Abstract
The bacterial flagellum and the virulence-associated injectisome are complex, structurally related nanomachines that bacteria use for locomotion or the translocation of virulence factors into eukaryotic host cells. The assembly of both structures and the transfer of extracellular proteins is mediated by a unique, multicomponent transport apparatus, the type III secretion system. Here, we discuss the significant progress that has been made in recent years in the visualization and functional characterization of many components of the type III secretion system, the structure of the bacterial flagellum, and the injectisome complex.
Flagella and the injectisomes that some bacteria use to deliver proteins into their hosts share a conserved export machinery. This is highly specific and allows rapid transport of components across the cell membrane.
Bacteria swim in their environment by rotating a rigid, helical filament, the flagellum (Berg and Anderson 1973). The flagellum is a sophisticated molecular nanomachine made of about 25 different proteins (Berg and Anderson 1973; Namba and Vonderviszt 1997; Macnab 2003; Kojima and Blair 2004; Chevance and Hughes 2008) (Fig. 1A). Closely related to the flagellum is the type III injectisome, another complex nanomachine that allows Gram-negative bacteria to deliver effector proteins into eukaryotic host cells (Galán and Collmer 1999; Cornelis and Van Gijsegem 2000; Cornelis 2006; Galán and Wolf-Watz 2006) (Fig. 1B).
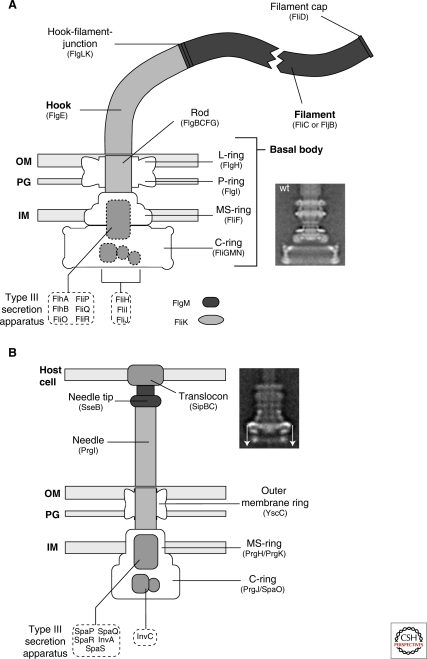
Figure 1.
Schematic comparison of the flagellum and type III injectisome of Salmonella. (A) Schematic overview of the flagellum of Salmonella. The structure of the flagellum consists of three parts: 1) a basal body with a flagellar-specific type III secretion system within the inner membrane ring; 2) a flexible hook acting as a universal joint to 3) the rigid filament. Dashed boxes illustrate proteins with functions in flagellar type III secretion. Also indicated are FlgM, the negative regulator of late substrate gene expression that is secreted after hook-basal-body secretion, and the hook-length regulator FliK that measures rod-hook length and ultimately determines the secretion substrate specificity switch. OM = outer membrane; PG = peptidoglycan; IM = inner membrane. The inlay EM (electron micrograph) picture shows an isolated hook-basal-body complex of Salmonella (Thomas et al. 2001). (B) Schematic overview of the SPI-1 injectisome of Salmonella. Many components of the flagellum and injectisome are structurally and/or functionally related. The injectisome structure can also be divided into three main parts: 1) the basal body with the type III secretion apparatus within the inner membrane ring; 2) a straight needle connecting the bacterial secretion system to 3) the translocon complex that forms a pore in the membrane of eukaryotic host cells. The inlay EM picture shows an injectisome of Salmonella enterica (Marlovits et al. 2004). (The arrows on this image served in the original paper to explain aspects of the reconstruction procedure.) (A [inlay], Reprinted, with permission, from Thomas et al. 2001 [© ASM]; B [inlay] reprinted, with permission, from Marlovits et al. 2004 [© AAAS].)
Most extracellular components of the flagellum and the injectisome are exported via a specific type III secretion system (T3SS). Importantly, the export apparatus of both the flagellum and the injectisome are closely related on structural and functional levels (Fig. 2 and Table 1).
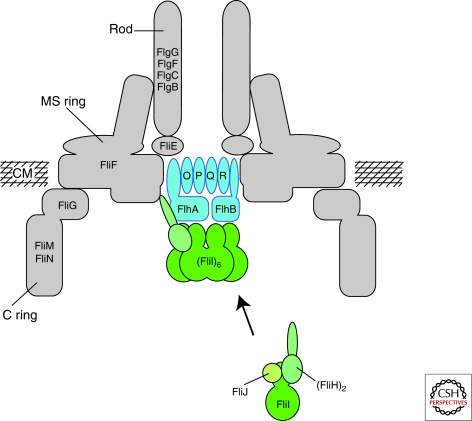
Figure 2.
Schematic of the flagellar type III secretion apparatus. FlhA, FlhB, FliO, FliP, FliQ, and FliR are integral membrane components of the flagellar type III secretion apparatus. The membrane components of the T3S apparatus are believed to assemble within a central pore of the MS-ring. FliI, the flagellar-specific ATPase, FliH, the regulator of FliI and FliJ, a general chaperone, are soluble proteins. FliI forms a heterotrimer together with the homodimer FliH in the cytoplasm. After docking to the membrane components of the T3S apparatus, FliI forms a functional hexamer. The protein translocation is dependent on the proton motive force and the substrates have to be secreted in an unfolded state through a narrow channel of about 2.0 nm in diameter. It has been proposed that FliI facilitates substrate unfolding in an ATP-dependent manner. CM = cytoplasmic membrane. (Reprinted, with permission, from Minamino et al. 2008a [Royal Society of Chemistry].)
Table 1.
Comparison of structurally and/or functionally related components of the flagellum and injectisome systems of Salmonella enterica, Yersinia ssp., Shigella ssp., and enteropathogenic Escherichia coli.
| Flagellum (S. enterica) | Injectisome (S. enterica SPI-1) | Injectisome (Yersinia ssp. Ysc) | Injectisome (Shigella ssp.) | Injectisome (enteropathogenic Escherichia coli EPEC) | Structure/Function |
|---|---|---|---|---|---|
| FliF | PrgH/PrgK | YscJ | MxiJ | MxiJ | MS-ring inner-membrane ring |
| FliI | InvC | YscN | Spa47 | EscN | ATPase |
| FliJ | YscO | T3S chaperone | |||
| FliGMN | PrgJ/SpaO | YscQ | Spa33 | C-ring cytoplasmic ring (HrcQ in Pseudomonas) | |
| FliP | SpaP | YscR | Spa24 | T3S apparatus inner-membrane protein | |
| FliQ | SpaQ | YscS | Spa33 | T3S apparatus inner-membrane protein | |
| FliR | SpaR | YscT | Spa29 | T3S apparatus inner-membrane protein | |
| FlhA | InvA | YscV | MxiA | T3S apparatus inner-membrane protein | |
| FlhB | SpaS | YscU | Spa40 | T3S apparatus inner-membrane protein | |
| N/A | InvG | YscC | MxiD | outer-membrane ring | |
| FlgE? | PrgI | YscF | MxiH | EscF | extracellular needle |
| N/A | SipBC | YopBD | IpaBC | EsoBD | translocation pore |
| FliC? | SseB | LcrV | IpaD | EspA | needle extension |
| FliH? | YscL | MxiN | ATPase regulator | ||
| FliK | InvJ | YscP | Spa32 | hook/needle length regulator |
STRUCTURAL AND FUNCTIONAL SIMILARITIES OF THE FLAGELLUM AND INJECTISOME
Electron micrographs (EM) of the injectisome and the flagellum revealed clear structural similarities (Fig. 1). However, in place of the flagellar hook and helical filament, injectisomes have usually a short, straight needle. In addition to the structural resemblance, there is strong conservation in the constituent proteins. Of the roughly 20 proteins needed to form the flagellar basal body, about half have clear counterparts in the injectisome (Table 1).
Importantly, most components of T3SS are conserved in both flagellar and virulence-associated T3SS and many proteins are homologous in sequence or function (Kubori et al. 1998; Blocker et al. 2003; Macnab 2004; Cornelis 2006) (Table 1).
Common features of all T3SS are the very fast operation (in case of the flagellar T3SS, almost 20 molecules of 55-kDa flagellin are transported per second in early stages of filament assembly [Iino 1974]) and stringent substrate discrimination (only a few proteins are exported, from among the many hundreds present in the cell). Additionally, substrates of T3SS feature an amino (N)-terminal peptide secretion signal that is usually structurally disordered (Namba and Vonderviszt 1997; Namba 2001) and distinct secretion chaperones stabilize or target their cognate secretion substrates to the secretion apparatus (Parsot et al. 2003). Recently, it has been unraveled that translocation of the substrates across the inner membrane is dependent on the proton motive force (PMF) (Minamino and Namba 2008; Paul et al. 2008) and is presumably coupled to ATP-dependent substrate release and unfolding (Akeda and Galán 2005).
Another interesting fact is that the flagellar T3SS is believed to be the ancestor of all T3SS. Flagellar T3SS are present in both Gram-positive and Gram-negative bacteria, and it has been proposed that type III secretion required for pathogenesis evolved from flagellar-specific T3SS (Hueck 1998; Macnab 2004). However, recent phylogenetic studies indicate that both the flagellum and the injectisome share a common ancestor (Gophna et al. 2003).
THE FLAGELLAR TYPE III SECRETION APPARATUS
The flagellar-specific T3S apparatus is believed to assemble within the MS-ring at the basis of the flagellar basal body and consists of six integral membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, FliR) and three cytoplasmic proteins (FliH, FliI, FliJ) (Minamino and Macnab 1999), which are essential for the export of rod-type, hook-type, and filament-type secretion substrates (Minamino 1999; Minamino 2008a). A schematic of the flagellar-specific T3S apparatus is displayed in Figure 2.
It is important to note that secretion substrates are translocated by the flagellar T3S apparatus across the cytoplasmic membrane through a narrow channel of about 2.0 nm in diameter (Yonekura et al. 2003); therefore, the proteins are most likely exported in an unfolded or partially folded state.
The soluble components FliH, FliI, and FliJ of the flagellar T3S system are thought to facilitate delivery and unfolding of secretion substrates to the export apparatus (Macnab 2003). FliI is an ATPase (Fan and Macnab 1996) that can form a hexameric ring-shaped structure (Claret et al. 2003) and that is presumed to couple ATP hydrolysis to some energy-utilizing step(s). FliI was thought to energize the transmembrane transport process, but other studies indicate that ATP hydrolysis energizes steps in substrate delivery instead, probably facilitating the unfolding of the substrate and its release from the chaperone (Akeda and Galán 2005).
Recent findings have shown that the transport process itself is energized by the proton motive force and does not require FliI (Minamino and Namba 2008; Paul et al. 2008). The two other soluble components of the T3S apparatus are FliH, which regulates the ATP-hydrolyzing activity of FliI, and FliJ, a general chaperone for flagellar secretion substrates.
The integral membrane components FliOPQR of the flagellar-specific T3SS are relatively small (FliO = 13.1 kDa; FliP = 26.8 kDa; FliQ = 9.6 kDa; FliR = 28.9 kDa) and are predicted to have one to eight membrane spanning helices. Little is known about possible functions of FliO, FliP, FliQ, and FliR respectively, but the proteins are essential for flagellar type III secretion. FlhA and FlhB are much larger integral membrane proteins (75 kDa and 42 kDa, respectively) and also have large cytoplasmic domains, where the proteins interact with the soluble components of the T3S apparatus, FliH, FliI, and FliJ (Minamino and Macnab 2000). FlhB is furthermore controlling the switch of substrate specificity to export late structural subunits upon completion of early flagellar structures (Ferris and Minamino 2006). Although the major players in export have probably all been identified, the molecular mechanism of flagellar T3SS is still poorly understood. A reasonable, general proposal for the mechanism is that FliH/FliI/FliJ complexes bind to substrate proteins and target them to the membrane-bound export apparatus. Subsequently, the membrane components of the apparatus then translocate the substrate across the membrane and into the channel, using energy from the electrochemical potential gradient of protons, which is the sum of the membrane potential Δψ and the proton concentration gradient ΔpH (Minamino and Namba 2008; Paul et al. 2008).
STRUCTURE AND ASSEMBLY OF THE BACTERIAL FLAGELLUM
The structure of the flagellum can be divided into three parts: I) the basal body as the rotary motor (Berg and Anderson 1973; Silverman and Simon 1974) and the type III export apparatus; II) the flexible hook that couples the rotary motor to III) the rigid filament (DePamphilis and Adler 1971a,b) (Fig. 1A). The filament is about 10 to 15 µm long, yet only 120–240 nm in diameter, and the rotary machine turns at hundreds of revolutions per second, utilizing both Δψ and ΔpH (Manson et al. 1977; Matsuura et al. 1977).
Flagellum assembly initiates with the formation of the MS-ring (consisting of FliF) in the inner membrane, followed by attachment of the rotor/switch complex (C-ring proteins FliG, FliM, and FliN) at the cytoplasmic face of the MS-ring. An overview of the assembly process is given in Figure 3. Upon completion of the C-ring, the motor proteins MotA and MotB are assembled in the inner membrane. MotA and MotB form the stator complex, which also forms the pathway for proton influx, and attach noncovalently to the peptidoglycan layer via the C-terminal periplasmic domain of MotB, whereas the rotor (FliG) is noncovalently attached to the MS-ring. Together, the stator and the rotor form the flagellar motor, whose rotation is energized by the proton motive force (Macnab 2003; Chevance and Hughes 2008). The periplasmic domain of MotB is thought to go through a relatively large conformational change to bind to the peptidoglycan layer and open the proton pathway upon assembly of the MotAB complex to the motor (Kojima et al. 2008).
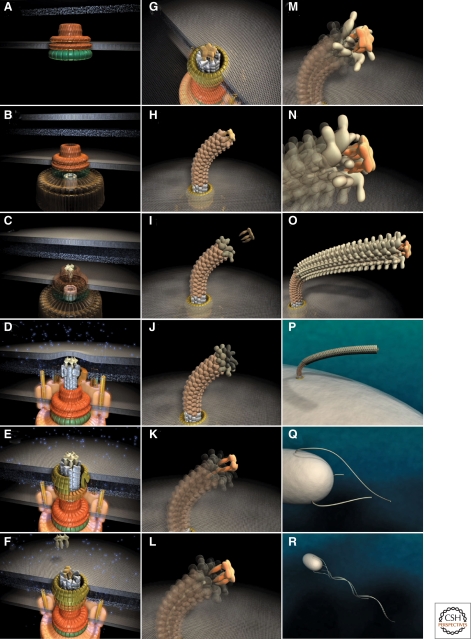
Figure 3.
Steps in assembly of the bacterial flagellum. The self-assembly process of the bacterial flagellum starts from the top left (A) and proceeds to the bottom right (R). Assembly starts with the formation of the MS-ring in the cytoplasmic membrane (A). Afterward, the cytoplasmic C-ring is attached to the MS-ring and the flagellar-specific type III secretion apparatus assembles within a central pore of the MS-ring (B). Flagellar secretion substrates are now secreted specifically via the T3S apparatus. Flagellar proteins that reach the distal end of the growing structure, self-assemble onto the existing structure with the help of distal cap proteins (shown as pentamers at the tips of the axial structures) (C–O). First, the rod acting as a driveshaft is assembled beneath the rod-cap, which is also a muramidase to allow penetration through the peptidoglycan layer (D). In addition, motor force generators that couple proton flow to torque generation assemble in the cytoplasmic membrane and interact with C-ring components (D). Rod assembly ends with the formation of the PL-ring bushing, outer membrane penetration, and the replacement of the rod scaffold with the hook scaffold (E–G). Afterward, hook subunits are secreted and the hook grows to a final length of 55 nm (Hirano et al. 1994), triggering a secretion specificity switch from rod-hook-type substrates to late-secretion substrates (H). Upon completion of the hook-basal-body complex, hook-filament junction proteins (I–J), the filament cap (K–L), and filament subunits are secreted, and the filament grows to a maximal length of about 10–15 µM (M–R). (Reprinted, with permission, from Minamino et al. 2008a [Royal Society of Chemistry].)
After assembly of the C-ring, the flagellar-specific T3SS (consisting of FlhA, FlhB, FliH, FliI, FliJ, FliO, FliP, FliQ, and FliR) assembles within the putative central pore of the MS-ring at the basis of the basal body (Aizawa 1996), facilitating the PMF-dependent export of most extra cytoplasmic components of the flagellum (Paul et al. 2008). Subsequently, the rod components FliE, FlgB, FlgC, FlgF (proximal rod), and FlgG (distal rod) are secreted by the T3S apparatus and progressively assembled (Minamino et al. 2000). The P-ring protein FlgI and the L-ring lipoprotein FlgH are probably exported by the Sec-dependent pathway (Homma et al. 1987), but the assembly of the periplasmic P-ring and the L-ring in the outer membrane is dependent upon the construction of the preceding flagellar rod structure. Upon completion of the rod and the rings, the hook (consisting of FlgE) is assembled to an approximate length of 55 nm, which is controlled by the hook-length regulator FliK (Patterson-Delafield et al. 1973; Hirano et al. 1994). The last steps of flagellum assembly include secretion of the anti-σ28 factor FlgM (Hughes et al. 1993), which results in σ28-dependent expression of flagellar genes that are under Class III promotor regulation, and the assembly of the hook-associated proteins FlgK, FlgL, and FliD (Homma et al. 1990). Subsequently, the filament is constructed with the subunit FliC or FljB (flagellin) as the last structural component of the flagellum, where the assembly of flagellin occurs at the distal end of the growing filament under the cap composed of FliD, as described later. A single filament can be constructed of as many as 20,000–30,000 flagellin subunits (Macnab 2003; Chevance and Hughes 2008), which represents a significant amount of the total protein mass of the cell.
STRUCTURE AND ASSEMBLY OF THE INJECTISOME
The injectisome is used by many Gram-negative pathogens to pump virulence-effector proteins into host cells (Kubori et al. 1998; Blocker et al. 2003; Journet et al. 2005; Cornelis 2006; Galán and Wolf-Watz 2006). Members of the injectisome family are found in many Gram-negative plant and animal pathogens, including Salmonella sp., Yersinia sp., enteropathogenic Escherichia coli (EPEC), Shigella sp., Chlamydia sp., and Erwinia carotovora (Blocker et al. 2003; Galán and Wolf-Watz 2006). Injected effector proteins alter host-cell functions and membrane cytoskeletons to promote invasion, survival, and growth of the bacterium, or in some cases are used to facilitate symbiosis.
The overall structure of the injectisome is very similar to the flagellum: I) a basal, cylindrical structure spanning the inner and outer membrane and containing the T3SS and II) an extracellular, hollow tubular structure (Tampakaki et al. 2004; Cornelis 2006) (Fig. 1B). The extracellular appendages have different structures and length, depending on the family of injectisomes; for example, a stiff needle with an approximate length of 58 nm in Yersinia enterocolitica, a filament with a length up to 600 nm in enteropathogenic Escherichia coli, or a Hrp pilus with a length of several µm in Pseudomonas syringae (Cornelis 2006).
Similar to the assembly of the flagellum, it has been assumed that the injectisome also assembles in a sequential manner. The basal structure of the injectisome includes the T3SS, a pair of rings in the inner and outer membranes, and the rod. In Salmonella ssp., the inner ring consists of the proteins PrgH and PrgK that are homologous to the MS-ring protein FliF (Kimbrough and Miller 2000; Kubori et al. 2000). In the flagellar system, the C-ring (FliGMN) is attached to the MS-ring on the cytoplasmic face. Although no EM pictures of injectisome show any attached C-ring-like structure, the proteins of the YscQ family share significant similarity to FliN and FliM (Cornelis 2006). The membrane-embedded components YscR, YscS, YscT, YscU, and YscV are located within the inner membrane ring of the injectisome T3SS. Although these inner-membrane components of the injectisome T3SS share high sequence homology to the inner-membrane proteins FliPQR and FlhAB of the flagellum, no further structural data is available. As it has been shown for the flagellar T3SS (Minamino and Namba 2008; Paul et al. 2008), the T3SS of Yersinia enterolitica is also dependent on the proton-motive force (Wilharm et al. 2004), indicating also a functional similarity. At the base of the inner membrane ring, the hexameric ATPase of the YscN family shares striking similarity to the flagellar ATPase FliI (Imada et al. 2007; Zarivach et al. 2007). It has been shown for the Salmonella homolog InvC that the ATPase energizes the secretion substrate release from its cognate cytoplasmic chaperone prior to PMF-dependent secretion (Akeda and Galán 2005).
A protruding rod structure assembles on top of the MS-ring in the flagellar basal body, and it has been suggested that the inner rod of the Salmonella injectisome is composed of PrgJ (Kubori et al. 2000; Marlovits et al. 2004). The last components of the basal structure are a pair of rings associated with the peptidoglycan layer and the outer membrane made of the YscC family of Secretin proteins (Koster et al. 1997; Kubori et al. 2000).
The extracellular structures of the Yersinia injectisome are exported via the membrane-embedded T3SS and can be divided into three parts: I) a needle, II) a needle-extension forming the tip complex, and III) the translocation pore that presumably forms a channel in the host cell membrane. The injectisome needle is made of approximately 100–150 molecules of the YscF family proteins that polymerize as a helical assembly into a filamentous structure (Kubori et al. 2000; Cornelis 2006). The molecular ruler YscP controls needle length in Yersinia to about 58 ±10 nm (Journet et al. 2003). At the top of the needle, the tip complex formed by LcrV is located (Mueller et al. 2005; Deane et al. 2006). It has been suggested that the tip complex forms a scaffold for the assembly of the translocator pore consisting of YopB and YopD, which will be inserted into the membrane of host cells (Cornelis 2006).
ELECTRON MICROGRAPH RECONSTRUCTIONS OF THE FLAGELLAR HOOK-BASAL BODY COMPARED TO THE NEEDLE COMPLEX STRUCTURE
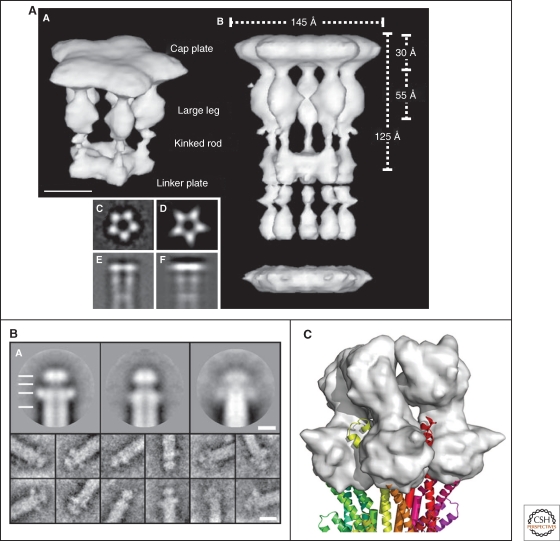
The core basal complexes of both the flagellar hook-basal-body and injectisome are structurally very similar. Figure 4 compares 3D-EM image reconstructions of both the hook-basal-body (Derosier 2006) and the needle complex (Marlovits et al. 2004) of Salmonella enterica serovar Typhimurium. The structure of the needle complex from Shigella has also been studied by 3D-EM image reconstruction (Hodgkinson et al. 2009). The 3D images of those purified needle complexes revealed a cylindrical basal body and a hollow, needle-like structure that spans both membranes with a central channel of 20–30 Å diameter (Blocker et al. 2003). The hook-basal-body structure of the flagellum also displays a cylindrical basal body with rings in the inner and outer membrane, as well as in the peptidoglycan layer. The main differences are the diameter of the cytoplasmic ring of the flagellar basal body, which is significantly wider (approx. 40 nm) (Macnab 2003) compared to the ring of the injectisomes (approx. 20 nm) (Marlovits et al. 2004; Hodgkinson et al. 2009), and the lack of a C-ring-like structure in the injectisome.
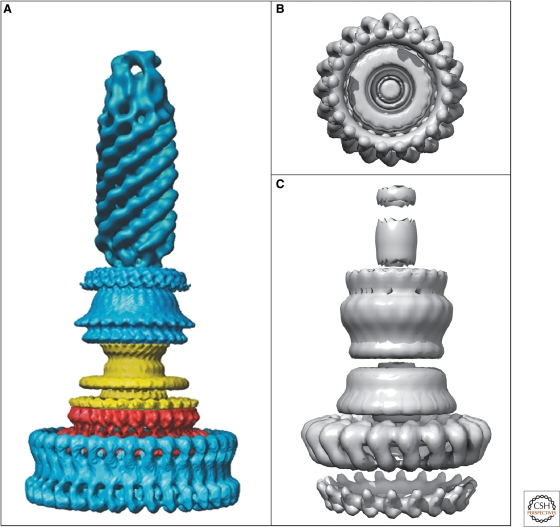
Figure 4.
EM reconstructions of basal bodies of the flagellum and type III injectisome of Salmonella. (A) Model of the bacterial hook-basal-body complex based on EM reconstructions (Derosier 2006). (B, C) Surface renderings of the injectisome basal body and needle structure based on EM reconstruction data EmDep Database accession number EMD1100 (Marlovits et al. 2004). (B) View of the injectisome basal body complex from the cytoplasm and (C) side-view of the injectisome basal body with attached needle. (A, Reprinted, with permission, from Derosier 2006 [© Elsevier]; B, C, based on Marlovits et al. 2004 [© AAAS].)
Although these EM studies provide an excellent overview of the architecture of both the flagellar and injectisome basal bodies, the resolution of these EM maps is too low to allow detailed studies of the localization of individual structural elements. Additionally, because of harsh sample preparation methods, the resulting structures lack essential components, like the soluble components of the T3SS and the C-ring homolog of the injectisome.
A POSSIBLE ATOMIC MODEL OF THE INNER MEMBRANE RING ESCJ OF THE INJECTISOME
The formation of a ring complex in the inner membrane seems to be one of the first steps in the assembly of both the flagellum and injectisome basal structure. The inner membrane ring is thought to act as a scaffold for the assembly of other sub-structures and houses the membrane-embedded components of the T3SS.
The inner membrane ring of the flagellum is made of a single protein, FliF, forming the MS-ring. In case of the injectisome, the inner membrane ring is composed of the highly conserved YscJ family (PrgK in Salmonella, EscJ in EPEC). Recently, the atomic structure of EscJ has been determined by X-ray crystallography, and a symmetrical 24-subunit model has been constructed (Yip et al. 2005) (Fig. 5). The overall dimensions of the modeled EscJ ring (diameter 180 Å, height 52 Å) are similar to previous estimations of the diameter of the inner ring based on EM reconstructions of the injectisome (Marlovits et al. 2004). Intriguingly, the EscJ ring model can be fitted into existing 3D-EM maps of the injectisome basal body of Salmonella enterica (Moraes et al. 2008).
Figure 5.
Model of the injectisome inner membrane ring, EscJ. Shown are ribbon and surface representations of the 24-subunit EscJ ring model. The sideview illustrates the two-layered exterior structure that closely resembles the flagellar MS-ring structure based on cryo-EM studies. The N-termini of the EscJ subunits are located at the wide face of the ring (also labeled N in the ribbon diagrams sideview). Scale bar 25 Å. (Reprinted, with permission, from Yip et al. 2005 [Nature Publishing Group].)
CRYSTAL STRUCTURES OF THE C-RING COMPONENTS FliN, FliG, AND HrcQ
At the cytoplasmic face of the flagellar MS-ring, a cytoplasmic ring (C-ring) composed of FliG, FliM, and FliN is assembled. The C-ring components of the flagellum are assumed to be part of the rotor and have functions in torque generation, switching of the rotational direction, and flagellar export (Vogler et al. 1991; Sockett et al. 1992; Lloyd et al. 1996). It has been estimated that about 26 copies of FliF and FliG, about 34 copies of FliM, and about 130 copies of FliN are located in the flagellar basal body structure (Thomas et al. 1999, 2001, 2006; Paul and Blair 2006).
The crystal structure of the middle and C-terminal domains of FliG is available (Brown et al. 2002) and importantly can be fitted into a 3D-EM structure of the C-ring (Brown et al. 2007), where both the middle and C-terminal domains of FliG can interact with FliM (Fig. 6A).
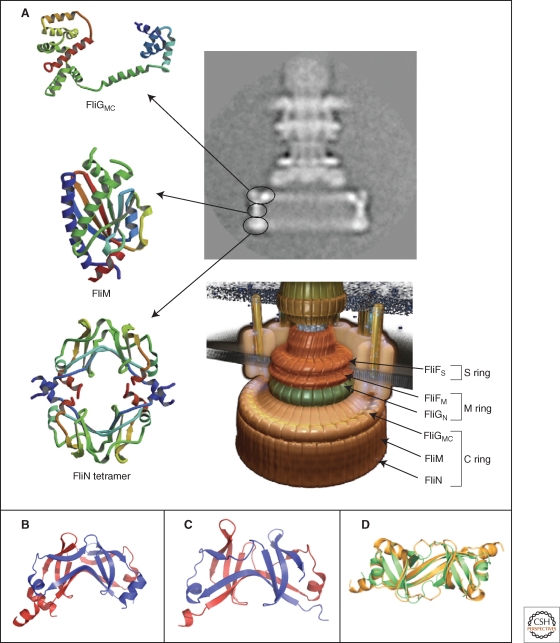
Figure 6.
Crystal structures of the components of the C-ring, FliGMC, FliM, and FliN. (A) Left panel: Ribbon representation of the crystal structures of FliGMC (middle and C-terminal domains of FliG), FliM, and the doughnut-like tetramer of FliN. Upper right panel: cyro-EM image of a hook-basal-body complex. The arrows illustrate the positions of the FliG, FliM, and FliN proteins in the C-ring structure. Lower right panel: Model of the flagellar basal body complex with proposed locations of FliF, FliG, FliM, and FliN forming the MS-ring and C-ring, respectively (Minamino et al. 2008b). (B,C) Comparison of the homologous structures of FliN and HrcQBC (Fadouloglou et al. 2004). (B) Crystal structure of the C-terminal fragment of FliN of Thermotoga maritima (PDB 1O6A). (C) Crystal structure of the C-terminal domain of HrcQBC of Pseudomonas syringae (PDB 1O9Y) (Fadouloglou et al. 2004). (D) Ribbon representation of the FliN dimer (gold) superimposed on half of the HrcQBC tetramer (green) illustrating the highly homologous structure (Brown et al. 2005). (A, reprinted, with permission, from Minamino et al. 2008b [© Elsevier]; B, Protein data base accession number 1O6A, based on Fadouloglou et al. 2004; C, Protein data base accession number 1O9Y, based on Fadouloglou et al. 2004; D, reprinted, with permission, from Brown et al. 2005 [© ASM].)
In the case of the injectisome, no EM images are available that prove the presence of a C-ring-like structure. However, biochemical evidence argues strongly toward the existence of a C-ring in injectisomes made of proteins of the YscQ family (Cornelis 2006). Additionally, the YscQ family shows significant sequence similarity to the flagellar C-ring components FliM and FliN, and high-resolution structures of both FliN with the N-terminal 22 residues truncated (Brown et al. 2005), and the C-terminal part of HrcQ of the YscQ family of Pseudomonas syringae (Fadouloglou et al. 2004) are available. The overall structures of HrcQ and FliN are remarkably similar and both proteins form dimers composed mostly of β-sheets (Fig. 6B–D). Two HrcQ dimers form a homotetramer that has been proposed to be the repeating unit forming a C-ring-like structure in the injectisome basal body (Fadouloglou et al. 2004), and a FliN tetramer can also be modeled to fit into the flagellar basal body C-ring (Paul and Blair 2006) (Fig. 6A).
COMPARISON OF THE STRUCTURES OF ATPase SUBUNITS OF THE T3SS, FliI, AND EscN
At the basis of the basal body of both the flagellum and the injectisome, the T3SS is localized. The T3SS is composed of several integral-membrane and soluble components. Soluble components are a T3SS-specific ATPase, regulators of the ATPase, and T3SS chaperones. As noted above, the ATPase subunit FliI of the flagellar T3SS is thought to unfold secretion substrates prior to the PMF-dependent export (Akeda and Galán 2005; Minamino et al. 2008a; Paul et al. 2008).
In the case of the injectisome, the YscN family of proteins represents the ATPase subunit. The ATPase subunit forms hexameric oligomers that are associated with the inner membrane as it has been shown for both FliI and HrcN, the ATPase of Pseudomonas syringae (Claret et al. 2003; Pozidis et al. 2003).
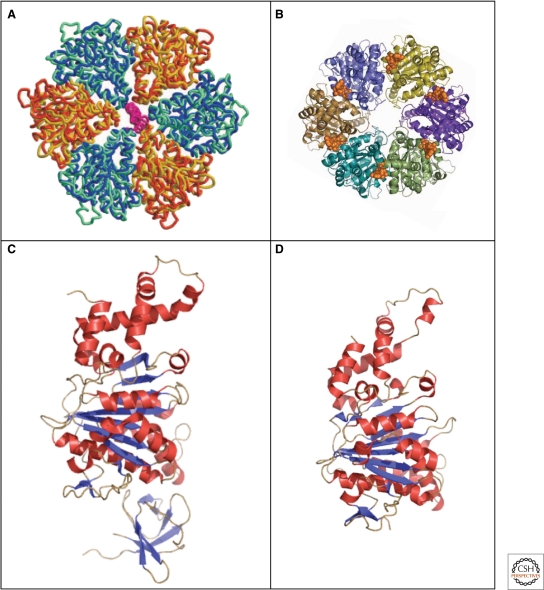
Recently, the atomic structures of EscN of EPEC (Zarivach et al. 2007) and FliI of Salmonella (Imada et al. 2007) have been determined (Fig. 7C,D). Although the probable function of the T3SS ATPase is to disassemble itself and secretion chaperones from their cognate secretion substrates, the high-resolution structures of both EscN and FliI reveal a structural homology to the α/β-subunit of F1-ATPase rather than to the AAA ATPase as previously suggested (Akeda and Galán 2005). Based on the atomic structures of EscN and FliI, respectively, hexameric models have been constructed that reveal the close structural homology to the α3/β3-heterohexamer of F1-ATPase (Fig. 7A,B).
Figure 7.
Crystal structures of the type III secretion ATPases, FliI, and EscN. (A) Model of the ATPase domain of the FliI hexamer (blue and yellow) superimposed onto the ATPase domain (blue-green) and (orange) of F1-ATPase (Imada et al. 2007). (B) Top-view of the EscN hexamer model; position of the ATP shown in a van-der-Waals representation in gold (Zarivach et al. 2007). (C) Crystal structure of the flagellar type III ATPase FliI (Δ1–18) missing the first 18 residues (PDB 2DPY). (D) Crystal structure of the C-terminal domain, residues 103–446 of the injectisome ATPase EscN (PDB 2OBL). (A, reprinted, with permission, from Imada et al. 2007 [© National Academy of Sciences]; B, reprinted, with permission, from Zarivach et al. 2007 [Nature Publishing Group]; C, Protein database accession number 2DPY, based on Imada et al. 2007; D, Protein database accession number 2OBL, based on Zarivach et al. 2007.)
As displayed in Figure 7, the overall structures of EscN and FliI are highly similar to each other. The EscN structure is composed of two domains corresponding to the ATPase and C-terminal domains of F1-ATPase, respectively, while the FliI structure contains all three domains of F1-ATPase. The N-terminal domain containing a six-stranded β-barrel might function in oligomerization (Okabe et al. 2009). The ATPase domain has Walker A/B motifs and the Rossman fold for nucleotide binding. The C-terminal domain shares less homology to that of F1-ATPase and might function in interaction with T3SS chaperones.
STRUCTURES OF EXTRACELLULAR APPENDAGES: THE FLAGELLAR HOOK, FILAMENT, AND INJECTISOME NEEDLE
The injectisome needle is a straight, hollow tube of approximately 60 nm in length and 7 nm in diameter and is made of 100–150 subunits of proteins of the YscF family (Cornelis 2006). YscF proteins are relatively small with about 9 kD, compared to the flagellar hook subunit FlgE (42 kDa) and filament subunit FliC (51 kDa).
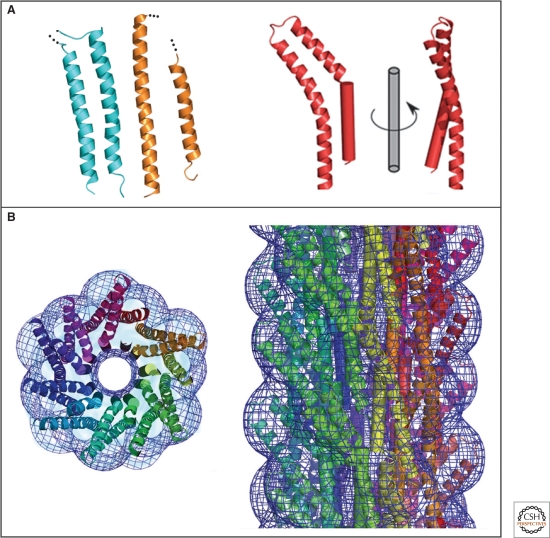
Although YscF proteins share no sequence homology to the flagellar filament or hook, the atomic resolution structure of MxiH, the needle subunit of Shigella ssp. (Deane et al. 2006), revealed structural features similar to the D0 domain of flagellin and EspA, the needle extension protein (Fig. 8A). MxiH consists of two antiparallel α-helices that are connected via a short turn, and the same helix bundles can be found in both EspA and flagellin. Furthermore, the atomic modeling of an injectisome needle was possible by docking the MxiH structure onto a 3D-EM reconstruction of the Shigella needle (Cordes et al. 2003; Deane et al. 2006) (Fig. 8B).
Figure 8.
Crystal structure and molecular model of a type III secretion needle. (A) Ribbon representation of the D0 domain of flagellin (cyan), the ordered (chaperone-bound) region of EspA (orange) and molecule A of MxiH (red). The modeled N-terminal helix of MxiH is also shown rotated by 90° about the long axis of the molecule. (B) Model of the type III secretion needle obtained by docking of the atomic model of MxiH into the EM density of the Shigella T3SS needle. Left panel: end-on view of a 40-Å-thick section of the assembled needle. Each MxiH monomer is colored differently. The EM density is shown as a blue mesh. Right panel: side-view of the assembled needle, with colors as in the left panel. Note that the needle models in the left and right panel are not shown at the same scale. (Reprinted, with permission, from Deane et al. 2006 [© National Academy of Sciences].)
Importantly, the overall helical properties of the needles isolated from Shigella are remarkably similar to the helical properties of the flagellar hook and filament (5.6 subunits per turn of the 1-start helix and a helical pitch of 2.4 nm), highlighting the structural similarities of both the flagellum and the injectisome (Cordes et al. 2003; Cornelis 2006).
The flagellar hook acts as a universal joint connecting the motor with the helical filament and thereby allows for transmission of torque even if the motor and the helical filament are not coaxially orientated (Berg and Anderson 1973). The hook is composed of about 120 subunits of FlgE and assembles, like the filament, into 11 circularly arranged protofilaments. Importantly, overall hook-length is relatively tightly controlled to a length of 55 nm±6 nm (Hirano et al. 1994; Macnab 2003; Chevance and Hughes 2008). The structure of the FlgE31 fragment, which is missing both terminal regions of the hook protein FlgE, has been solved (Samatey et al. 2004). The FlgE31 structure consists of two domains, D1 and D2, each having an oval shape, and is predominantly made of β-sheets. Although the structure of the hook subunit FlgE is very different compared to that of the filament subunit flagellin, their helical symmetries are strikingly similar to each other as stated above (Morgan et al. 1993; Mimori et al. 1995).
A partial atomic hook model was obtained by docking the crystal structure of the FlgE31 fragment into a 3D-EM density map of a straight hook (Samatey et al. 2004; Shaikh et al. 2005) (Fig. 9A). By deforming the helical lattice of the straight hook it was possible to model a curved hook, as shown in Figure 9B,C. Using this curved hook model, the molecular mechanism of the universal joint function of the flagellar hook has been studied in detail by molecular dynamics simulations (Samatey et al. 2004). This curved hook model revealed that, while the hook subunits are tightly packed in the inner core domains, the outer two domains are loosely packed in the axial intersubunit interactions in the protofilament structure.
Figure 9.
Model of the supercoiled flagellar hook and crystal structure of the hook subunit fragment, FlgE31. (A) Docking of the atomic model of the crystal structure of the hook subunit fragment FlgE31 into the outer two domains of the hook obtained by cyro-EM. Left panel: end-on-view; right panel: side-view. The hook EM density is shown as a purple mesh. (B) Atomic model of the supercoiled hook and a schematic diagram of the basal body complex. OM, outer membrane; PG, peptidoglycan layer; CM, cytoplasmic membrane. (C) Magnified, atomic model of the supercoiled hook shown in (B). The innermost and the outermost protofilaments are displayed on the left and right, respectively. Dotted gray lines represent the central channel. (Reprinted, with permission, from Samatey et al. 2004 [Nature Publishing Group].)
The bacterial flagellar filament is much longer than the injectisome needle and grows up to 10–15 µm long, consisting of about 20,000–30,000 subunits of flagellin per filament (Namba and Vonderviszt 1997; Berg 2003). The filament structure is composed of 11 protofilaments, each of which can be in either a left-handed (L-type) or a right-handed (R-type) conformation (Asakura 1970). Only when all protofilaments are in the same conformation, the filament morphology is straight (Kamiya et al. 1979), and accordingly the filament can adopt a variety of helical morphologies by the supercoiling of the filament structure due to the mixture of the two protofilament conformations in different number ratios (Asakura 1970; Calladine 1975, 1978). The left-handed supercoiled filaments allow the formation of a bundle that propels the cells through the media. During a process called chemotaxis, the transition to a few different types of right-handed supercoiled forms upon reversal of the motor rotation results in disruption of the bundle and tumbling of the cell (Macnab and Ornston 1977).
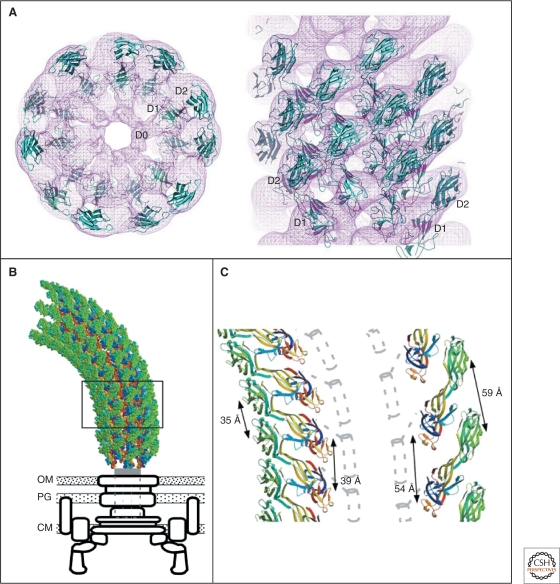
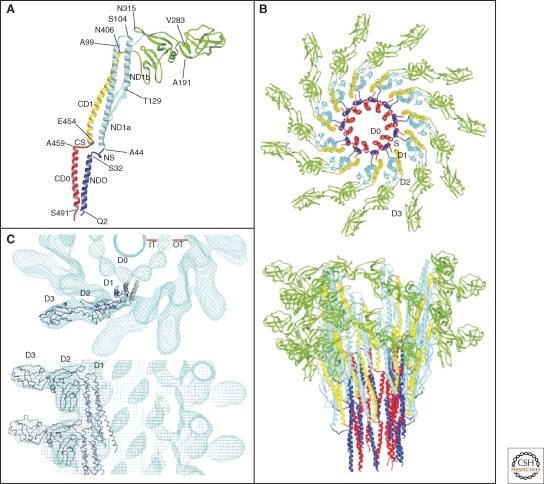
The complete atomic structure of the filament and the individual flagellin subunit has been solved by cryo-EM image analysis and X-ray crystallography (Samatey et al. 2001; Yonekura et al. 2003) (Fig. 10). The structure of the filament has a diameter of about 23 nm with a central channel of about 2 nm (Fig. 10B,C). The filament structure is stabilized by interaction of the α-helical coiled-coils of the D0 and D1 domains of flagellin in the inner core of the filament structure, in which the structure of the D0 domain is strikingly similar to the anti-parallel α-helices of the needle protein MxiH (Deane et al. 2006) (Fig. 8A).
Figure 10.
Crystal structure and model of the flagellar filament. (A) Atomic model of the flagellar filament obtained by cyro-EM. Ribbon representation of the filament subunit (Yonekura et al. 2003). (B) Ribbon representation of the model of the flagellar filament. Upper panel: end-on-view from the distal end of the filament displaying 11 subunits. Lower panel: side-view from outside of the filament (Yonekura et al. 2003). (C) Docking of a protofilament into the electron density map of the filament. Upper panel: end-on-view from the top. Bottom panel: Side-view. D0, D1, D2, and D3 indicate domains of the flagellin protein (Samatey et al. 2001). (A, B, Reprinted, with permission, from Yonekura et al. 2003; C, reprinted, with permission, from Samatey et al. 2001 [all Nature Publishing Group].)
CAP STRUCTURES, THE FILAMENT TIP FliD, AND THE NEEDLE TIP LrcV
The flagellar filament grows at its distal end by self-assembly of flagellin subunits that have to be transported in an unfolded conformation through the narrow channel of the filament. At the very end of the filament, an essential cap structure made of the FliD protein promotes flagellin self-assembly and polymerization by a rotation mechanism. EM studies revealed that the pentameric cap is attached at the distal end of the filament via its five leg-shaped anchor domains (Yonekura et al. 2000) (Fig. 11A). The five domains are separated from each other, thereby forming five differently sized gaps. One significantly larger gap is thought to represent the exit site as well as the binding site for individual flagellin subunits during the assembly process. Insertion of a flagellin subunit presumably forces the legs of the cap subunits to transition into the next stable conformation, thereby providing another gap for the subsequent assembly of the following flagellin subunit. This conformational rearrangement sequence has been described as a walk of the pentameric cap along the helical steps of the filament end. Accordingly, the assembly of approximately 55 flagellin subunits results in a complete rotation of the cap (Yonekura et al. 2000; Maki-Yonekura et al. 2003).
Figure 11.
EM images of the flagellar filament cap, needle tip and model of the needle-tip interaction. (A) 3D-EM image reconstruction of the flagellar cap pentamer (labeled A in the image) and decamer (labeled B in the image). On the lower left side of the image are shown: averaged end-on-view (labeled C); projection of the 3D reconstruction along the 5-fold axis (labeled D); averaged side-view (labeled E), and projection of the 3D reconstruction perpendicular to the 5-fold axis (labeled F) (Maki-Yonekura et al. 2000). (B) Upper three panels: averaged EM images of the needle tip complexes formed by LcrV (left; resolution 1.5 nm), PcrV (center; resolution 1.5 nm), and AcrV (right; resolution 2.5 nm). Also visible is the central channel of both the needle and tip complex. Lower images on the bottom display typical single images (Mueller et al. 2005). (C) The LcrV tip complex modeled onto the distal end of a injectisome needle. The LcrV tip complex is displayed in surface representation (gray) (Deane et al. 2006). (A, Reprinted, with permission, from Maki-Yonekura et al. 2003 [© National Academy of Sciences]; B, reprinted, with permission, from Mueller et al. 2005 [© AAAS]; C, reprinted, with permission, from Deane et al. 2006 [© National Academy of Sciences].)
In the case of the injectisome needle, the tip proteins are thought to be responsible for the correct insertion of the translocon complex that forms a pore in eukaryotic cell membranes. In Yersinia, LrcV forms the tip complex at the distal end of the needle (Mueller et al. 2005). It has been suggested that LrcV functions as a scaffold or platform for the proteins of the translocation pore (Cornelis 2006).
However, it cannot be ruled out that the LrcV tip complex also promotes the assembly of individual needle subunits during needle growth, similarly to the function of the filament cap described above. The overall structure of the needle tip complex and the filament cap are indeed comparable. Similar to the filament cap, the LrcV tip complex seems to form a bell-shaped structure at the distal end of the needle, as visualized by EM studies of Yersinia needles (Mueller et al. 2005) (Fig. 11B). A pentameric complex has been suggested by superimposing the LrcV structure with the C-terminal helices of the needle subunit MxiH (Deane et al. 2006). A maximum of five LrcV molecules could be placed at the tip of the needle, and the resulting bell-shaped structures are strikingly reminiscent of the flagellar cap complex (Yonekura et al. 2003; Deane et al. 2006) (Fig. 11C).
CONCLUSIONS AND FUTURE PERSPECTIVES
Significant progress has been made in the visualization of various components of both the flagellum and the type III injectisome. Importantly, high-resolution structures of several key components of the highly homologous systems, like the cytoplasmic ATPase, the inner membrane ring, the flagellar C-ring proteins, the injectisome needle, the flagellar hook, and the flagellar filament are now available. Together with the 3D-EM image reconstructions of both the flagellar hook-basal-body and the injectisome basal body complex, we now have a relatively comprehensive picture of the overall structure of these fascinating nanomachines. However, high-resolution 3D images that allow the visualization of secondary structures are still essential to build more reliable atomic models to obtain deeper insights into their mechanisms.
The other main challenges in the field are the structure determination of several missing components like the flagellar rod and C-ring of the injectisome. Importantly, no structural information about the membrane components of the T3SS is available. Recent studies emphasized the importance of the membrane components in the PMF-dependent protein transport process, and obtaining structural information about how proton influx is coupled to the protein export process remains the most challenging goal.
ACKNOWLEDGMENTS
This work was supported by PHS grant GM056141 from the National Institutes of Health. We thank the Hughes lab for useful comments and discussions of the manuscript. M.E. gratefully acknowledges scholarship support of the Boehringer Ingelheim Fonds.
Footnotes
Editors: Lucy Shapiro and Richard M. Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Aizawa SI 1996. Flagellar assembly in Salmonella typhimurium. Mol Microbiol 19: 1–5 [DOI] [PubMed] [Google Scholar]
- Akeda Y, Galán JE 2005. Chaperone release and unfolding of substrates in type III secretion. Nature 437: 911–915 [DOI] [PubMed] [Google Scholar]
- Asakura S 1970. Polymerization of flagellin and polymorphism of flagella. Adv Biophys 1: 99–155 [PubMed] [Google Scholar]
- Berg HC, Anderson RA 1973. Bacteria swim by rotating their flagellar filaments. Nature 245: 380–382 [DOI] [PubMed] [Google Scholar]
- Berg H 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72: 19–54 [DOI] [PubMed] [Google Scholar]
- Blocker A, Komoriya K, Aizawa S 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci U S A 100: 3027–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Hill CP, Blair DF 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J 21: 3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, Mathews M, Joss L, Hill C, Blair D 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J Bacteriol 187: 2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Terrazas M, Paul K, Blair DF 2007. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol 189: 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calladine CR 1975. Construction of bacterial flagella. Nature 255: 121–124 [DOI] [PubMed] [Google Scholar]
- Calladine CR 1978. Change of waveform in bacterial flagella: the role of mechanics at the molecular level. J Mol Biol 457–479 [Google Scholar]
- Chevance FFV, Hughes KT 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Micro 6: 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret L, Calder SR, Higgins M, Hughes C 2003. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol 48: 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J Biol Chem 278: 17103–17107 [DOI] [PubMed] [Google Scholar]
- Cornelis GR 2006. The type III secretion injectisome. Nat Rev Microbiol 4: 811–825 [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Van Gijsegem F 2000. Assembly and function of type III secretory systems. Annu Rev Microbiol 54: 735–774 [DOI] [PubMed] [Google Scholar]
- Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WL, Blocker AJ, Lea SM 2006. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci USA 103: 12529–12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Adler J 1971a. Purification of intact flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 105: 376–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Adler J 1971b. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 105: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosier D 2006. Bacterial flagellum: visualizing the complete machine in situ. Curr Biol 16: R928–930 [DOI] [PubMed] [Google Scholar]
- Fadouloglou VE, Tampakaki AP, Glykos NM, Bastaki MN, Hadden JM, Phillips SE, Panopoulos NJ, Kokkinidis M 2004. Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc Natl Acad Sci U S A 101: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Macnab RM 1996. Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem 271: 31981–31988 [DOI] [PubMed] [Google Scholar]
- Ferris HU, Minamino T 2006. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol 14: 519–526 [DOI] [PubMed] [Google Scholar]
- Galán JE, Collmer A 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Galán JE, Wolf-Watz H 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Gophna U, Ron EZ, Graur D 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312: 151–163 [DOI] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 176: 5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson JL, Horsley A, Stabat D, Simon M, Johnson S, da Fonseca PC, Morris EP, Wall JS, Lea SM, Blocker AJ 2009. Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol 16: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Komeda Y, Iino T, Macnab RM 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol 169: 1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, DeRosier DJ, Macnab RM 1990. Flagellar hook and hook-associated proteins of Salmonella typhimurium and their relationship to other axial components of the flagellum. J Mol Biol 213: 819–832 [DOI] [PubMed] [Google Scholar]
- Hueck C 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262: 1277–1280 [DOI] [PubMed] [Google Scholar]
- Iino T 1974. Assembly of Salmonella flagellin in vitro and in vivo. J Supramol Struct 2: 372–384 [DOI] [PubMed] [Google Scholar]
- Imada K, Minamino T, Tahara A, Namba K 2007. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc Natl Acad Sci USA 104: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P, Cornelis GR 2003. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Journet L, Hughes KT, Cornelis GR 2005. Type III secretion: a secretory pathway serving both motility and virulence (review). Mol Membr Biol 22: 41–50 [DOI] [PubMed] [Google Scholar]
- Kamiya R, Asakura S, Wakabayashi K, Namba K 1979. Transition of bacterial flagella from helical to straight forms with different subunit arrangements. J Mol Biol 131: 725–742 [DOI] [PubMed] [Google Scholar]
- Kimbrough TG, Miller SI 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci U S A 97: 11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int Rev Cytol 233: 93–134 [DOI] [PubMed] [Google Scholar]
- Kojima S, Furukawa Y, Matsunami H, Minamino T, Namba K 2008. Characterization of the periplasmic domain of MotB and implications for its role in the stator assembly of the bacterial flagellar motor. J Bacteriol 190: 3314–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster M, Bitter W, de Cock H, Allaoui A, Cornelis GR, Tommassen J 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol 26: 789–797 [DOI] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán JE, Aizawa SI 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280: 602–605 [DOI] [PubMed] [Google Scholar]
- Kubori T, Sukhan A, Aizawa SI, Galán JE 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A 97: 10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SA, Tang H, Wang X, Billings S, Blair DF 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol 178: 223–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab R 2003. How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100 [DOI] [PubMed] [Google Scholar]
- Macnab RM 2004. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta 1694: 207–217 [DOI] [PubMed] [Google Scholar]
- Macnab RM, Ornston MK 1977. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol 112: 1–30 [DOI] [PubMed] [Google Scholar]
- Maki-Yonekura S, Yonekura K, Namba K 2003. Domain movements of HAP2 in the cap-filament complex formation and growth process of the bacterial flagellum. Proc Natl Acad Sci U S A 100: 15528–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson M, Tedesco P, Berg H, Harold F, Van der Drift C 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci 74: 3060–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306: 1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura A, Shioi JI, Imae Y 1977. Motility in Bacillus subtilis driven by an artificial protonmotive force. FEBS Lett 82: 187–190 [DOI] [PubMed] [Google Scholar]
- Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K 1995. The structure of the R-type straight flagellar filament of Salmonella at 9 A resolution by electron cryomicroscopy. J Mol Biol 249: 69–87 [DOI] [PubMed] [Google Scholar]
- Minamino T, Macnab RM 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J Bacteriol 181: 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol Microbiol 37: 1494–1503 [DOI] [PubMed] [Google Scholar]
- Minamino T, Namba K 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451: 485. [DOI] [PubMed] [Google Scholar]
- Minamino T, Yamaguchi S, Macnab RM 2000. Interaction between FliE and FlgB, a proximal rod component of the flagellar basal body of Salmonella. J Bacteriol 182: 3029–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K 2008a. Mechanisms of type III protein export for bacterial flagellar assembly. Mol Biosyst 4: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K 2008b. Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18: 693–701 [DOI] [PubMed] [Google Scholar]
- Moraes TF, Spreter T, Strynadka NC 2008. Piecing together the type III injectisome of bacterial pathogens. Curr Opin Struct Biol 18: 258–266 [DOI] [PubMed] [Google Scholar]
- Morgan DG, Macnab RM, Francis NR, DeRosier DJ 1993. Domain organization of the subunit of the Salmonella typhimurium flagellar hook. J Mol Biol 229: 79–84 [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR 2005. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310: 674–676 [DOI] [PubMed] [Google Scholar]
- Namba K 2001. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6: 1–12 [DOI] [PubMed] [Google Scholar]
- Namba, Vonderviszt 1997. Molecular architecture of bacterial flagellum. Q Rev Biophys 30: 1–65 [DOI] [PubMed] [Google Scholar]
- Okabe M, Minamino T, Imada K, Namba K, Kihara M 2009. Role of the N-terminal domain of FliI ATPase in bacterial flagellar protein export. FEBS Lett 583: 743–748 [DOI] [PubMed] [Google Scholar]
- Parsot C, Hamiaux C, Page AL 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol 6: 7–14 [DOI] [PubMed] [Google Scholar]
- Patterson-Delafield J, Martinez RJ, Stocker BA, Yamaguchi S 1973. A new fla gene in Salmonella typhimurium – flaR – and its mutant phenotype-superhooks. Archiv für Mikrobiologie 90: 107–120 [DOI] [PubMed] [Google Scholar]
- Paul K, Blair DF 2006. Organization of FliN subunits in the flagellar motor of Escherichia coli. J Bacteriol 188: 2502–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT 2008. Energy source of flagellar type III secretion. Nature 451: 489–492 [DOI] [PubMed] [Google Scholar]
- Pozidis C, Chalkiadaki A, Gomez-Serrano A, Stahlberg H, Brown I, Tampakaki AP, Lustig A, Sianidis G, Politou AS, Engel A, et al. 2003. Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J Biol Chem 278: 25816–25824 [DOI] [PubMed] [Google Scholar]
- Samatey, Imada, Nagashima, Vonderviszt, Kumasaka, Yamamoto M, Namba K 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410: 331–337 [DOI] [PubMed] [Google Scholar]
- Samatey FA, Matsunami H, Imada K, Nagashima S, Shaikh TR, Thomas DR, Chen JZ, Derosier DJ, Kitao A, Namba K 2004. Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism. Nature 431: 1062–1068 [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Thomas DR, Chen JZ, Samatey FA, Matsunami H, Imada K, Namba K, Derosier DJ 2005. A partial atomic structure for the flagellar hook of Salmonella typhimurium. Proc Natl Acad Sci USA 102: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M, Simon M 1974. Flagellar rotation and the mechanism of bacterial motility. Nature 249: 73–74 [DOI] [PubMed] [Google Scholar]
- Sockett H, Yamaguchi S, Kihara M, Irikura VM, Macnab RM 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol 174: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampakaki A, Fadouloglou V, Gazi A, Panopoulos N, Kokkinidis M 2004. Conserved features of type III secretion. Cell Microbiol 6: 805–816 [DOI] [PubMed] [Google Scholar]
- Thomas DR, Morgan DG, DeRosier DJ 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci U S A 96: 10134–10139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Morgan DG, DeRosier DJ 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J Bacteriol 183: 6404–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188: 7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AP, Homma M, Irikura VM, Macnab RM 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol 173: 3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilharm G, Lehmann V, Krauss K, Lehnert B, Richter S, Ruckdeschel K, Heesemann J, Trülzsch K 2004. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect Immun 72: 4004–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas NA, Pfuetzner RA, Frey EA, Finlay BB, Miller SI, Strynadka NC 2005. Structural characterization of the molecular platform for type III secretion system assembly. Nature 435: 702–707 [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki S, Morgan D, DeRosier D, Vonderviszt F, Imada K, Namba K 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Yonekura K, Maki-Yonekura S, Namba K 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424: 643–650 [DOI] [PubMed] [Google Scholar]
- Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC 2007. Structural analysis of a prototypical ATPase from the type III secretion system. Nature Struct & Mol Biol 14: 131–137 [DOI] [PubMed] [Google Scholar]