 14298–14307 [DOI] [PubMed] [Google Scholar]
- Levit MN, Stock JB (2002) Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J Biol Chem 277: 36760–36765 [DOI] [PubMed] [Google Scholar]
- Li G, Weis RM (2000) Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100: 357–365 [DOI] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL (2004) Cellular stoichiometry of the components of the chemotaxis signalling complex. J Bacteriol 186: 3687–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL (2005) Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol 56: 1617–1626 [DOI] [PubMed] [Google Scholar]
- Macnab RM, Koshland DE Jr (1972) The gradient-sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA 69: 2509–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L (1993) Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259: 1717–1723 [DOI] [PubMed] [Google Scholar]
- Manson MD, Boos W, Bassford PJ Jr, Rasmussen BA (1985) Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J Biol Chem 260: 9727–9733 [PubMed] [Google Scholar]
- Manson MD, Kossmann M (1986) Mutations in tar suppress defects in maltose chemotaxis caused by specific malE mutations. J Bacteriol 165: 34–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall E, Newman EB (1996) Amino acids as carbon sources. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds), pp 358–379. Washington, D.C: ASM Press [Google Scholar]
- Mello BA, Tu Y (2005) An allosteric model for heterogeneous receptor complexes: understanding bacterial chemotaxis responses to multiple stimuli. Proc Natl Acad Sci USA 102: 17354–17359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello BA, Tu Y (2007) Effects of adaptation in maintaining high sensitivity over a wide range of backgrounds for Escherichia coli chemotaxis. Biophys J 92: 2329–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesibov R, Adler J (1972) Chemotaxis toward amino acids in Escherichia coli. J Bacteriol 112: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesibov R, Ordal GW, Adler J (1973) The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. Weber law and related phenomena. J Gen Physiol 62: 203–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowbray SL, Koshland DE Jr (1987) Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell 50: 171–180 [DOI] [PubMed] [Google Scholar]
- Ordal GW, Adler J (1974) Properties of mutants in galactose taxis and transport. J Bacteriol 117: 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Wolanin PM, Yuzbashyan EA, Lin H, Darnton NC, Stock JB, Silberzan P, Austin R (2003) Influence of topology on bacterial social interaction. Proc Natl Acad Sci USA 100: 13910–13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR (1993) Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57: 543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman H, Libchaber A (2007) A concentration-dependent switch in the bacterial response to temperature. Nat Cell Biol 9: 1098–1100 [DOI] [PubMed] [Google Scholar]
- Schellenberg GD, Furlong CE (1977) Resolution of the multiplicity of the glutamate and aspartate transport systems of Escherichia coli. J Biol Chem 252: 9055–9064 [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2000) Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol 37: 740–751 [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2002) Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA 99: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC (2004) Functional interactions between receptors in bacterial chemotaxis. Nature 428: 437–441 [DOI] [PubMed] [Google Scholar]
- Sourjik V, Vaknin A, Shimizu TS, Berg HC (2007) In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol 423: 365–391 [DOI] [PubMed] [Google Scholar]
- Strange PG, Koshland DE Jr (1976) Receptor interactions in a signalling system: competition between ribose receptor and galactose receptor in the chemotaxis response. Proc Natl Acad Sci USA 73: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghmai R, Hazelbauer GL (1993) Strategies for differential sensory responses mediated through the same transmembrane receptor. EMBO J 12: 1897–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeppenfeld T, Larisch C, Lengeler JW, Jahreis K (2000) Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICB(Glc) and induction behavior of the ptsG gene. J Bacteriol 182: 4443–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gardina PJ, Kuebler AS, Kang HS, Christopher JA, Manson MD (1999) Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc Natl Acad Sci USA 96: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.