Abstract
Dosage compensation is a strategy to deal with the imbalance of sex chromosomal gene products relative to autosomes and also between the sexes. The mechanisms that ensure dosage compensation for X-chromosome activity have been extensively studied in mammals, worms, and flies. Although each entails very different mechanisms to equalize the dose of X-linked genes between the sexes, they all involve the co-ordinate regulation of hundreds of genes specifically on the sex chromosomes and not the autosomes. In addition to chromatin modifications and changes in higher order chromatin structure, nuclear organization is emerging as an important component of these chromosome-wide processes and in the specific targeting of dosage compensation complexes to the sex chromosomes. Preferential localization within the nucleus and 3D organization are thought to contribute to the differential treatment of two identical homologs within the same nucleus, as well as to the chromosome-wide spread and stable maintenance of heterochromatin.
X-chromosome inactivation/up-regulation ensures balanced expression of sex-linked genes both between the sexes and relative to autosomal genes. Subnuclear localization and compartmentalization mechanisms help coordinately regulate the hundreds of genes involved.
Species that have evolved a chromosomally based sex determination system face the problem of dosage compensation of sex chromosomes between males and females. As the non-recombining chromosome degenerates, the heterogametic sex is left with only one functional copy of genes on the sex chromosomes compared with the double dose that exists in the homogametic sex. Diverse dosage compensation strategies have evolved in different species to deal with this problem and have been most extensively studied in flies (D. melanogaster), worms (C. elegans), and mammals. In flies, males (XY) up-regulate their single X to achieve expression levels equivalent to the two copies present in females (reviewed in Gelbart and Kuroda 2009). Increased transcriptional output of the X chromosome is also observed in worms (Gupta et al. 2006) and mammals (Nguyen and Disteche 2006; Lin et al. 2007). However, in these latter cases, hyper-expression is not specific to the heterogametic sex, as it occurs in both males and females, necessitating secondary compensation mechanisms in females who would otherwise over-express X-linked genes. In C. elegans, hermaphrodites (XX) down-regulate transcription from both their X chromosomes by half, whereas in female mammals, one of the two X chromosomes is chosen for inactivation (reviewed in Vicoso and Bachtrog 2009). Therefore, it is through the co-ordinate regulation of hundreds of genes across the chromosome that these compensation mechanisms restore not only the balance between sexes but also the ratio of gene dosage between the sex chromosomes and the autosomes. How dosage compensation complexes are targeted specifically to the sex chromosomes on a chromosome-wide scale is still not fully understood in any of these organisms. Changes in chromatin structure and higher-order chromosome architecture clearly participate, but it is also becoming increasingly clear that in all of these dosage compensation systems, nuclear organization may play a role.
NUCLEAR ORGANIZATION AND DOSAGE COMPENSATION
The eukaryotic nucleus is a highly organized organelle consisting of multiple compartments. In addition to more stable structures, such as the nuclear envelope and the nucleolus, it also contains numerous specialized functional compartments, where processes such as RNA transcription and processing, as well as DNA replication and repair, take place (reviewed in Spector 2003; Misteli 2007). Chromosomes themselves also exhibit a distinct 3D organization and can adopt non-random positions within the nucleus, both with respect to each other and with respect to specific nuclear landmarks. It is becoming clear that this spatial organization likely plays an important role in the regulation and expression of the genome and is believed to be a key factor in the chromosome-wide regulation that occurs during dosage compensation. In particular, the interphase organization of X-chromosome sequences may facilitate their differential activity compared to autosomes or between the sexes. Similarly, preferential localization in specialized nuclear sub-compartments may contribute to specific processes that affect the sex chromosomes more than the autosomes. In the case of mammals, nuclear compartmentalization is likely to facilitate the differential treatment of the two X chromosomes within the same nucleus.
Drosophila melanogaster
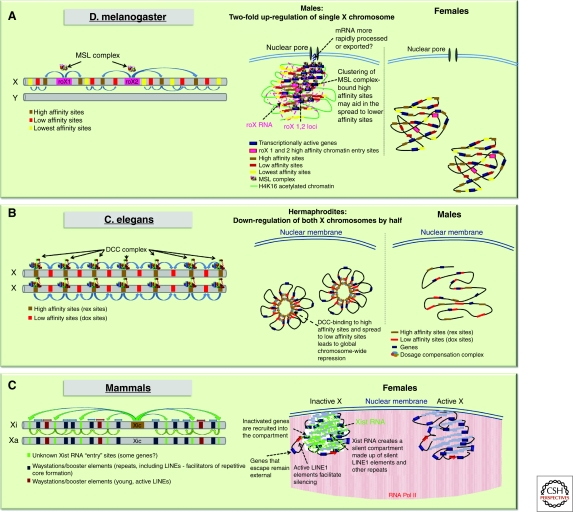
In Drosophila, dosage compensation is mediated by a ribonucleoprotein complex that binds hundreds of sites on the single X chromosome in males, and induces a two-fold up-regulation of X-chromosomal genes (Fig. 1A). The complex, known as the male-specific lethal (MSL) complex, is made up of five protein subunits (MSL1, MSL2, MSL3, MOF, MLE) and two non-coding RNAs (roX1, roX2) (reviewed in Gelbart and Kuroda 2009; Hallacli and Akhtar 2009). It can only form in male cells, due to the male-specific expression of the male-specific-lethal 2 (MSL2) protein. MSL2 is required to stabilize components of the complex such as MSL1 and roX RNAs. MOF and MLE are enzymatic components, MLE being an RNA/DNA helicase and MOF a histone acetyltransferase, responsible for acetylation of histone H4 at lysine 16 (H4K16Ac). The H4K16Ac chromatin modification is believed to be a key component in the transcriptional up-regulation across the X chromosome in males (Bone et al. 1994). MSL1 and MSL2 form the core of the complex and are thought to be the main targeting components; however, the full complex is required for complete localization to the X chromosome (Gelbart and Kuroda 2009). Targeting to the X chromosome is thought to occur through co-transcriptional assembly of the complex at the sites of roX1 and roX2 transcription on the X chromosome (Kelley et al. 1999). From these assembly sites, the complex then spreads to other high affinity binding sites on the X or chromatin entry sites (CES), and is thought to occur, at least partially, through the recognition of specific DNA sequence motifs (Alekseyenko et al. 2008; Straub et al. 2008) (Fig. 1A). Nevertheless, the identified DNA motifs alone are unable to unequivocally predict MSL complex binding since many motifs are not associated with MSL binding; therefore, other factors must also be involved. It has been proposed that nuclear organization and higher order chromatin structure may play a role in this regard, perhaps facilitating the spread of the complexes to the actively transcribed portions of the chromosome. In fact, the X chromosome has been found to adopt a male-specific 3D conformation with “clustering” of the high affinity MSL complex binding sites occurring in males, but not females (Grimaud and Becker 2009). In addition, MSL proteins co-purify with components of the nuclear pore complex, Nup153 and Megator (Mtor), and RNAi knockdowns have shown that these factors are essential for Msl-mediated dosage compensation in male cells (Mendjan et al. 2006). Although the nuclear periphery is often associated with heterochromatin and transcriptional silencing, there are sub-regions at the nuclear envelope, particularly at nuclear pores, that are associated with transcriptional activity (Taddei et al. 2004; Akhtar and Gasser 2007). Recently, the genome-wide chromatin binding profiles of Nup153 and Megator have been reported (Vaquerizas et al. 2010). These Nucleoporin-Associated Regions (NARs) represent regions of open chromatin as they are enriched in markers for active transcription such as RNA polymerase II and histone H4K16 acetylation. Interestingly, the male hyperactive X chromosome is particularly enriched in NARs and relies on the interaction with nucleoporins for its peripheral localization (Vaquerizas et al. 2010). The link between the MSL complex, nuclear pore components, and the nuclear periphery suggests that the X chromosome in males specifically localizes to nuclear sub-domains through its interactions with the nuclear pore and that this might contribute to its two-fold up-regulation. Thus, the enhanced expression of the X chromosome in males may be not only transcriptional via chromatin-mediated effects, but also post-transcriptional, perhaps through the more rapid processing and export of messenger RNA, thanks to the association with the nuclear pore.
Figure 1.
Diverse strategies for dosage compensation in different organisms. (A) In D. melanogaster, males (XY) up-regulate two-fold their single X chromosome. The dosage compensation complex has been found to be associated with nuclear pore proteins, which may indicate a participation of nuclear localization in this up-regulation. (B) C. elegans hermaphrodites (XX) down-regulate both X chromosomes by one half. This may be linked to a particular organization of the two Xs with the dosage compensation machinery. (C) Female mammals inactivate one of their two X chromosomes. The X chromosomes often reside at the nuclear periphery or at the nucleolus. The inactive X is organized as a silent, repetitive compartment by Xist RNA, into which genes are located as they become silenced.
Caenorhabditis elegans
In C. elegans, dosage compensation involves a two-fold reduction in X-chromosomal gene activity in hermaphrodites (Fig. 1B). The worm dosage compensation complex (DCC) consists of a hermaphrodite-specific core of SDC (sex determination and dosage compensation) proteins, required for the stabilization and targeting of the complex to the X chromosome, as well as a condensin subcomplex, which somehow induces the chromatin changes necessary for dosage compensation. How the DCC imposes a precise two-fold down-regulation of X-linked gene expression in a chromosome-wide manner remains unclear. The DCC is targeted to X chromosomes by initial binding to a number of recruiting elements, followed by dispersal or spreading to secondary sites. However, DCC binding does not correlate with DCC-mediated repression, implying that the complex acts in a chromosome-wide manner, rather than on a gene-by-gene basis. Indeed, it has been proposed that the DCC does not repress genes by direct binding, but rather acts at a distance and may involve looping of chromatin fibers to bring regulated genes to the vicinity of DCC bound sites (Jans et al. 2009). The implication of condensins suggests that X-chromosome down-regulation may involve similar mechanisms to mitotic chromosome condensation (Chuang et al. 1996; Lieb et al. 1996; Lieb et al. 1998; Tsai et al. 2008; Csankovszki et al. 2009). The DCC may alter higher order chromosome organization by inducing partial chromosome condensation at interphase, and this might in turn lead to decreased transcription. This is supported by the fact that the DCC condensin subunit differs from the mitotic condensin I complex by only a single subunit. Although the exact molecular mechanisms of condensin-mediated X chromosome down-regulation are still unclear, the consensus is that this process must involve some higher-order reorganization of the X chromosomes within the nucleus. Whether nuclear compartmentalization or localization also plays a role remains to be seen.
NUCLEAR ORGANIZATION OF THE INACTIVE X CHROMOSOME IN FEMALE MAMMALS
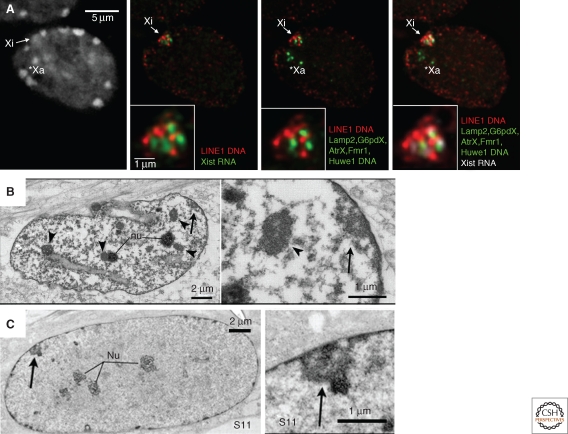
In mammals, the presence of an inactive X (Xi) chromosome in female cells was first noted at the cytological level by Barr in 1949 as a heteropycnotic body that was often at the nuclear periphery or within the perinucleolar region (Barr and Bertram 1949). These initial observations were already suggestive of a possible role for nuclear compartmentalization in the formation/maintenance of the inactive state (Comings 1968). The nuclear and perinucleolar compartments are indeed often associated with heterochromatin (de Wit and van Steensel 2009), and in the case of the mammalian X chromosome, this might promote the inactive state by facilitating access to heterochromatic factors or restricting access to transcription factors. The nucleolar localization of the Xi (Zhang et al. 2007; Rego et al. 2008) has been proposed to be important for faithful replication of the Xi’s epigenetic state, as the nucleolar periphery is enriched in factors required for replication of heterochromatin, such as Snf2h (Zhang et al. 2007). In addition to its preferential locations in the nucleus, the inactive X chromosome also appears to adopt a very distinctive 3D organization. In both mouse and humans, fluorescence in situ hybridization (FISH) studies have shown that the inactive X chromosome is coated by the non-coding Xist RNA, which triggers the initiation of X inactivation. Xist induces global changes in chromatin structure, and also mediates changes in the global organization of the chromosome, both of which are thought to play roles in chromosome-wide silencing. Xist RNA initially coats a central core of the X-chromosome territory that is made up largely of repetitive DNA (Chaumeil et al. 2006; Chow et al. 2010). X-linked genes are mostly excluded from this inner region, particularly at the early stages of inactivation and instead reside outside, or at the periphery, of this repetitive compartment (Chaumeil et al. 2006). However, as X inactivation proceeds and genes become silenced, they move into the Xist RNA coated, silent compartment. There is thus a progressive nuclear reorganization of the X chromosome as it becomes inactivated and heterochromatic, ultimately resulting in a condensed structure with a repetitive core and silent genes embedded into its periphery (Clemson et al. 2006) (Figs. 1C, 2A). The degree to which this particular 3D organization of the Xi is due to a compartmentalizing function of Xist RNA, or to the chromatin changes that are induced during X chromosome inactivation (XCI) or to the repetitive nature of the mammalian X chromosome, still remains to be seen.
Figure 2.
Nuclear organization of the inactive X chromosome in female mammals. (A) Combined RNA-DNA fluorescence in-situ hybridization in day 8 differentiated female ES cells shows the distinctive organization of the silent inactive X (Xi) compared to the active X (Xa). Both the Xa and the Xi often reside at the periphery of the nucleus. However, on the Xi, Xist up-regulation and coating results in the formation of a transcriptionally silent compartment made up of a central core of repetitive sequences, including LINE1 elements. Genes transcriptionally silenced at this stage (Lamp2, G6pdX, AtrX, Fmr1, and Huwe1) have been recruited into the compartment. All images shown are single slices taken from the same Z plane. (B,C) Electron micrographs of single 200nm sections from female WI-38 human fibroblasts. The inactive X chromosome (arrow) has an ultrastructure distinct from the chromocenters (arrowheads), adopting a looser, less compact organization. Adapted, with permission, from Rego et al. 2008, Journal of Cell Science.
HETEROGENEOUS HETEROCHROMATIN ON THE INACTIVE X CHROMOSOME
Inactivation of the X chromosome involves the acquisition of many chromatin changes that clearly distinguish it from its active homolog. Immunofluorescence studies in mitotic and/or interphase cells show that several histone marks typical of heterochromatin appear to be enriched on the Xi, including H3K27me3, H3K9me2, H3K9me3, H4K20me1, H2AK119Ub, and the histone variant macroH2A (reviewed in Heard and Disteche 2006). Conversely, euchromatic marks such as H3K4me2/3 and H3 and H4 acetylated lysines are largely depleted. However, these modifications are not uniformly distributed across the chromosome, suggesting that the inactive X chromosome is made up of several different types of heterochromatin. In human somatic cell lines, at least two different heterochromatic states have been characterized by immunofluorescence: One is defined by the presence of XIST RNA, macroH2A, H3K27me3, H4K20me1, and H2AK119Ub, and the other is defined by features more reminiscent of constitutive heterochromatin, including later replication timing, H3K9me3, H4K20me3, and association with HP1. These different heterochromatin domains appear to remain spatially distinct from each other, both during metaphase and interphase (Chadwick and Willard 2004; Chadwick 2007). Ultra-structural analyses of the H3K27me3-enriched portion of the Xi by light and electron microscopy in mouse and human fibroblasts (Rego et al. 2008) have shown that this portion of the heterochromatic X has a distinctive structure, being made up of densely packed fibers with intervening spaces and a mean chromatin compaction higher than euchromatin, but less than constitutive, H3K9me3-enriched heterochromatin (Fig. 2B, C). The significance of these different types of heterochromatin and whether they reflect differences in the epigenetic mechanisms underlying the initiation and/or maintenance of the inactive state is still unknown. The relative proportions of these two heterochromatic signatures varies considerably between species. In mouse, enrichment of HP1 and H3K9me3 on the Xi is much less distinctive than has been described in humans (Peters et al. 2002; Kohlmaier et al. 2004). Nevertheless, it should be noted that in both of these species, the inactive X appears to be similarly organized in the nucleus, with genes surrounding a silent repetitive core (Chaumeil et al. 2006; Clemson et al. 2006). In marsupials, although the Xi is clearly depleted for active marks as on the mouse and human Xi (Koina et al. 2009; Mahadevaiah et al. 2009), the degree of enrichment on the Xi for histone modifications characterizing heterochromatin (H3K9me2, H3K27me3, H3K9me3, H4K20me3) is less clear-cut, with some studies reporting no enrichment on the Xi at metaphase (Koina et al. 2009) and others reporting some enrichment of H3K27me3 at interphase (Mahadevaiah et al. 2009). Given that marsupials lack the XIST gene (Duret et al. 2006), differences in histone modification patterns on the Xi might be expected, since Xist is required for the recruitment of many of the chromatin marks in mouse. The situation in monotremes appears to be even more complex with females having five pairs of XX chromosomes and males having five XY pairs. Although there does appear to be some degree of dosage compensation, no systematic analysis has been performed to determine the heterochromatic status of the X chromosomes in females, and dosage compensation appears to be largely incomplete and variable between genes (Grutzner et al. 2004; Rens et al. 2004; Deakin et al. 2008).
A more detailed view of the chromatin content of the eutherian inactive X chromosome is starting to be obtained by higher resolution epigenomic analyses, such as ChIP-chip and allele-specific ChIP (Brinkman et al. 2006; Valley et al. 2006; Valley and Willard 2006; Chadwick 2007; Marks et al. 2009; Mietton et al. 2009). ChIP-chip for macroH2A1 in mouse liver cells reveals an approximately 1.5-fold enrichment in females versus males. As this enrichment is distributed uniformly across the entire X chromosome, this implies that macroH2A1 may influence global chromatin structure, rather than directly inhibiting transcription at promoters of genes (Mietton et al. 2009). The distribution of H3K27me3 has also been examined on the inactivating X chromosome in differentiating mouse ES cells by ChIP-Seq (Marks et al. 2009). Allele-specific mapping of ChIP-seq tags showed that the H3K27me3 mark is deposited on the inactive X chromosome in female ES cells, particularly in gene-rich, active regions, which is in agreement with the results observed in human somatic cells by immunofluorescence (Chadwick 2007; Marks et al. 2009).
DNA methylation of promoters of X-linked genes is another hallmark of XCI in eutherian somatic cells. Global microarray analyses involving methylated DNA immunoprecipitation (MeDIP) were used to assess the DNA methylation status of the Xi relative to the active X (Xa) in human primary cells. This study revealed that although CpG islands are hypermethylated on the Xi, the overall levels of methylation on the X chromosome are in fact lower in females compared to males, especially in gene-poor regions (Weber et al. 2005). Hellman and Chess (2007) analyzed the DNA methylation status of more than 1000 X-linked loci and found that on the active X, DNA methylation is concentrated in gene bodies, confirming previous studies (for review, see Heard et al. 1997). Although the function of methylation within transcribed genes is unknown, it is a general feature of active genes in many organisms (for example, Zhang et al. 2006). The lack of gene-body DNA methylation on the inactive X chromosome at least partly explains its globally hypomethylated status, with only promoters of genes being hyper-methylated.
In summary, the inactive X chromosome contains many different epigenetic marks, with some variations in distribution between different mammals. The degree to which the different signatures on the Xi reflect differences in the higher-order structure and/or nuclear organization of the inactive X should become more clear with the advent of techniques such as chromosome conformation capture, which measures physical interaction within the chromatin fiber (Simonis et al. 2007; van Berkum and Dekker 2009).
INITIATING X INACTIVATION: XIST RNA REGULATION AND FUNCTION
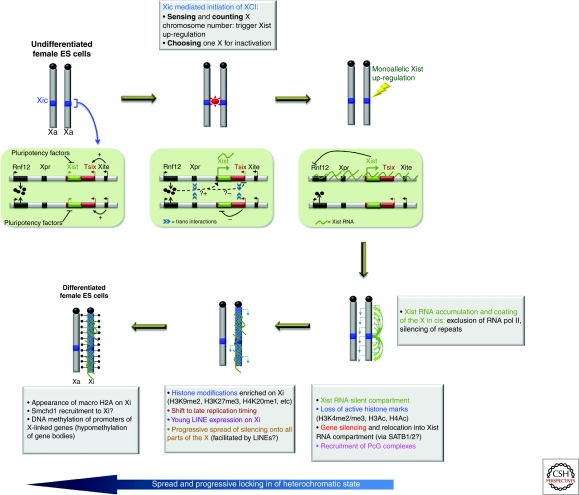
The evolution of the Xist gene is believed to have been a key event in the evolution of the stable, chromosome-wide X-inactivation process that exists in eutherian mammals. Xist RNA coating of the prospective inactive X chromosome appears to be the key trigger for the X-inactivation cascade. Deletion analyses have shown that Xist is essential for silencing and likely has a role at multiple levels in the process (Penny et al. 1996; Marahrens et al. 1997; Csankovszki et al. 1999; Wutz et al. 2002; Hoki et al. 2009). The regulation of Xist during development is a complex process, ensured by multiple long-range cis elements and trans-acting factors. Xist is in fact located in a region of the X chromosome known as the X-inactivation center (Xic) that ensures the appropriate initiation of X inactivation in female cells (reviewed in Barakat et al. 2010) (Fig. 3). The Xic contains sequence elements that ensure that Xist is only up-regulated in XX and not XY cells. The Xic also ensures that in XX cells, Xist up-regulation occurs on only one and not both X chromosomes. Recent insights into this process have revealed that the presence of two or more copies of certain X-linked loci can activate Xist. This may work at several levels. For example, a double dose of the X-linked Rnf12 gene, which encodes a ubiquitin ligase, can trigger Xist up-regulation (Jonkers et al. 2009). Trans interactions (pairing) between Xics may also be involved in triggering the expression of Xist and in ensuring its monoallelic regulation in female cells (Bacher et al. 2006; Xu et al. 2006; Augui et al. 2007). These transient Xic pairing events may facilitate the coordination of reciprocal Xist expression patterns on the future active and inactive X chromosomes during the initiation of X inactivation (Augui et al. 2007; Nicodemi and Prisco 2007; Xu et al. 2007). Thus, the initiation of monoallelic Xist expression in female cells involves a dynamic series of events at the Xic loci, integrating developmental triggers, dosage-dependent proteins factors, interactions in trans, and non-coding RNAs (Fig. 3).
Figure 3.
Random X chromosome inactivation is a multi-stage process. During the initiation stage, developmental factors as well as a complex interplay of elements within the X inactivation center (Xic) ensure that monoallelic Xist up-regulation occurs only when more than one X chromosome is present per diploid autosome set. Once up-regulated, Xist RNA coats the chromosome in cis and triggers chromosome-wide gene silencing. The spread of silencing occurs progressively with the acquisition of many different epigenetic changes that together serve to create an extremely stable, heterochromatic state.
Once Xist RNA has become monoallelically up-regulated (at around day 2 in differentiating female ES ells), it begins the process of chromosome-wide gene silencing. Xist RNA induces X inactivation during an early differentiation time window, and recent studies have implicated the SATB1 and SATB2 (Special AT-rich Binding Protein) proteins in enabling Xist function during this early differentiation time window and also in some cancer cells (Agrelo et al. 2009). The 17,000–19,000nt-long, spliced, poly-adenylated Xist transcript consists of multiple different functional domains (Wutz et al. 2002). The most conserved region of Xist is a set of tandem repeats at the 5′ end of the gene, called the A-repeats, that are required for its gene silencing function (Wutz et al. 2002; Hoki et al. 2009). Inducible Xist transgenes deleted for A-repeats are unable to induce gene silencing, but can still coat the chromosome in cis and recruit polycomb group proteins, which are involved in the recruitment of repressive chromatin changes including H3K27me3 to the X chromosome (Wutz et al. 2002; Plath et al. 2003; Kohlmaier et al. 2004). Thus, other portions of Xist RNA are required for its capacity to coat the X chromosome in cis and to recruit polycomb and macroH2A (Wutz et al. 2002). The Xist A-repeat region also produces an independent 1.6-kb transcript, RepA, and although its function is not entirely clear, this transcript may play a role in regulating Xist itself (Zhao et al. 2008; Hoki et al. 2009).
Although Xist RNA may be involved in directly or indirectly recruiting chromatin-modifying enzymes, including polycomb group proteins, so far there is no evidence that this accounts for its gene-silencing role. However, there is increasing evidence pointing to Xist RNA having an architectural role that may be important for the specific 3D-organization of the inactive X (Chaumeil et al. 2006; Clemson et al. 2006; Rego et al. 2008), which may impact both the initiation and maintenance of the inactive state. One of the earliest events following Xist up-regulation and coating of the future inactive X chromosome is the formation of a nuclear compartment that is depleted for RNA polymerase, transcription factors, and euchromatic marks (Chaumeil et al. 2006). Based on the analysis of Xist and PcG mutant ES cell lines, gene silencing and the recruitment of polycomb group proteins can be functionally dissociated from the formation of the Xist silent nuclear compartment (Plath et al. 2003; Kohlmaier et al. 2004; Chaumeil et al. 2006; Schoeftner et al. 2006). The silent compartment initially encompassed by Xist RNA is in fact largely constituted of X-chromosome repeat sequences, rather than genes. Prior to their inactivation, X-linked genes actually reside outside or at the periphery of this Xist RNA compartment. As differentiation proceeds and X inactivation occurs, genes move into this Xist RNA domain as they become silenced (Chaumeil et al. 2006). The mechanisms underlying this relocalization of genes are currently unknown. The Xist A-repeat region, which is essential for gene silencing, may have a role since genes do not become relocated into the silent repetitive compartment in Xist A-repeat mutants. Furthermore, given the implication of the SATB1 and SATB2 factors in Xist RNA’s competence to induce gene silencing during early differentiation (Agrelo et al. 2009), and the fact that these DNA-binding proteins facilitate the organization of chromatin structure during T-cell development (Cai et al. 2006), they may similarly play a role in gene relocation into the silent Xist repetitive compartment during X inactivation. So far, no physical association between Xist A-repeats and SATB1/2 has been demonstrated, but their window of action and their known function in nuclear organization make them key candidates for a role in Xist-mediated gene silencing and relocation. Importantly, recent findings demonstrate that Xist RNA can actually induce gene silencing outside an early developmental context, in some cancer cells (Hall et al. 2002; Chow et al. 2007), and in adult progenitor stem cells (Savarese et al. 2006). Agrelo et al. (2009) show that this reacquired capacity for Xist-induced silencing in tumor cells is associated with the re-expression of the SATB1 protein. This finding reinforces the notion that the nuclear and epigenetic plasticity found in some cancer cells may be similar to that found during early embryogenesis.
SPREADING INACTIVATION ACROSS THE X CHROMOSOME
Early studies using inducible Xist genes in male ES cells demonstrated that Xist had a developmentally restricted function, limited to the first few days of differentiation (Wutz and Jaenisch 2000). X-inactivation was thus originally thought to be a rather concerted process, occurring in the space of just a few cell divisions. However, recent studies in mouse pre-implantation embryos (Patrat et al. 2009) and differentiating female ES cells (Lin et al. 2007; Chow et al. 2010) have revealed that inactivation of the X chromosome in its entirety can take much longer. Some genes are silenced very quickly (in a matter of days or even hours after Xist RNA accumulation), while others are silenced much later (days, or >1 week), well outside of the time window in which Xist RNA is thought to act (Lin et al. 2007; Patrat et al. 2009). This suggests either that Xist’s silencing function is not limited to early differentiation for some loci, or else that the changes induced by Xist are propagated more slowly into some regions compared to others. Such heterogeneity suggests that the kinetics of inactivation are controlled by region-specific processes, presumably due to different sequence environments. In addition, several X-linked genes are known to escape the X-inactivation process either partly or fully (Yang et al. 2010). Some of these genes (e.g., Jarid1c/Kdm5c) have the capacity to escape from X inactivation, whatever their location on the X chromosome (Li and Carrel 2008), implying that they have specific features preventing them from being efficiently silenced. Importantly, all escapees tend to be located outside the Xist RNA compartment, suggesting that part of the mechanism enabling escape may be linked to their capacity to resist internalization into the silent, repetitive compartment. However, the exact sequence and/or chromatin signatures that underlie the capacity of some genes to escape and those that underlie the variability in X-inactivation kinetics between genes, are still not well understood. Identifying such genomic and/or chromatin features has turned out to be a challenge since multiple actors are likely to be at work and account for the diversity of X-inactivation profiles. Genes that can escape from X inactivation may be flanked by insulator elements that help form domains that prevent the spread of heterochromatinization and/or recruitment of a gene into the silent repetitive Xist compartment. CTCF-binding sites have been hypothesized to play such a role as they have been identified between inactivation and escape domains (Filippova et al. 2005).
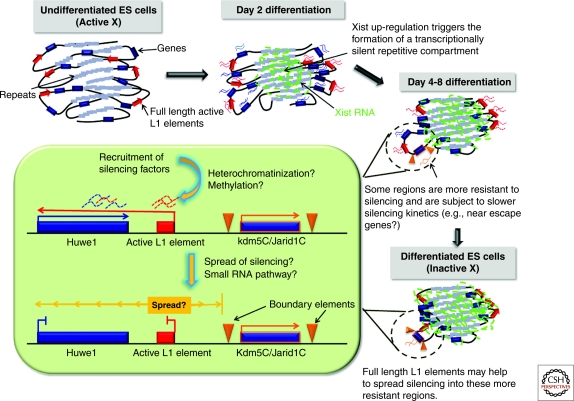
Sequences that facilitate the propagation of X inactivation have also been proposed to exist, termed “way-stations” or “booster elements” (Gartler and Riggs 1983). Lyon proposed that possible candidates for such booster elements could include non-LTR retrotransposon LINE1 repeats, as they are enriched on the X chromosome and their density correlates well with the spread of silencing into autosomal sequences in X: autosome translocations (Lyon 1998). Furthermore, LINE1 elements are depleted in the immediate vicinity of escape genes (Bailey et al. 2000; Carrel et al. 2006). More recently, it has been shown that LINEs may facilitate the propagation of X inactivation at at least two different levels. Truncated, ancient LINEs may participate in nucleating the heterochromatic repetitive compartment induced by Xist RNA early in X inactivation (Chow et al. 2010). On the other hand, younger, more active LINEs, which are actually transcribed from the inactive X during development, may participate in the local propagation of X inactivation into regions that would otherwise be prone to escape (Chow et al. 2010). One such region that has been studied in detail contains the Huwe1 gene, which has a full length LINE at its 3′ end. Huwe1 displays very slow X-inactivation kinetics, initiating only after day 2 of differentiation (Patrat et al. 2009; Chow et al. 2010). The slow silencing of Huwe1 may be due to its proximity to an escapee, Jarid1c, which might render its surrounding region, including Huwe1, more resistant to X inactivation. Huwe1 is, however, eventually inactivated, possibly due to the presence of nearby full-length LINE1 elements. Intriguingly, these full length LINEs are transcribed from the inactive X chromosome during the exact window of time that X inactivation is occurring and their transcription is accompanied by the production of small RNAs (Chow et al. 2010) (Fig. 4). In the case of Huwe1, it has been proposed that transient LINE1 transcription may drive antisense transcription into the gene and this may help to induce its silencing. Antisense transcription has increasingly emerged as an important regulator of gene expression in a diverse number of ways (Su et al. 2010). Active LINE1 elements/promoters may therefore represent a significant source of antisense transcription and play regulatory roles in the genome, particularly on the X chromosome, where they are enriched, at least in eutherian mammals. In summary, LINEs may participate in generating heterochromatin in more than one way, with young, full-length LINEs acting as local boosters to facilitate silencing of certain regions of the X chromosome; for example, by driving the transient production of natural antisense transcripts on the X chromosome undergoing inactivation; while older, truncated LINEs may facilitate creation of the silent repetitive core of the X chromosome, into which genes are recruited as they are inactivated (Chaumeil et al. 2006; Chow et al. 2010).
Figure 4.
Diverse, region-specific elements contribute to the spread and stability of silencing and result in the heterogeneous inactivation kinetics observed across the chromosome. The X-linked Huwe1 gene is X-inactivated relatively late during development. This resistance to silencing may be due to the influence of the nearby escapee domain containing the Jarid1C/Kdm5c gene. The presence of a full length LINE1 element 3′ of Huwe1 may help to eventually spread silencing into the Huwe1 gene.
Although it is clear that Xist RNA is essential for the recruitment and propagation of chromatin modifications as well as the specific reorganization of the chromosome, how these changes result in later changes, such as the shift to late replication timing, macroH2A enrichment, and DNA methylation at promoters, remains unclear. A recent new player in X inactivation that may be involved in some of the downstream changes is the Smchd1 protein (Blewitt et al. 2008). Smchd1 mutant female embryos display both placental and extra-embryonic defects and die prior to stage E13.5. This relatively late lethality is suggestive of a role in the maintenance, rather than the initiation of XCI. Indeed, promoter DNA hypomethylation on the Xi was detected in mutants, suggesting that Smchd1 may affect the DNA methylation deposition step (Blewitt et al. 2008). Smchd1 is an SMC-like protein with homology to components of cohesin and condensin complexes. Interestingly, condensins are implicated in the down-regulation of both X chromosomes in C. elegans hermaphrodites (Chuang et al. 1994), providing an exciting potential link between these two forms of dosage compensation.
CONCLUSIONS AND PERSPECTIVES
Although very different mechanisms have evolved in different organisms to ensure chromosome-wide gene regulation during dosage compensation, a common theme is the necessity for recruitment of dosage compensation complexes across the length of the X chromosome in a concerted fashion. In flies and worms, specific recruitment or entry sites are beginning to be identified, and should help to unravel the exact mechanisms by which the dosage compensation complexes are recruited and spread in these organisms. However, chromatin structure, non-coding RNAs, and nuclear organization may also play important roles. Mammals present the unusual situation whereby two genetically identical X chromosomes have to be differentially treated within the same nucleus and nuclear organization may contribute to this process. Coating of one of the two X chromosomes by the non-coding Xist RNA triggers formation of a silent nuclear compartment, into which genes are recruited as they become inactivated, as well as the recruitment of repressive complexes. The cis-limited action of Xist RNA still remains mysterious, as do the proteins or sequences to which it binds. The diversity of X-inactivation kinetics and chromatin patterns found for different regions of the inactive X chromosome suggest that there may be more than one way of inducing heterochromatin during X inactivation. Furthermore, different mammals exploit different mechanisms to achieve chromosome-wide silencing, with variations in chromatin marks. The extent to which nuclear organization and spatial segregation of the X chromosomes represent a universal feature of dosage compensation strategies will remain an interesting question for the future.
ACKNOWLEDGMENTS
Our work is supported by grants from the CNRS, FRM (Equipe FRM), ANR, and Canceropole (GepiG), as well as HEROIC (Highthroughput Epigenetic Regulatory Organization in Chromatin), an integrated project funded by the European Union, and the ERC.
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. 2009. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell 16: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517 [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E 2007. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science 318: 1632–1636 [DOI] [PubMed] [Google Scholar]
- Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol 8: 293–299 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A 97: 6634–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat TS, Jonkers I, Monkhorst K, Gribnau J 2010. X-changing information on X inactivation. Exp Cell Res 316: 679–687 [DOI] [PubMed] [Google Scholar]
- Barr ML, Bertram EG 1949. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676. [DOI] [PubMed] [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669 [DOI] [PubMed] [Google Scholar]
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev 8: 96–104 [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Roelofsen T, Pennings SW, Martens JH, Jenuwein T, Stunnenberg HG 2006. Histone modification patterns associated with the human X chromosome. EMBO Rep 7: 628–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Carrel L, Park C, Tyekucheva S, Dunn J, Chiaromonte F, Makova KD 2006. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet 2: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP 2007. Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis. Chromosoma 116: 147–157 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF 2004. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci U S A 101: 17450–17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969 [DOI] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ 2007. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc Natl Acad Sci U S A 104: 10104–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ 1994. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474 [DOI] [PubMed] [Google Scholar]
- Chuang PT, Lieb JD, Meyer BJ 1996. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science 274: 1736–1739 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB 2006. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci U S A 103: 7688–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comings DE 1968. The rationale for an ordered arrangement of chromatin in the interphase nucleus. Am J Hum Genet 20: 440–460 [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet 22: 323–324 [DOI] [PubMed] [Google Scholar]
- de Wit E, van Steensel B 2009. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma 118: 25–36 [DOI] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JA 2008. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet 4: e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM 2005. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell 8: 31–42 [DOI] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD 1983. Mammalian X-chromosome inactivation. Annu Rev Genet 17: 155–190 [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Becker PB 2009. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev 23: 2490–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O’Brien PC, Jones RC, Ferguson-Smith MA, Marshall Graves JA 2004. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432: 913–917 [DOI] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B 2006. Global analysis of X-chromosome dosage compensation. J Biol 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB 2002. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U S A 99: 8677–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallacli E, Akhtar A 2009. X chromosomal regulation in flies: when less is more. Chromosome Res 17: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P 1997. X-chromosome inactivation in mammals. Annu Rev Genet 31: 571–610 [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A 2007. Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Hoki Y, Kimura N, Kanbayashi M, Amakawa Y, Ohhata T, Sasaki H, Sado T 2009. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136: 139–146 [DOI] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J 2009. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell 139: 999–1011 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522 [DOI] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JA 2009. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res 17: 115–126 [DOI] [PubMed] [Google Scholar]
- Li N, Carrel L 2008. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci U S A 105: 17055–17060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ 1998. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92: 265–277 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ 1996. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 274: 1732–1736 [DOI] [PubMed] [Google Scholar]
- Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O’Neill LP, Turner BM 2007. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol 5: e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF 1998. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet 80: 133–137 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM 2009. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19: 1478–1484 [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166 [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG 2009. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res 19: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823 [DOI] [PubMed] [Google Scholar]
- Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L, Grange T, Wutz A, Dimitrov S 2009. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol 29: 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T 2007. Beyond the sequence: cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53 [DOI] [PubMed] [Google Scholar]
- Nicodemi M, Prisco A 2007. Symmetry-breaking model for X-chromosome inactivation. Phys Rev Lett 98: 108104. [DOI] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E 2009. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci U S A 106: 5198–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30: 77–80 [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS 2008. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci 121: 1119–1127 [DOI] [PubMed] [Google Scholar]
- Rens W, Grutzner F, O’Brien PC, Fairclough H, Graves JA, Ferguson-Smith MA 2004. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc Natl Acad Sci U S A 101: 16257–16261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A 2006. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol Cell Biol 26: 7167–7177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4: 895–901 [DOI] [PubMed] [Google Scholar]
- Spector DL 2003. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72: 573–608 [DOI] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB 2008. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4: e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WY, Xiong H, Fang JY 2010. Natural antisense transcripts regulate gene expression in an epigenetic manner. Biochem Biophys Res Commun 396: 177–181 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM 2004. The function of nuclear architecture: a genetic approach. Annu Rev Genet 38: 305–345 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev 22: 194–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valley CM, Pertz LM, Balakumaran BS, Willard HF 2006. Chromosome-wide, allele-specific analysis of the histone code on the human X chromosome. Hum Mol Genet 15: 2335–2347 [DOI] [PubMed] [Google Scholar]
- Valley CM, Willard HF 2006. Genomic and epigenomic approaches to the study of X chromosome inactivation. Curr Opin Genet Dev 16: 240–245 [DOI] [PubMed] [Google Scholar]
- van Berkum NL, Dekker J 2009. Determining spatial chromatin organization of large genomic regions using 5C technology. Methods Mol Biol 567: 189–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6: e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res 17: 585–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174 [DOI] [PubMed] [Google Scholar]
- Xu N, Donohoe ME, Silva SS, Lee JT 2007. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat Genet 39: 1390–1396 [DOI] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT 2007. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]