Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT
HSF1, the key transcription factor of the heat shock response, plays a major role in the control of polyQ aggregation via heat shock proteins and so far uncharacterised non-heat shock proteins. This study identifies the transcription factor NFATc2 as a key downstream target of HSF1 that cooperates with HSF1 in regulating proteostasis.
Keywords: heat shock, HSF, NFAT, polyglutamine, protein homeostasis
Abstract
Heat shock transcription factor 1 (HSF1) is an important regulator of protein homeostasis (proteostasis) by controlling the expression of major heat shock proteins (Hsps) that facilitate protein folding. However, it is unclear whether other proteostasis pathways are mediated by HSF1. Here, we identified novel targets of HSF1 in mammalian cells, which suppress the aggregation of polyglutamine (polyQ) protein. Among them, we show that one of the nuclear factor of activated T cells (NFAT) proteins, NFATc2, significantly inhibits polyQ aggregation in cells and is required for HSF1-mediated suppression of polyQ aggregation. NFAT deficiency accelerated disease progression including aggregation of a mutant polyQ-huntingtin protein and shortening of lifespan in R6/2 Huntington's disease mice. Furthermore, we found that HSF1 and NFAT cooperatively induce the expression of the scaffold protein PDZK3 and αB-crystallin, which facilitate the degradation of polyQ protein. These results show the first mechanistic basis for the observation that HSF1 has a much more profound effect on proteostasis than individual Hsp or combination of different Hsps, and suggest a new pathway for ameliorating protein-misfolding diseases.
Introduction
Cellular protein homeostasis or proteostasis involves controlling the concentration, conformation, binding interaction, and location of individual proteins, and is maintained by a network of pathways that influence protein synthesis, folding, translocation, assembly/disassembly, and degradation (Balch et al, 2008). Loss of cellular proteostasis results in many systemic and neurodegenerative disorders, termed protein-misfolding diseases or protein conformational diseases, caused by inherited misfolding-prone proteins such as mutant huntingtin, tau, and superoxide dismutase-1, or by metabolic/environmental stress-mediated misfolding of cellular proteins (Powers et al, 2009). Models have been established in yeast, Drosophila, and Caenorhabditis elegans that express misfolding proteins to identify regulators of proteostasis. These regulators include proteins that are involved in RNA metabolism, protein synthesis, protein degradation, ubiquitin-dependent protein catabolism, and lipid metabolism (Fernandez-Funez et al, 2000; Kazemi-Esfarjani and Benzer, 2000; Willingham et al, 2003; Nollen et al, 2004; Bilen and Bonini, 2007). Prominent among them are molecular chaperones and their stress-inducible response (Bukau et al, 2006; Ron and Walter, 2007; Morimoto, 2008).
The heat shock response is characterized by the expression of a set of molecular chaperones termed heat shock proteins (Hsps) that facilitate the folding of proteins and maintain protein homeostasis (Parsell and Lindquist, 1993), and is regulated mainly at the transcriptional level by heat shock transcription factors (HSFs) (Wu, 1995). A single HSF, HSF1, regulates this response in yeast, Drosophila, and C. elegans, whereas four members of the HSF family (HSF1–4) have been characterized in vertebrates, and the expression of major Hsps is regulated mainly by HSF1 in mammals (Morimoto, 1998; Fujimoto et al, 2010). Gain of HSF1 function significantly ameliorates disease progression in C. elegans models of neurodegenerative disorders such as polyglutamine (polyQ) diseases (Hsu et al, 2003; Morley and Morimoto, 2004) and Alzheimer's disease (Cohen et al, 2006), whereas loss of HSF1 function accelerates it through the reduced expression of major Hsps. In mammals, this HSF1-mediated induction of Hsp expression is correlated with the acquisition of thermotolerance (McMillan et al, 1998) and the protection of cells from various pathological conditions such as neurodegenerative disorders (Fujimoto et al, 2005; Steele et al, 2008). However, HSF1 regulates the expression of not only major Hsps, products of classical heat shock genes, during heat shock, but also tremendous numbers of what are called non-classical heat shock genes in yeast, Drosophila, and mammalian cells (Hahn et al, 2004; Trinklein et al, 2004; Birch-Machin et al, 2005). Furthermore, HSF1 and other members of the HSF family are involved in the development and maintenance of neuronal, reproductive, and sensory tissues, which are associated with the expression of not only Hsps, but also development-related genes (Akerfelt et al, 2007; Nakai, 2009). Even though HSF1 regulates the expression of numerous genes, it is believed to maintain proteostasis by regulating the expression of major Hsps.
Interestingly, the expression of classical heat shock genes is regulated by HSF3 in chicken cells (Nakai et al, 1995; Tanabe et al, 1998), whereas chicken HSF1 (cHSF1) is dispensable for their expression (Inouye et al, 2003). However, cHSF1 and mouse HSF3 can suppress polyQ aggregation without the expression of classical heat shock genes (Inouye et al, 2003; Fujimoto et al, 2010). Therefore, we hypothesized that HSFs might regulate non-classical heat shock genes involved in proteostasis. Here, we identified target genes of human HSF1 in HeLa cells using a DNA microarray, and examined whether they regulate proteostasis. We showed a novel HSF1-mediated proteostasis pathway, which is unrelated to the expression of major Hsps. Identification of the non-chaperone pathway explains why HSF1 has a great impact on protein homeostasis.
Results
Identification of HSF1-target genes that suppress polyQ aggregation
To analyse target genes of human HSF1, we generated two independent HeLa clones (RDT1 and RDT2) expressing an actively mutated hHSF1 (hHSF1ΔRDT) (Fujimoto et al, 2005), as well as a HeLa clone expressing cHSF1 (HeLa/cHSF1). We first confirmed that protein and mRNA levels of major Hsps were increased in RDT1 and RDT2 cells compared with those in HeLa cells, whereas those were constant or even decreased in HeLa/cHSF1 cells (Supplementary Figure 1A and B). We then carried out a DNA microarray, and found 62 genes that showed a more than two-fold increase in both RDT1 and RDT2 cells (Supplementary Figure 1C; Supplementary Table 1). Among them, 29 genes were confirmed by RT–PCR to be increased in both RDT1 and RDT2 cells (Figure 1A; Supplementary Figure 1D). The expression of 18 genes (62%) was also increased in HeLa/cHSF1 cells (class a), whereas the expression of the other 11 genes (38%) was constant (class b) or decreased (class c). The 29 genes were temporarily induced in heat-shocked HeLa cells or in cells in which hHSF1ΔRDT was expressed under the control of tetracycline-responsive element, strongly suggesting that they are targets of hHSF1 (Supplementary Figure 2). The gene products included various kinds of proteins such as transcription factors and cytokines (Figure 1A).
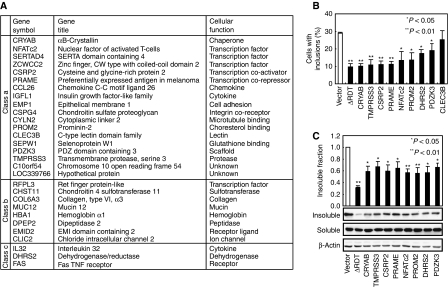
Figure 1.
Identification of non-chaperone HSF1-target genes that suppress polyQ aggregation. (A) Summary of 29 novel HSF1-target genes, expression of which was increased in RDT1 and RDT2 lines. The expression of 18 genes (62%) was also increased in HeLa/cHSF1 cells (class a), whereas the expression of the other 11 genes (38%) was constant (class b) or decreased (class c). (B) Percentage of cells with polyQ inclusions in MEF cells co-infected with the retrovirus expressing each indicated gene and adenovirus expressing polyQ81-GFP. A retrovirus expressing an active hHSF1 (hHSF1ΔRDT) was used as a positive control. The averages of five experiments with the mean+s.d. are shown. (C) Soluble and insoluble fractions of polyQ81-GFP were analysed by western blotting (lower), and the insoluble polyQ81-GFP was quantified (upper). The averages of four experiments are shown. Error bars shows the mean+s.d. Statistical significance (P-value) was determined with an unpaired t-test.
To search genes that regulate proteostasis, we established a procedure using MEF cells, in which each HSF1-target gene was expressed using a retrovirus and then a pathologic polyQ 81 fused to GFP (polyQ81-GFP) was expressed using an adenovirus (Fujimoto et al, 2005). We counted the number of cells with inclusion bodies at 24 and 36 h after the adenoviral infection, and found eight HSF1-target genes that inhibited aggregation at both time points similar to an active hHSF1 gene (Figure 1B; Supplementary Figure 3). One of these eight was a chaperone gene, αB-crystalline (CRYAB), but the rest were non-chaperone genes: transmembrane serine protease 3 (TMPRSS3), cysteine and glycine-rich protein 2 (CSRP2) encoding an actin-interacting and transcriptional co-activator, preferentially expressed antigen of melanoma (PRAME) encoding a transcriptional co-repressor, nuclear factor of activated T cell (NFATc2) encoding a transcription factor, prominin-2 (PROM2) encoding a membrane protein, dehydrogenase/reductase (SDR family) member 2 (DHRS2), and PDZ domain-containing 3 (PDZK3) encoding a scaffold protein, and were different from the genes that had a suppressive effect on polyQ aggregation in C. elegans and Drosophila, except for αB-crystalline (Nollen et al, 2004; Bilen and Bonini, 2007). These genes partially suppressed the accumulation of NP40-insoluble polyQ81-GFP (Figure 1C), and the reduction in mitochondrial activity (data not shown). The expression of these genes was also increased in HeLa/cHSF1 cells, except that of DHRS2 (Supplementary Figure 1D). The results showed for the first time that non-classical heat shock genes activated by HSF1 can prevent polyQ from forming aggregates. It is noticeable that overexpression of an active HSF1 gene suppressed polyQ aggregation more than that of any HSF1-target gene.
Increased aggregation in HSF1-null cells is reversed by overexpression of HSF1-target genes
As a deficiency of HSF1 greatly accelerates the polyQ aggregation in C. elegans (Hsu et al, 2003; Morley and Morimoto, 2004) and mouse cells (Homma et al, 2007), we analysed the contribution of the newly identified HSF1 targets in HSF1-null cells. We first confirmed in our MEF assay the increase in the number of cells forming polyQ inclusions and the level of NP40-insoluble polyQ protein in HSF1-null cells (Figure 2A). Furthermore, the level of ubiquitylated cellular protein was markedly increased in HSF1-null cells (Figure 2A; Supplementary Figure 4A and B), indicating disruption of the ubiquitin-proteasome system (Bennett et al, 2007). In the HSF1-null cells, levels of major Hsps such as Hsp110, Hsp90, Hsp70, Hsp40, and Hsp27 are the same as those in wild-type cells (Supplementary Figure 4C). In contrast, the expression of four of the newly identified genes, NFATc2, PDZK3, CSRP2, and CRYAB, was decreased in HSF1-null cells (Figure 2B). Therefore, we re-expressed the genes in HSF1-null cells, and found a significant reduction in the number of cells with polyQ inclusions, the amount of insoluble polyQ protein, and the accumulation of NP40-insoluble ubiquitylated cellular proteins, similar to the overexpression of Hsp70-1 and Hsp27 genes (Figure 2C). These results suggested that the impaired proteostasis in HSF1-null MEF cells is at least in part because of the down-regulation of the four HSF1 targets.
Figure 2.
Overexpression of target genes improves aggregate formation in HSF1-null cells. (A) Percentage of cells having polyQ inclusions (left), accumulation of insoluble polyQ protein (middle), and accumulation of insoluble ubiquitylated protein in WT and HSF1-null MEF cells. Error bars show the mean+s.d. (B) mRNA levels of novel HSF1-target genes in HSF1-null MEF cells relative to the levels in WT cells. The averages of three experiments are shown. Error bars show the mean+s.d. (C) Re-expression of NFATc2, PDZK3, CSRP2, or CRYAB in HSF1-null MEF cells restored HSF1 deficiency, such as overexpression of Hsp70-1, Hsp27, or three Hsps containing Hsp70-1, Hsp27, and Hsp40 (Hsp mix). Percentage of cells with polyQ inclusions (left), accumulation of insoluble polyQ protein (middle), and accumulation of insoluble ubiquitylated protein (right) are shown. The averages of three experiments are shown. Error bars show the mean +s.d. Statistical significance (P-value) was determined with an unpaired t-test.
NFATc2 is required for HSF1-mediated suppression of polyQ aggregation
Among the four targets of HSF1, we focused on the transcription factor NFATc2 (NFAT1), a member of the NFAT family (Hogan et al, 2003), as it strongly suppressed polyQ aggregation and its expression greatly depended on HSF1 in MEF cells (Figure 2B and C). The expression of NFATc2 was decreased in HSF1-null cells, and heat shock treatment markedly increased the amount of NFATc2 in wild type, but not in HSF1-null cells (Figure 3A). Overexpression of hHSF1 restored the expression of NFATc2 mRNA in the HSF1-null cells, whereas overexpression of hHSF1 mutants, hHSF1R176P and hHSF1ΔAB that cannot form a trimer (Inouye et al, 2007), did not because of an inability to bind the promoter (Figure 3B; Supplementary Figure 5A). The mouse NFATc2 gene contains two alternative 5′ exons, IA and IB (Vihma et al, 2008). Both exons were transcribed in the cerebral cortex, whereas only exon IA was transcribed in MEF cells (Supplementary Figure 5B). The expression of the exon IA transcript was induced during heat shock and was decreased in HSF1-null MEF cells and in the HSF1-null cerebral cortex (Supplementary Figure 5C). We performed a reporter analysis of each upstream sequence, the P1 (2.0 kb) or P2 (3.0 kb) promoter, in HEK293 cells, and found that the reporter activity of the P1 promoter increased two-fold after heat shock such as that of the Hsp70 promoter, whereas the reporter activity of the P2 promoter was unchanged (Figure 3C). Deletion of region 6 (−1500 to −2000) or mutation of the HSE3 sequence in the region 6 abolished both the constitutive and the heat-induced reporter activity of the P1 promoter. A ChIP analysis showed that HSF1 binds to region 6 in both unstressed and heat-shocked cells (Supplementary Figure 5D). These results indicate that a trimeric HSF1 binds to region 6 through HSE3 and activates the NFATc2 gene under normal and heat shock conditions.
Figure 3.
NFATc2 has a major function in HSF1-mediated suppression of polyQ aggregation. (A) Western blot of NFATc2 protein in WT and HSF1-null MEF cells. Cells were heat shocked at 42°C for 30 min and recovered for 9 h. (B) RT–PCR analysis of NFATc2 mRNA levels in WT and HSF1-null MEF cells expressing GFP, hHSF1, or hHSF1 mutants (hHSF1R176P and hHSF1ΔAB) that cannot form trimers. (C) Reporter analysis of the mouse NFATc2 promoter under normal and heat-stressed conditions. The promoter was divided into six regions (region 1–6). HSE2 and HSE3 in region 6 were mutated in pP1-luc-m1 and pP1-luc-m2, respectively. Error bars show the mean+s.d. The averages of three experiments are shown with P-values. (D) Percentage of cells with polyQ inclusions (left), accumulation of insoluble polyQ protein (middle), and that of insoluble ubiquitylated protein (right) in WT and NFATc2-null MEF cells. Error bars show the mean+s.d. The averages of three experiments are shown. (E) Percentage of cells with polyQ inclusions in WT, NFATc2-null, and HSF1-null MEF cells infected with Ad-hHSF1ΔRDT or Ad-NFATc2 (1.0, 0.1, and 0.01 × 106 pfu/ml) (upper). Levels of HSF1 and NFATc2 proteins are shown by western blotting (middle). The accumulation of insoluble polyQ protein, accumulation of insoluble ubiquitylated protein, and expression of Hsp70 and β-actin are also shown (lower). Error bars show the mean + s.d. The averages of three experiments are shown. (F) Accumulation of insoluble ubiquitylated protein in WT, NFATc2-null, and HSF1-null MEF cells expressing GFP (cont.), hHSF1ΔRDT, or NFATc2. Error bars show the mean +s.d. The averages of three experiments are shown. Statistical significance (P-value) was determined with an unpaired t-test through Figure 3.
We next examined polyQ aggregation in NFATc2-null cells and found increased numbers of cells with polyQ inclusions and increased amounts of NP40-insoluble polyQ81-GFP in the cells (Figure 3D). Furthermore, the accumulation of insoluble ubiquitylated cellular proteins was significantly increased in these cells. Considering that NFATc2 is a downstream target of HSF1, we examined whether NFATc2 has a critical function in HSF1-mediated proteostasis or not. Overexpression of NFATc2 in HSF1-null cells efficiently reduced the number of cells with polyQ inclusions, associated with a decrease in the amount of NP40-insoluble polyQ81-GFP (Figure 3E). In marked contrast, overexpression of hHSF1ΔRDT in NFATc2-null cells suppressed the polyQ aggregation only partially, although the expression of Hsp70 was well induced. Similarly, overexpression of NFATc2 in HSF1-null cells significantly suppressed the accumulation of insoluble ubiquitylated proteins, whereas overexpression of hHSF1ΔRDT in NFATc2-null cells suppressed it only partially (Figure 3F). These results clearly show that NFATc2 is required for HSF1-mediated suppression of polyQ aggregation.
Shortening of lifespan in both HSF1-null and NFATc2-null Huntington's disease mice
To investigate the impact of HSF1 and NFATc2 on polyglutamine disease in vivo, we used a model of Huntington's disease, the R6/2 mice (Mangiarini et al, 1996), which was transgenic for the human huntingtin gene carrying fewer CAG repeats (95–97-fold). A single nuclear polyQ-huntingtin aggregate per cell was observed in the striatum at 38 weeks (Figure 4I and J), whereas several small aggregates were observed in the nucleus at 8 weeks (Figure 4A, B, E, and F). Remarkably, numbers of cells having the aggregates in the striatum were increased and each aggregate in both the nucleus and cytoplasm was more evident in HSF1-null and NFATc2-null R6/2 mice at 8 weeks than in wild-type R6/2 mice (Figure 4C, D, G, H, and K; Supplementary Figure 6A). Furthermore, the accumulation of polyQ-huntingtin protein (Supplementary Figure 6B) and the formation of its highly insoluble aggregates examined by filter trap assay (Figure 4L) were increased in the HSF1-null and NFATc2-null brain compared with that in the wild-type R6/2 brain, although deficiency of HSF1 or NFATc2 did not affect mRNA level of polyQ-huntingtin (Supplementary Figure 6C). PolyQ-huntingtin aggregates did not appear in other tissues such as the skeletal muscle at 38 weeks (data not shown) (Fujimoto et al, 2005).
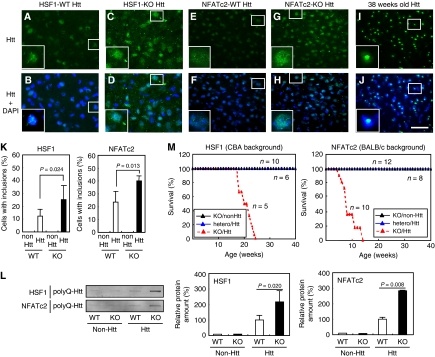
Figure 4.
Shortening of lifespan in HSF1-null and NFATc2-null Huntington's disease mice. PolyQ-huntingtin protein aggregates were detected by immunohistochemistry using a goat anti-huntingtin antibody in the striatum of 8-week-old R6/2 mice (95–97 CAG repeats) with or without HSF1 (CBA background) (A–D) and in that of 8-week-old R6/2 mice with or without NFATc2 (BALB/c background) (E–H). Typical inclusions in R6/2 mice at 38 weeks old are shown (I, J). Boxed regions are magnified in the insets. Bar, 50 μm. (K) Percentage of cells with inclusions in (A–H). The averages of three mice are shown with P-values calculated using an unpaired t-test. Error bars show the mean +s.d. (L) Filter trap assay of polyQ-huntingtin protein in the brain of HSF1-null and NFATc2-null R6/2 mice. SDS-insoluble aggregates were trapped on a cellulose acetate membrane, and immunoblotting was performed using a goat anti-huntingtin antibody (left). The intensity of the signals was quantified, and the averages of three mice in each genotype are shown with P-values using an unpaired t-test (right). Error bars show the mean+s.d. (M) The lifespan of R6/2 mice with or without HSF1 or NFATc2.
We also examined lifespan. First, we generated HSF1-null R6/2 (CAG95–97) mice with the C57BL/6 background, but found that they died quite early (within 6 weeks), whereas 80% of control R6/2 (CAG95–97) mice lived for up to 40 weeks (data not shown). Therefore, we generated HSF1-null R6/2 mice against the CBA background and NFATc2-null R6/2 mice against the BALB/c background. The R6/2 mice with the CBA or BALB/c background lived for up to 40 weeks, whereas the HSF1-null R6/2 mice with the CBA background and NFATc2-null R6/2 mice with the BALB/c background died in 25 and 13 weeks, respectively (Figure 4M). In addition, both HSF1-null and NFATc2-null R6/2 mice showed neurological symptoms much earlier than R6/2 mice. In fact, clasping appeared at 8–11 weeks in HSF1-null R6/2 mice (n=5) and at 5–8 weeks in NFATc2-null R6/2 mice (n=10), whereas it appeared at 27 weeks in HSF1-hetero-R6/2 mice (n=10) and at 32 weeks in NFATc2-hetero-R6/2 mice (n=12). Taken together, both HSF1 deficiency and NFATc2 deficiency caused accelerated polyQ protein expression and aggregation in the brain, and resulted in a marked shortening of lifespan.
Both HSF1 and NFATc2 are required for marked activation of PDZK3 and CRYAB
As HSF1 deficiency and NFATc2 deficiency showed similar phenotypes in cultured cells and in mice, we examined whether HSF1 and NFATc2 target the same genes. Using HeLa cells expressing NFATc2-HA under the control of a tetracycline-responsive promoter, we found that the expression of PDZK3 and CRYAB, which are targets of HSF1, was induced in response to elevated levels of NFATc2 (Figure 5A). Conversely, expression of the two genes as well as CSRP2 was decreased in NFATc2-null MEF cells (Figure 5B). ChIP assays revealed that HSF1 and NFATc2 directly bind to region 2 (−298 to −179) and region 3 (−500 to −294) of the PDZK3 promoters, respectively (Figure 5C; Supplementary Figure 7). Similarly, HSF1 and NFATc2 directly bind to region 2 (−297 to −91) and region 1 (−93 to −69) of the CRYAB promoters, respectively (Figure 5D; Supplementary Figure 7). Heat shock induced binding of not only HSF1, but also NFATc2 to both promoters. HSF1 or NFATc2 binding to the promoters was not affected by the other factor under control and heat shock conditions. These results showed that HSF1 and NFATc2 independently bind to the PDZK3 and CRYAB genes before and after heat shock.
Figure 5.
Both HSF1 and NFATc2 are required for marked activation of PDZK3 and CRYAB. (A) RT–PCR of novel HSF1-target genes in HeLa cells expressing tetracycline-off NFATc2-HA. Expression levels relative to those in control cells are shown. (B) RT–PCR of novel HSF1-target genes in NFATc2-null MEF cells. Expression levels are shown as in (A). The averages of three experiments are shown with P-values using an unpaired t-test. Error bars show the mean + s.d. in (A) and (B). (C) In vivo binding of HSF1 and NFATc2 in the PDZK3 promoter. ChIP of control and heat-shocked (42°C, 15 min) WT, HSF1-null (left), and NFATc2-null (right) MEF cells was performed using a preimmune serum (p.i.), an antiserum for HSF1, or that for NFATc2. DNA fragments of three regions (R1, R2, R3) were amplified by PCR using primers listed in Supplementary Table 4. (D) In vivo binding of HSF1 and NFATc2 in the CRYAB promoter. ChIP assay was performed as above. DNA fragments of two regions (R1, R2) were amplified by PCR. (E) RT–PCR of PDZK3 and CRYAB mRNA in wild-type, HSF1-null, and NFATc2-null MEF cells overexpressing hHSF1ΔRDT, NFATc2, or both (upper). Levels of HSF1 and NFATc2 proteins are shown (lower). Error bars show the mean +s.d.
Remarkably, the level of PDZK3 mRNA in NFATc2-null cells overexpressing only an active hHSF1 was similar to that in wild-type cells, whereas it was significantly increased in the same cells overexpressing both an active hHSF1 and NFATc2 (Figure 5E). Although the expression of CRYAB mRNA was slightly induced by overexpressing an active hHSF1 in NFATc2-null cells, it was markedly induced by overexpression of both factors. The cooperative effect on the expression of PDZK3 and CRYAB mRNA was also observed when we overexpressed an active hHSF1 and/or NFATc2 in HSF1-null cells. These results suggested that both HSF1 and NFATc2 may be required for efficient expression of PDZK3 and CRYAB.
We next examined the expression of target genes in mice. The expression of PDZK3 and CRYAB mRNAs was decreased in both HSF1-null and NFATc2-null mice in the cerebral cortex, heart, skeletal muscle, and spleen (Supplementary Figure 8A–E). The expression of NFATc2 mRNA was also decreased in various tissues of HSF1-null mice, though not in the skeletal muscle. Furthermore, the expression of NFATc2 and PDZK3 mRNA was increased in the skeletal muscle of HSF1 transgenic mice (Supplementary Figure 8F) (Fujimoto et al, 2005). Thus, the expression of these genes is under the control of HSF1 in vivo in mice.
PDZK3 and CRYAB promote degradation of polyQ protein
We next examined whether the overexpression of PDZK3 and CRYAB can reverse the accelerated aggregation in NFATc2-null cells and found that it greatly inhibited polyQ aggregation, similar to the overexpression of NFATc2 (Figure 6A). Furthermore, the knockdown of PDZK3 mRNA resulted in a marked increase in polyQ aggregates and that of CRYAB mRNA a moderate increase (Figure 6B). These results clearly indicated that both HSF1 and NFATc2 suppress polyQ aggregation in part by up-regulating the expression of PDZK3 and CRYAB genes.
Figure 6.
PDZK3 and CRYAB suppress polyQ aggregation by promoting degradation of polyQ protein. (A) Re-expression of NFATc2, PDZK3, and CRYAB in NFATc2-null MEF cells reversed the effects of NFATc2 deficiency, more than that of hHSF1ΔRDT. At 48 h after infection with a retrovirus expressing each indicated gene, NFATc2-null cells were infected with Ad-polyQ-GFP for 24 h. The percentage of cells with polyQ inclusions (left), accumulation of insoluble polyQ protein (middle), and accumulation of insoluble ubiquitylated protein (right) are shown. The averages of three experiments are shown. P-values are calculated using an unpaired t-test. Error bars show the mean+s.d. (B) Knockdown of PDZK3 and CRYAB promotes polyQ aggregation. MEF cells were infected with each Ad-shRNA (Supplementary Table 5) for 2 h, and then maintained with normal medium for 46 h. After re-plated on new dishes for 24 h, the cells were infected with Ad-polyQ81-GFP and Ad-HA-Ub for 2 h, and then maintained with normal medium for 22 h. RT–PCR of the transcript was shown (left). The percentage of cells with polyQ inclusions, accumulation of insoluble polyQ protein, and accumulation of insoluble ubiquitylated protein are shown (right). The averages of three experiments are shown. P-values are calculated using an unpaired t-test. Error bars show the mean ± s.d. (C) PDZK3 and CRYAB form a complex with polyQ protein. HEK293 cells transfected with expression vectors pClneoMyc-PDZK3, and EGFP-N1 or polyQ81-EGFP-N1. After treatment with MG132 for 4 h, the cells were harvested, and immunoprecipitation was performed using an antibody for GFP (αGFP) or a non-specific antibody (αNS). Precipitated proteins were subjected to western blotting using each indicated antibody. (D) Knockdown of PDZK3 and CRYAB stabilizes polyQ protein. MEF cells infected with Ad-shRNA for PDZK3 or CRYAB for 46 h were co-infected with Ad-polyQ81-GFP protein. At 6 h after the infection, cells were treated with cycloheximide for the periods indicated. PolyQ-GFP protein and β-actin were detected by western blotting (lower), and polyQ-GFP levels were quantified (upper). The averages of three experiments are shown with P-values calculated by ANOVA. Error bars show the mean±s.d.
PDZK3, called PDZD2 or PAPIN, has six PDZ domains (Deguchi et al, 2000; Yeung et al, 2003). As PDZ domains are protein–protein recognition modules (Harris and Lim, 2001), PDZK3 could be a scaffold of polyQ protein or ubiquitylated cellular proteins for degradation, such as the PDZ-containing PDLIM2 (Tanaka et al, 2007). Furthermore, αB-crystallin, the product of the CRYAB, is a component of the E3 ubiquitin ligase complex (Lin et al, 2006). We found that CRYAB bound to both GFP and polyQ-GFP, and PDZK3 bound only to polyQ-GFP (Figure 6C). To clarify whether this complex is required for the degradation of polyQ protein, we investigated the amounts of polyQ protein after the treatment of cells with cycloheximide. We found that the degradation of polyQ protein was markedly inhibited in cells, in which PDZK3 or αB-crystallin expression was down-regulated (Figure 6D). These results showed that PDZK3 and CRYAB form a complex with polyQ protein and are required for the degradation of polyQ protein.
Discussion
Individual Hsps suppress polyQ-induced protein misfolding and the resultant stress on proteostasis, whereas the combined overexpression of Hsps such as Hsp70 and Hsp40 has a synergistic effect (Muchowski and Wacker, 2005; Williams and Paulson, 2008). Therefore, it would be reasonable for gain of HSF1 function to suppress polyQ aggregation and the resultant stress. In fact, knockdown of Hsp70 (hsp-1) or a small Hsp (hsp-16.1) had only modest effects on polyQ aggregates and lifespan in C. elegans, whereas knockdown of HSF1 resulted in a remarkable acceleration of polyQ aggregation and reduction of lifespan (Hsu et al, 2003; Morley and Morimoto, 2004). In R6/2 mice, overexpression of Hsp70 or Hsp27 had little effect on polyQ aggregation in the brain, neuronal phenotype, or lifespan (Hay et al, 2004; Zourlidou et al, 2007). In contrast, overexpression of an active HSF1 even in non-neuronal tissues such as heart and muscle tissues resulted in a marked reduction in polyQ aggregates in these tissues and lengthening of lifespan (Fujimoto et al, 2005). Inversely, lack of Hsp70 had a modest effect on the lifespan of R6/2 mice (Wacker et al, 2009), whereas HSF1 deficiency resulted in a marked reduction in lifespan (Figure 4). These observations would be consistent with the idea that HSF1 suppresses protein misfolding and the resultant stress on proteostasis by up-regulating the expression of a set of classical heat shock genes (Morimoto, 2008). Here, we showed for the first time that non-classical heat shock genes suppress polyQ aggregation (Figure 1). Remarkably, 8 (28%) of the 29 HSF1-target genes that we examined had a profound effect on polyQ aggregates, and NFATc2 was required for HSF1-mediated suppression of polyQ aggregation (Figures 2 and 3). These observations explain why HSF1 has a much stronger effect on protein misfolding than individual or multiple Hsps (Fujimoto et al, 2005).
The NFAT family consists of five members (Hogan et al, 2003). The primordial family member is NFAT5, the only NFAT-related protein (DmNFAT) present in Drosophila. DmNFAT was identified as a suppressor of polyQ aggregation in Drosophila (Bilen and Bonini, 2007), but is functionally different from the other four members. It is identical to mammalian tonicity element-binding protein and responsive to hypotonic stress (López-Rodríguez et al, 1999). In contrast, the remaining four NFAT proteins are regulated by calcineurin, a Ca2+-dependent phosphatase. Upon an increase in calcium, they are dephosphorylated and translocated to the nucleus, in which they activate target genes. In addition to their developmental functions in immune and non-immune tissues (Hogan et al, 2003), the NFAT proteins function in cellular adaptations to environmental changes (Horsley and Pavlath, 2002). However, the molecular mechanisms of these adaptations are poorly understood. Here, we showed that NFATc2 has a significant function in the maintenance of proteostasis. An FOXO-family transcription factor DAF-16, which is a component of the insulin-like signalling pathway that regulates lifespan, inhibits polyQ aggregates, indicating that ageing and proteostasis are highly related (Hsu et al, 2003; Morley and Morimoto, 2004; Cohen et al, 2006, 2009). Likewise, our results showed a relationship between regulation of calcium signalling and proteostasis, which was suggested in folding of proteins in the endoplasmic reticulum and lysosome (Egan et al, 2002; Mu et al, 2008).
NFATc2 is known to regulate the expression of many cytokine genes (Hogan et al, 2003). Unexpectedly, here we found that NFATc2 also regulates the expression of PDZK3 and CRYAB, which suppress polyQ aggregation (Figure 5). Catenin-related proteins interact with PDZ domain-containing PDZK3, suggesting it to be a scaffold protein connecting components of cell–cell junctions (Ohno et al, 2002). On the other hand, PDLIM2 containing a PDZ domain was recently shown to form an E3 ubiquitin ligase complex and degrades the p65 subunit of NF-κB (Tanaka et al, 2007), implying that PDZK3 could be a scaffold of polyQ protein for degradation. As was expected, we found that polyQ protein, PDZK3, and CRYAB form a complex, and PDZK3 and CRYAB are required for degradation of polyQ protein (Figure 6). These results show that NFATc2 controls proteostasis in part by regulating the degradation of polyQ protein, and suggest that PDZK3 could be a scaffold of some proteins for degradation.
One promising therapeutic strategy for protein-misfolding disorders such as polyQ diseases is to induce the expression of Hsps (Powers et al, 2009), and novel small molecules that activate HSF1 were recently identified and shown to ameliorate disease progression in vivo (Westerheide et al, 2004; Neef et al, 2010). However, the ectopic overexpression of an active HSF1 in the testis caused germ cell apoptosis, in part through the expression of a death-related gene (Nakai et al, 2000; Hayashida et al, 2006). Consistently, we could not generate transgenic mice or Xenopus highly expressing the active HSF1 in the brain (data not shown) (Dirks et al, 2010). These results suggest the activation of HSF1 at unnecessary levels to be detrimental to cells. Furthermore, HSF1 promotes tumour initiation and is required for maintaining proliferation and survival of the transformed cells (Dai et al, 2007; Min et al, 2007). Hsps induced by HSF1 can also potentiate oncogenesis in a variety of ways such as influencing the apoptotic pathways (Mosser and Morimoto, 2004). These observations imply that HSF1 can be detrimental to organisms in some cases. Therefore, it is of great value to distinguish the HSF1-mediated pathways that regulate proteostasis. Our results suggest that pharmacological regulation of NFATc2, which is activated by calcineurin, would be a new therapeutic strategy for protein-misfolding diseases.
Materials and methods
Screening of HSF1-target genes
HeLa cells were maintained in DMEM containing 10% FBS. Stable HeLa clones were generated using pcDNA3.1/cHSF1 and pcDNA3.1/hHSF1ΔRDT (Fujimoto et al, 2005). Total RNA was prepared from control HeLa cells, a HeLa clone expressing cHSF1, or two independent clones expressing hHSF1ΔRDT. Microarray analyses were carried out as described (Takii et al, 2010), and the results confirmed by RT–PCR. Primers used are listed in Supplementary Tables 2 and 3.
Constructions of retroviral and adenoviral vectors
An XhoI/NotI fragment of polyQ81-GFP (Fujimoto et al, 2005) was inserted into the vector pShuttle-CMV (Stratagene) at SalI/NotI sites. Viral DNA containing cDNA for polyQ81-GFP was generated with the AdEasy adenoviral vector system (Stratagene) according to the instructions supplied. Viruses were infected into HEK293 cells, and the viral particles were enriched by CsCl gradient centrifugation and stored at −80°C until used.
To generate retroviruses, we used the vector tgLS(+)HyTK-CMV (Li et al, 1997). cDNA for HSF1-target genes was amplified by RT–PCR using total RNA isolated from MEF or HeLa cells, with the exception of the cDNA for NFATc2, CSPG4, PROM2, and PDZK3 (gifts from Drs J Redondo, A Nishiyama, D Corbeil, and H Hata, respectively) and were inserted at BamHI/ClaI or ClaI sites of the vector. Retroviral vectors were transfected using Lipofectamine 2000 (Invitrogen) into Plat-E cells (a gift from T Kitamura, University of Tokyo). Supernatants containing retroviruses were collected at 48 h after medium change and stored at −80°C.
Analysis of polyQ aggregation
To search genes suppressing polyQ aggregation, MEF cells were infected with a retrovirus expressing each HSF1-target gene in medium supplemented with 8 μg/ml of polybrene for 24 h, and then the culture medium was replaced. At 48 h after the infection, the adenovirus expressing polyQ81-GFP or GFP was infected. Cells with inclusion were counted at 24 h after the adenoviral infection (Fujimoto et al, 2005).
Detection of soluble and insoluble polyQ81-GFP protein
Cell extract was prepared in NP40 lysis buffer and centrifuged for 10 min at 15 000 r.p.m. (Carra et al, 2008). The supernatant was used as the soluble fraction. The resulting pellet was washed five times with the NP40 lysis buffer, sonicated in the same buffer in a volume equivalent to that of the soluble fraction, and used as the insoluble fraction. Same volumes of soluble and insoluble fractions were subjected to western blotting, and signals were quantified using NIH Image.
Detection of ubiquitinated protein
A KpnI/XhoI fragment of pCAGGS-HA-Ub, an expression vector for HA-tagged ubiquitin (a gift from Dr K Iwai, Osaka University), was inserted into the vector pShuttle-CMV (Stratagene) at the same sites. A viral DNA containing HA-Ub cDNA and an Ad-HA-Ub virus were generated as described above. To detect ubiquitylated protein, HA-tagged ubiquitin was expressed in cells by infecting them with Ad-HA-Ub, and cell extracts were subjected to western blotting using an antibody for HA.
Establishment of HSF1- and NFATc2-null R6/2 mice
The transgenic mouse line R6/2, which was originally transgenic for the human huntingtin gene carrying 154 CAG repeats (Mangiarini et al, 1996), was obtained from Jackson Laboratory (Bar Harbor, ME). After maintaining R6/2 mice by ovary transplantation for several years (Fujimoto et al, 2005), we found that the number of CAG repeats decreased 95–97-fold in one line, which showed a decrease in the severity of the neurological phenotype. The R6/2 male mice (CBA × C57BL/6 background) were crossed with HSF1+/− female mice (CBA background) (Inouye et al, 2003) and HSF1+/− R6/2 mice were generated. To generate HSF1−/− R6/2 mice, the HSF1+/− R6/2 male mice were crossed again with HSF1+/− female mice (CBA background). Thus, the resulting mice with different six genotypes including HSF1KO/non-Htt, HSF1KO/Htt, and HSF1-hetero/Htt had mixed background of CBA × C57BL/6 (87.5:12.5%). We examined the lifespan of all mice having six genotypes. NFATc2−/− R6/2 mice were similarly generated using NFATc2+/− female mice (BALB/c background) (a gift from Dr G Pavlath, Emory University and Dr L Glimcher, Harvard Medical School) (Hodge et al, 1996), and the resulting mice had the background of BALB/c × CBA × C57BL/6 (75:12.5:12.5%). All experimental protocols related with these mice were reviewed by the Committee for Ethics on Animal Experiments of Yamaguchi University Graduate School of Medicine.
Immunofluorescence
Immunofluorescence microscopy was performed as described previously (Fujimoto et al, 2005). Briefly, the mouse brain was dissected and immediately frozen in OCT compound. Sections 10-μm thick were cut using a CM1900 cryostat (Leica). Immunohistochemistry was carried out by using a goat anti-huntingtin polyclonal antibody (N-18, Santa Cruz).
Filter trap assay
The brains from 8-week-old mice were homogenized with RIPA buffer (50 mM Tris–HCl, pH8.0, 150 mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitors. The extracts were incubated with 1 × SDS-sample buffer at 95°C for 5 min, and then applied onto a cellulose acetate membrane (0.2 μm pore, Whatman, Germany) prewashed three times with wash buffer (10 mM Tris–HCl, pH8.0, 150 mM NaCl, 50 mM DTT) containing 0.1% SDS by using a slot blot manifold (Bio-Dot SF, Bio-Rad) (Wacker et al, 2009). Membranes were washed three times with the same wash buffer, and subjected to immunoblotting by using a goat anti-huntingtin polyclonal antibody (N-18, Santa Cruz).
Immunoprecipitation
HEK293 cells plated in 100-mm dishes were co-transfected for 44 h with expression vectors pClneoMyc-PDZK3 and EGFP-N1 or polyQ81-EGFP-N1 using Lipofectamine 2000 (Invitrogen). After treatment with MG132 (20 μM) for 4 h, the cells were washed with PBS, suspended in NP-40 lysis buffer, and centrifuged (Fujimoto et al, 2010). The supernatant (500 μl) was added to 1 μl of a rabbit polyclonal antibody for GFP (MBL, Nagoya, Japan) or a rabbit non-specific antibody at 4°C for 1 h, and mixed with 40 μl of protein A-Sepharose beads (1:1 suspension in PBS, Amersham Biosciences, Piscataway, NJ) by rotating at 4°C for 1 h. The complexes were washed with NP-40 lysis buffer, suspended in SDS-sample buffer, and boiled. The samples were subjected to western blotting.
Statistical analysis
Data were analysed with Student's t-test and ANOVA. Error bars represent the s.d. for more than three independent experiments.
Accession number
Gene Expression Omnibus: full microarray data are deposited under the accession number GSE19797.
Supplementary Material
Acknowledgments
We are grateful to Drs T Kitamura, K Iwai, G Pavlath, L Glimcher, A Rao, J Redondo, A Nishiyama, D Corbeil, H Hata, and H Kampinga for reagents. This work was supported in part by Grants-in-aid for Scientific Research and on Priority Area-a Nuclear System of DECODE, from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Yamaguchi University Research Project on STRESS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akerfelt M, Trouillet D, Mezger V, Sistonen L (2007) Heat shock factors at a crossroad between stress and development. Ann NY Acad Sci 1113: 15–27 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448: 704–708 [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM (2007) Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet 3: 1950–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S (2005) Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol 6: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J (2008) HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macro- autophagy. J Biol Chem 283: 1437–1444 [DOI] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A (2006) Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A (2009) Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S (2007) Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi M, Iizuka T, Hata Y, Nishimura W, Hirao K, Yao I, Kawabe H, Takai Y (2000) PAPIN. A novel multiple PSD-95/Dlg-A/ZO-1 protein interacting with neural plakophilin- related armadillo repeat protein/delta-catenin and p0071. J Biol Chem 275: 29875–29880 [DOI] [PubMed] [Google Scholar]
- Dirks RP, van Geel R, Hensen SM, van Genesen ST, Lubsen NH (2010) Manipulating heat shock factor-1 in Xenopus tadpoles: neuronal tissues are refractory to exogenous expression. PLoS One 5: e10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ME, Glöckner-Pagel J, Ambrose C, Cahill PA, Pappoe L, Balamuth N, Cho E, Canny S, Wagner CA, Geibel J, Caplan MJ (2002) Calcium-pump inhibitors induce functional surface expression of Delta F508-CFTR protein in cystic fibrosis epithelial cells. Nat Med 8: 485–492 [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, McCall A, Canal I, Orr HT, Zoghbi HY, Botas J (2000) Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408: 101–106 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Hayashida N, Katoh T, Oshima K, Shinkawa T, Prakasam R, Tan K, Inouye S, Takii R, Nakai A (2010) A novel mouse HSF3 has the potential to activate non-classical heat shock genes during heat shock. Mol Biol Cell 21: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A (2005) Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem 280: 34908–34916 [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BZ, Lim WA (2001) Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci 114: 3219–3231 [DOI] [PubMed] [Google Scholar]
- Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP (2004) Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet 13: 1389–1405 [DOI] [PubMed] [Google Scholar]
- Hayashida N, Inouye S, Fujimoto M, Tanaka Y, Izu H, Takaki E, Ichikawa H, Rho J, Nakai A (2006) A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J 25: 4773–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge MR, Ranger AM, Charles de la Brousse F, Hoey T, Grusby MJ, Glimcher LH (1996) Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 4: 397–405 [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Gene Dev 17: 2205–2232 [DOI] [PubMed] [Google Scholar]
- Homma S, Jin X, Wang G, Tu N, Min J, Yanasak N, Mivechi NF (2007) Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J Neurosci 27: 7974–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Pavlath GK (2002) NFAT: ubiquitous regulator of cell differentiation and adaptation. J Cell Biol 156: 771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C (2003) Regulation of aging and age-related disease by daf-16 and heat-shock factor. Science 300: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A (2007) Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem 282: 33210–33217 [DOI] [PubMed] [Google Scholar]
- Inouye S, Katsuki K, Izu H, Fujimoto M, Sugahara K, Yamada S, Shinkai Y, Oka Y, Katoh Y, Nakai A (2003) Activation of heat shock genes is not necessary for protection by heat shock transcription factor 1 against cell death due to a single exposure to high temperatures. Mol Cell Biol 23: 5882–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S (2000) Genetic suppression of polyglutamine toxicity in Drosophila. Science 287: 1837–1840 [DOI] [PubMed] [Google Scholar]
- Li LP, Schlag PM, Blankenstein T (1997) Transient expression of SV 40 large T antigen by Cre/LoxP-mediated site-specific deletion in primary human tumor cells. Hum Gene Ther 8: 1695–1700 [DOI] [PubMed] [Google Scholar]
- Lin DI, Barbash O, Kumar KG, Weber JD, Harper JW, Klein-Szanto AJ, Rustgi A, Fuchs SY, Diehl JA (2006) Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF(FBX4-alphaB crystallin) complex. Mol Cell 24: 355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Rodríguez C, Aramburu J, Rakeman AS, Rao A (1999) NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA 96: 7214–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87: 493–506 [DOI] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ (1998) Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem 273: 7523–7528 [DOI] [PubMed] [Google Scholar]
- Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF (2007) Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26: 5086–5097 [DOI] [PubMed] [Google Scholar]
- Morimoto RI (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Morimoto RI (2008) Proteotoxic stress and inducible chaperone networks in neuro-degenerative disease and aging. Genes Dev 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Morimoto RI (2004) Molecular chaperones and the stress of oncogenesis. Oncogene 23: 2907–2918 [DOI] [PubMed] [Google Scholar]
- Mu TW, Fowler DM, Kelly JW (2008) Partial restoration of mutant enzyme homeostasis in three distinct lysosomal storage disease cell lines by altering calcium homeostasis. PLoS Biol 6: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL (2005) Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci 6: 11–22 [DOI] [PubMed] [Google Scholar]
- Nakai A (2009) Heat shock transcription factors and sensory placode development. BMB Rep 42: 631–635 [DOI] [PubMed] [Google Scholar]
- Nakai A, Kawazoe Y, Tanabe M, Nagata K, Morimoto RI (1995) The DNA-binding properties of two heat shock factors, HSF1 and HSF3 are induced in the avian erythroblast cell line HD6. Mol Cell Biol 15: 5168–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Suzuki M, Tanabe M (2000) Arrest of spermatogenesis in mice expressing an active heat shock transcription factor 1. EMBO J 19: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Turski ML, Thiele DJ (2010) Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol 8: e1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH (2004) Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci USA 101: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Hirabayashi S, Iizuka T, Ohnishi H, Fujita T, Hata Y (2002) Localization of p0071-interacting proteins, plakophilin-related armadillo-repeat protein-interacting protein (PAPIN) and ERBIN, in epithelial cells. Oncogene 21: 7042–7049 [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE (2009) Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem 78: 959–991 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Steele AD, Hutter G, Jackson WS, Heppner FL, Borkowski AW, King OD, Raymond GJ, Aguzzi A, Lindquist S (2008) Heat shock factor 1 regulates lifespan as distinct from disease onset in prion disease. Proc Natl Acad Sci USA 105: 13626–13631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takii R, Inouye S, Fujimoto M, Nakamura T, Shinkawa T, Prakasam R, Tan K, Hayashida N, Ichikawa H, Hai T, Nakai A (2010) Heat shock transcription factor 1 inhibits induction of IL-6 through inducing activation transcription factor 3. J Immunol 184: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Tanabe M, Kawazoe Y, Takeda S, Morimoto RI, Nagata K, Nakai A (1998) Disruption of the HSF3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance. EMBO J 17: 1750–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Grusby MJ, Kaisho T (2007) PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol 8: 584–591 [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM (2004) The role of heat shock transcription factor 1 in the Genome-wide regulation of the mammalian heat shock response. Mol Biol Cell 15: 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihma H, Pruunsild P, Timmusk T (2008) Alternative splicing and expression of human and mouse NFAT genes. Genomics 92: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker JL, Huang SY, Steele AD, Aron R, Lotz GP, Nguyen Q, Giorgini F, Roberson ED, Lindquist S, Masliah E, Muchowski PJ (2009) Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J Neurosci 29: 9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Bosman JD, Mbadugha BN, Kawahara TL, Matsumoto G, Kim S, Gu W, Devlin JP, Silverman RB, Morimoto RI (2004) Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem 279: 56053–56060 [DOI] [PubMed] [Google Scholar]
- Williams AJ, Paulson HL (2008) Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 31: 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ (2003) Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302: 1769–1772 [DOI] [PubMed] [Google Scholar]
- Wu C (1995) Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Yeung ML, Tam TS, Tsang AC, Yao KM (2003) Proteolytic cleavage of PDZD2 generates a secreted peptide containing two PDZ domains. EMBO Rep 4: 412–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourlidou A, Gidalevitz T, Kristiansen M, Landles C, Woodman B, Wells DJ, Latchman DS, de Belleroche J, Tabrizi SJ, Morimoto RI, Bates GP (2007) Hsp27 overexpression in the R6/2 mouse model of Huntington's disease: chronic neurodegeneration does not induce Hsp27 activation. Hum Mol Genet 16: 1078–1090 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.