SUMMARY
Much of the dynamics information is lost in bulk measurements because of the population averaging. Single-molecule methods measure one molecule at a time; they provide knowledge not obtainable by other means. In this article, we review the application of the two most widely used single-molecule methods—fluorescence resonance energy transfer (FRET) and force versus extension measurements—to several RNA reactions. First, we discuss folding/unfolding studies on a hairpin ribozyme that revealed multiple conformations of the RNA with distinct kinetics, and on a series of RNA pseudoknots, whose mechanical stabilities were found to show a strong correlation with their frameshifting efficiency during translation. We also discuss several RNA-related molecular motors. Single-molecule experiments revealed detailed mechanisms for the interaction of HIV reverse transcriptase and nucleic acid helicases (NS3 and RIG-1) with their substrates. Optical tweezers studies showed that translation of a single messenger RNA by a ribosome occurs by successive translocation-and-pause cycles. Single-molecule FRET experiments yielded important information on ribosome conformational changes and tRNA dynamics during translation. Overall, single-molecule experiments have been very valuable for understanding RNA reactions.
Single-molecule studies, such as FRET and force-extension measurements, reveal how RNA folds and interacts with helicases, reverse transcriptase, and components of the ribosome.
1. INTRODUCTION
In the usual bulk, or ensemble, measurements, the properties of all the molecules in the sample, contribute to the signal. For example, a fluorescence signal from the solution is a consequence of the different absorbance and emission properties of all the molecules with their varied dynamics. In ensemble measurements, the averaging over all conformations and species obscures the effects of minor contributions to the signal; the major contributors dominate the spectrum. However, single-molecule methods allow the measurement of the properties and reactions of one molecule at a time. By observing the conformational changes of each single molecule over time, one can learn about the distributions of properties and their dynamics, not just averages.
Because of the stochastic nature of kinetics, single-molecule methods are especially advantageous for studying reactions. Each molecule has a probability of reacting, but when it will actually react is not predictable. Thus, in bulk, reactions do not remain synchronized, and the resulting population average hides many kinetic details; intermediates may be difficult to detect. However, following the progression of a single molecule from reactant to product can reveal each intermediate; a detailed mechanism can be obtained. The kinetics and thermodynamics of the reaction can be obtained from the lifetimes of each conformation.
Any property that can be measured for one molecule at a time can be used to characterize molecules and their reactions. The two most widely used single-molecule methods applied to RNA structures and functions are fluorescence resonance energy transfer (FRET), and force versus extension measurements. A unique capability of studying one molecule at a time is to apply force to the molecule while not perturbing the rest of the solution. In such experiments, the molecule is attached to two beads controlled by optical tweezers, or it is attached to a surface and an atomic force microscope cantilever; the force on the molecule and the distance between the attached points are measured. Force becomes a thermodynamic variable, like temperature or pressure, that can influence a reaction. Force affects the equilibrium if there is a change in the length of the molecule during the reaction. Similarly, force affects rates of reactions depending on the distances to the transition states. Thus, force can be used to study thermodynamics and kinetics of reactions—such as unfolding of RNA—that would otherwise only occur at high temperatures, or in the presence of a denaturant. Unfolding and refolding of an RNA can be studied in the presence of proteins, other RNAs, ligands, and even mixtures approximating the contents of biological cells. Although the process is not the same as what occurs for RNA molecules in cells, it should be a better approximation than the conventional unfolding and folding studies in high concentrations of urea, or by thermal melting curves. The reversible mechanical work (force times distance) for unfolding an RNA is equal to the Gibbs free energy of unfolding. The temperature dependence of this work gives the enthalpy, and thus the entropy of the process.
In this article, we briefly describe methods used in studying single molecules, and review the applications of these methods to RNA reactions, including mechanical unfolding and folding of RNA secondary and tertiary structures, interactions with HIV reverse transcriptase, unfolding by helicases, and translation of messenger RNAs.
2. METHODS
2.1. Force
A standard method to apply force to a molecule using optical tweezers is shown in Figure 1 (Liphardt et al. 2001; Tinoco et al. 2006; Li et al. 2008). There, we see an RNA flanked by RNA•DNA-handles attached to beads by biotin-streptavidin or digoxigenin-antidigoxigenin. The motion of a helicase or a ribosome on the RNA is monitored by measuring the change of distance between the beads while the force is kept constant. As the helicase unwinds the hairpin (Dumont et al. 2006), or the ribosome translates the mRNA (Wen et al. 2008), the distance between the beads increases as base pairs become single stranded. Alternatively, the folding and unfolding of an RNA species (hairpin [Li et al. 2007], pseudoknot [Chen et al. 2007], ribozyme [Onoa et al. 2003], kissing complex [Li and Tinoco 2009], riboswitch [Greenleaf et al. 2008], and so forth) is studied by increasing, then decreasing, the force on the molecule. As each element of the structure unfolds or refolds, there is an abrupt increase or decrease in the end-to-end distance.
Figure 1.
Optical tweezers assay for studying NS3 helicase unwinding or ribosome translation. (A) The RNA has a short single-stranded region for loading NS3 helicase or ribosome, followed by a hairpin region to be unwound or translated. Note that NS3 translocates 3′ to 5′, whereas the ribosome translates mRNA 5′ to 3′. The two ends of the RNA are attached to long handles for separating the RNA/enzyme construct from the beads. The ends are then held between two micron-size polystyrene beads coated with antidigoxigenin antibody and streptavidin. Drawings are schematic and not to scale. (B, C) Representative traces of extension versus time for NS3 unwinding (B; force is maintained at 15 pN) and ribosome translation (C; force is maintained at 20 pN). Adapted from Wen et al. 2008 and Dumont et al. 2006.
The kinetics of reactions are studied by measuring the lifetimes of each species in the reaction; for example, the time a ribosome spends at each codon, or the lifetimes of intermediates in an unfolding reaction. Every time the reaction is repeated for a single molecule, the lifetime of each species will be different because of the stochastic nature of kinetics. Several steps in a reaction can occur without changing the measured property of the species. The distribution of lifetimes for a species tells you about the number of such kinetic steps that occur during the lifetime. For a single kinetic step, the distribution of lifetimes is exponential. The mean lifetime <τ> is the reciprocal of the single-step rate constant, k.
| 1 |
For more than one step contributing to the lifetime, the distribution has a maximum that does not occur at zero time and the mean lifetime depends on all the rate constants involved. The force dependence of the kinetics characterizes the position of the transition state along the reaction coordinate.
The Gibbs free energy change, ΔG, of a reaction is obtained from the reversible mechanical work done. Mechanical work is the integral of force times distance; if the force remains constant during the process, it is just the force times the change in distance.
| 2 |
An unfolding/folding reaction is reversible if it occurs at the same force in the forward process and the reverse process; there is no hysteresis. However, even for a nonreversible reaction, the reversible work can be obtained from the distribution of nonreversible work values. There are two ways this can be done. The intersection of the distribution of work values for an unfolding reaction and its refolding reaction corresponds to the reversible work—the Gibbs free energy (Crooks 1999). The unfolding of a three-helix junction from Escherichia coli ribosomal RNA that binds S15 ribosomal protein was analyzed this way (Collin et al. 2005). If the distribution of nonreversible work values for unfolding, or for refolding, but not both, is measured, the free energy is obtained from the exponential average of the distribution of work, w, values (Jarzynski 1997)
| 3 |
with k the Boltzmann constant and T the absolute temperature. Equation (3) is exact in the limit of averaging over infinite repetitions of the experiment. This method was tested on the unfolding of the P5abc three-helix junction domain from the Tetrahymena thermophila group I intron (Liphardt et al. 2002), where it was shown that a few hundred measurements of nonreversible unfolding accurately provided the free energy of unfolding.
The unfolding of single RNA molecules can also be studied using atomic force microscopy (AFM). However, up to now, nearly all AFM unfolding studies have been on proteins (Carrion-Vazquez et al. 1999; Cecconi et al. 2005; Walther et al. 2007).
2.2. Single-molecule Fluorescence Resonance Energy Transfer (smFRET)
Single-molecule fluorescence techniques allow the dissection of biochemical reactions and their molecular mechanisms in great detail (Selvin and Ha 2008; Roy et al. 2008). In FRET experiments, molecules are labeled with a donor fluorophore and an acceptor fluorophore at two specific positions. The distance between the donor and acceptor is monitored by a technique based on the 1948 theory of Förster, who showed that electronic excitation energy can be efficiently transferred through transition dipole–dipole interactions. The efficiency of energy transfer depends on the reciprocal of the distance to the sixth power, (1/R)6, the optical properties of the fluorophores, and on an orientation factor. The distance, Ro, at which energy transfer is 50% efficient, is specific to a given set of donor and acceptor molecules. The efficiency of energy transfer is given by
| 4 |
in which R is the distance between donor and acceptor. The largest variation in FRET signal occurs when R and Ro are equal, therefore donor-acceptor pairs of fluorophores are chosen to maximize the sensitivity of the experiment for the distance of interest. Usually, distances in the range of 2 nm to 10 nm are measurable by FRET.
FRET values can be obtained from donor and acceptor fluorescence intensities, and then used to calculate the distance between donor and acceptor using Equation (4). Measuring accurate FRET values, thus accurately determining R, requires knowledge of quantum yields of donor and acceptor. In practice, FRET values are often approximated as:
| 5 |
in which ID and IA are fluorescence intensities of donor and acceptor, respectively. Equation (5) suffices for identifying changes in the donor–acceptor distance. As a protein or RNA unfolds, or a molecular motor moves, the distance between donor and acceptor changes and FRET changes; in principle, FRET can also change if Ro changes, for example, if the orientation factor changes (Iqbal et al. 2008).
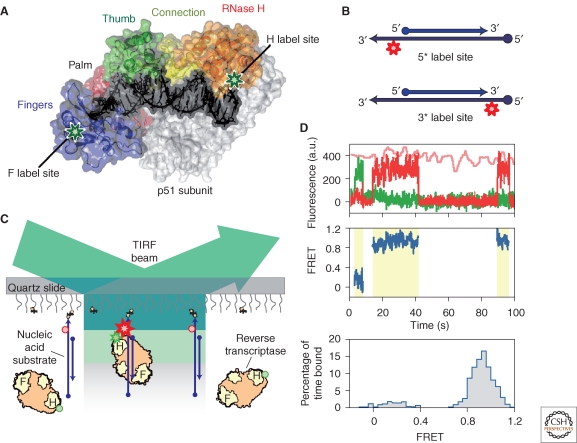
The usual single-molecule FRET (smFRET) method involves excitation of a donor by the evanescent wave in a total internal reflection fluorescence (TIRF) microscope, as shown in Figure 3C. Only molecules very close to the surface (usually within 100 nm) are excited by the evanescent laser light; data can be collected from hundreds of individual molecules simultaneously. Each molecule can be studied during multiple transitions or reactions, until one of the fluorophores undergoes photobleaching. Single-molecule FRET has been applied to translation of mRNA (Blanchard et al. 2004b; Fei et al. 2008; Cornish et al. 2009), unfolding by helicases (Rasnik et al. 2006), protein-mediated folding (Stone et al. 2007), ribozyme dynamics (Pereira et al. 2008), etc.
Figure 3.
Single-molecule FRET assay for probing the binding mechanism of HIV-1 reverse transcriptase (RT) on nucleic acid substrates. (A) The structure of HIV-1 RT bound to a substrate. RT is labeled with Cy3 at one of the two labeling sites (green stars). (B) The substrate consists of a 19–21-nt primer strand (DNA or RNA) annealed to a 50-nt DNA template strand to mimic the different binding substrates that RT encounters in vivo. The template is labeled with a Cy5 dye (red star) at either the 5′ or 3′ end. (C) One end of the substrate is immobilized on the coverslip surface for single-molecule detection. The RT is free to diffuse in solution. Under 532 nm excitation wavelength in a total internal reflection fluorescence (TIRF) microscospe, no fluorescence is detected without RT binding; when an RT binds to the substrate, the donor dye is excited by the evanescent laser illumination and the FRET signal of the dye pair can be observed until the RT dissociates from the substrate or the dyes photobleach. The stars and spheres indicate dyes that do and do not emit fluorescence, respectively. (D) An example of smFRET analysis. Top: fluorescence time traces from Cy3 (green) and Cy5 (red) under excitation at 532 nm, and that from Cy5 (pink) under excitation at 635 nm (to confirm that Cy5 has not photobleached). Middle: FRET value calculated over the duration of the binding events (yellow shaded regions). Bottom: FRET distribution histogram created for the binding events. The observed FRET value and its change over time report the RT binding orientation, and the dynamics of RT orientation change, respectively. The same experiment was repeated using different FRET labeling sites on the RT and/or substrate to strengthen the conclusions. (Reprinted, with permission, from Abbondanzieri et al. 2008 [© MacMillan].)
3. FOLDING, UNFOLDING, AND FUNCTION
RNA function depends on its folded structure and on dynamic fluctuations between functional states. Single-molecule methods are ideal for following the dynamics of folding and unfolding of the elements of RNA secondary and tertiary structure. Knowledge of the stabilities and dynamics of the folded structures, of intermediates in their formation, and of how these properties are affected by the binding of ligands or protein cofactors, facilitates better understanding of their functions and their structures. Many single-molecule experiments have been done in this line of research; here, we discuss only two systems as examples.
3.1. Hairpin Ribozyme
The hairpin ribozyme derived from tobacco ring spot virus satellite RNA is a self-cleaving four-helix-junction RNA structure containing two internal loops (Wilson et al. 2005). The tertiary structure of this ribozyme involves docking loop A with loop B, which facilitates the site-specific reversible cleavage and ligation reactions in loop A. This ribozyme has been extensively studied by smFRET (Ha 2004; Bokinsky and Zhuang 2005). A minimal hairpin ribozyme, with loop A and loop B domains connected by a six-nucleotide (nt) bulge, was made for both bulk and single-molecule studies. The kinetics were obtained from the lifetime distributions of undocked and docked conformations (Ditzler et al. 2008). By fitting the lifetime distributions to multiple exponential functions, smFRET revealed that there were multiple populations of molecules with distinct undocking kinetics (Ditzler et al. 2008). Surprisingly, the molecules with relatively slower undocking kinetics retained their undocking rates even after separation of the two strands of the minimal hairpin ribozyme by denaturing gel electrophoresis. The authors proposed that the intra-strand structure of the S-turn or loop E motif within loop B might remain through extensive denaturation and annealing processes (Ditzler et al. 2008). It is also possible that subpopulations of hairpin ribozymes are kinetically trapped in misfolded secondary structures in other parts of the ribozyme. Secondary structure dynamics can be followed by characteristic fluorescent properties that depend directly on self-cleavage/ligation reactions of the hairpin ribozyme (Nahas et al. 2004; Liu et al. 2007).
Directly monitoring RNA global conformational changes can be facilitated by constructing multiple FRET pairs at different positions. Three-color smFRET methods have been developed to probe global structural dynamics of a Holliday junction (Hohng et al. 2004). Sequential and simultaneous multiple-color smFRET experiments will be useful in directly revealing the hidden folding dynamics of hairpin ribozymes and other larger RNA molecules (Qu et al. 2008; Steiner et al. 2008).
Mechanical force has also been applied to perturb and map the docking/undocking pathways and kinetics, and thus provide new insight into molecular heterogeneity and catalytic function. Using a combined instrument of optical tweezers and smFRET, the kinetics of the conformational switch of a Holliday junction was found to be affected by mechanical force of less than 0.5 pN (Hohng et al. 2007). Applying forces in different directions revealed structures of the transient species during the conformation changes of the Holliday junction. The presence of native and near-native “quasi-docked” tertiary structures of a hairpin ribozyme may be revealed by their distinct force-dependent undocking kinetics. Force may also be used to directly test whether the heterogeneity observed in the hairpin ribozyme persists throughout mechanical denaturation.
3.2. Pseudoknots and Frameshifting
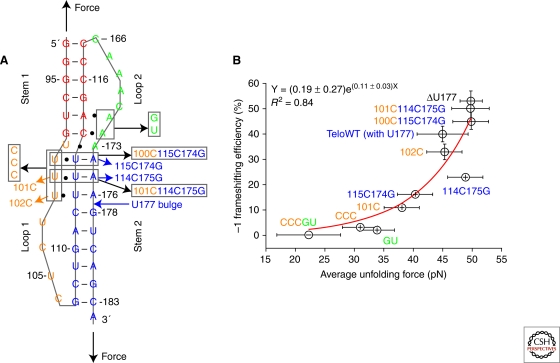
All the secondary and tertiary structures in the coding region of an mRNA have to be unfolded to single strand to be translated by the ribosome. Pseudoknot structures (see Fig. 2A for a typical pseudoknot containing two stems and two loops) stimulate programmed –1 ribosomal frameshifting at an upstream slippery sequence of the form X XXY YYZ (0 frame) to XXX YYY Z (–1 frame), in which X is any three identical nucleotides, Y is either AAA or UUU, and in eukaryotes Z is usually not G. With the mRNA slippery sequence located at the aminoacyl (A) and peptidyl (P) ribosome sites, a downstream pseudoknot structure provides resistance to ribosomal helicase activity and stimulates –1 ribosomal frameshifting.
Figure 2.
Pseudoknots and –1 ribosomal frameshifting. (A) Pseudoknots used for bulk frameshift assays and single-molecule studies. The pseudoknots contain stem 1 (red), loop 1 (yellow), stem 2 (blue), and loop 2 (green). All the mutants were made based on ΔU177. In mutant CCCGU, all base triples are disrupted (two in stem 1-loop 2 and three in stem 2-loop 1). In TeloWT, the single nucleotide bulge U177 in stem 2 is added. The directions of applied mechanical force by optical tweezers are shown with black arrows. (B) Correlation observed between bulk –1 frameshifting efficiency and average unfolding force. Error bars are standard deviations from bulk and single-molecule experiments. Adapted from Chen et al. 2009.
Mechanical unfolding of mRNA structures using optical tweezers (Fig. 1A) may mimic the helicase activity of ribosomes and provide insight into the molecular determinants of pseudoknot mechanical stability and –1 ribosomal frameshifting. Mechanical unfolding of frameshifting stimulatory pseudoknots from infectious bronchitis virus (IBV) revealed that mechanical stability and frameshifting efficiency are affected by the sequence and length of their stems (Hansen et al. 2007; Green et al. 2008).
Bulk frameshifting assays suggested that in addition to Watson-Crick base pairing interactions, minor-groove stem 1-loop 2 interactions (Fig. 2A) are also important in stimulating –1 frameshifting in some IBV pseudoknots (Liphardt et al. 1999). A crystal structure of a pseudoknot in beet western yellows virus (BWYV) revealed the presence of extensive minor-groove base triples important for stimulating –1 frameshifting (Kim et al. 1999; Su et al. 1999).
More recently, both major-groove and minor-groove base triples were found to enhance mechanical stability and increase –1 frameshifting efficiency in 11 pseudoknots studied (Fig. 2) (Chen et al. 2009). Excellent correlation was found between mechanical stability and frameshifting efficiency. The results indicate that –1 frameshifting is stimulated by: (1) stabilizing stem 1 by forming minor-groove stem 1-loop 2 base triples; and (2) increasing torsional resistance to unfolding by forming major-groove stem 2-loop 1 base triples. The combined mutational and single-molecule studies suggested that the folding intermediate pseudoknot structures (with partial formation of stem 2 and no base triples formed) unfold in one step or two steps at low force, whereas native pseudoknots unfold in one step at high force. The folding intermediate pseudoknot structures probably do not induce high-efficiency –1 frameshifting, because the unfolding force is typically below 35 pN (see Fig. 2B) (Chen et al. 2009).
Mechanical unfolding allows direct measurement of how base triple formation stabilizes a Watson-Crick duplex structure. Remarkably, codon-anticodon recognition is enhanced by two base triples formed at the minor-groove of the first two Watson-Crick pairs of the decoding A-site codon-anticodon duplex (Ogle et al. 2001). It will be interesting to see how –1 frameshifting is affected by the A-site base triples and other interactions within the ribosome complex.
4. MOLECULAR MOTORS
Molecular motors include a wide range of biological devices usually powered by ATP or GTP. Some motors deal with proteins, such as myosins, which walk along actin, and kinesins, which carry cargo along microtubules. Other motors are vital in gene replication, transcription, and translation (Seidel and Dekker 2007). Much detailed information on the dynamics of molecular motors has been obtained by following the motors’ action in real time. We will concentrate here on four motors with RNA as their substrates.
4.1. HIV Reverse Transcriptase
The infection of human immunodeficiency virus (HIV) relies on the conversion of its single-stranded RNA genome into double-stranded DNA, which is later incorporated into the host genome. This process involves three major steps: RNA-directed DNA synthesis (the synthesis of minus-strand DNA using the HIV RNA genome as template), DNA-directed RNA hydrolysis (the cleavage of the RNA template at multiple places), and DNA-directed DNA synthesis (the synthesis of the plus-strand DNA using the minus-strand DNA as the template). Amazingly, these different reactions are done by a single enzyme, the HIV reverse transcriptase (RT). RT consists of two subunits, one of which contains DNA polymerase and RNase H domains. The different nucleic acid substrates regulate the RT activities, but the mechanism of the regulation was poorly understood. The DNA directed DNA synthesis activity of RT has been studied by single-molecule experiments with or without force (Lu et al. 2004; Ortiz et al. 2005; Kim et al. 2007b).
Using smFRET techniques (see Fig. 3 caption for experimental details) (Abbondanzieri et al. 2008), Zhuang and her colleagues found that RT binds to the DNA template with a DNA or RNA primer in two opposite orientations, with either the DNA polymerase domain or the RNase H domain close to the 3′ end of the primer. This correlates nicely with RT's function of DNA-directed DNA synthesis or DNA-directed RNA hydrolysis on such substrates. RT has both DNA polymerase and RNase H function on other substrates, such as the chimeric DNA/RNA primer, a primer with two special 15-nt RNA purine sequences called polypurine tracts (PPT), and the primer with PPT and a few nucleotides of DNA extension. On such substrates, RT was observed to occupy both the DNA-polymesase-competent and RNase-H-competent orientations and to dynamically flip between the two orientations. The measured rate of primer extension correlated with the fraction of time for which the RT enzyme bound in the polymerase-competent orientation. Thus, these experiments suggest that RT can distinguish between different substrates and binds differently, and that the binding orientation determines the subsequent function. Small-molecule ligands, specifically dNTP and two clinically approved anti-HIV drugs (nevirapine and efavirenz), were also studied and found to greatly affect the equilibrium and flipping dynamics of RT binding orientation, and therefore, to regulate the RT activity.
RT has very low processivity for DNA synthesis (a few to a few hundred nucleotides), but the whole HIV genome is ∼10 kb long. Thus, having a highly efficient searching mechanism to locate the target site is crucial for RT function. Using smFRET, the Zhuang group observed that RT slides between the two ends of the substrate when the substrate is considerably longer than 19 nt (the length that RT covers on binding) (Liu et al. 2008). And this sliding motion was shown to be thermally driven. Very interestingly, it was also observed that when RT locates the target site on the substrate, it can flip its binding orientation to place the correct functional domain (DNA polymerase domain for DNA synthesis or the RNase H domain for RNA hydrolysis) close to the target site. The combination of sliding and flipping provides a very efficient searching mechanism. Furthermore, the nontemplate strand displacement capability of RT during DNA synthesis was investigated. It was found that in RNA strand displacement synthesis, RT was able to extend the primer with a few nucleotides before termination; but in DNA strand displacement synthesis, RT was able to complete a long stretch of primer extension before termination. The observed difference is consistent with the fact that DNA•RNA hybrids are usually more thermodynamically stable than DNA•DNA duplexes. This also shows that RT is not a very powerful motor in regard to strand displacement.
4.2. RNA Helicases
RNA helicases use nucleotide triphosphates (NTP) to unwind RNA duplexes and are involved in many viral and cellular RNA metabolism processes (Jankowsky and Fairman 2007; Pyle 2008). Some RNA helicases actually function as translocases without unwinding RNA structures. Understanding how RNA unwinding/translocating correlates with NTP binding, hydrolysis, and helicase conformational change is central to the understanding of the mechanism of helicase function. Even though single-molecule techniques have been successfully applied to many DNA helicases (Hopfner and Michaelis 2007) and other nucleic acid motors (Seidel and Dekker 2007), there have been only a few single-molecule studies of RNA helicases (Dumont et al. 2006; Marsden et al. 2006; Cheng et al. 2007; Yang et al. 2007; Myong et al. 2009).
Marsden et al. (Marsden et al. 2006) used AFM to stretch a single RNA hairpin in the presence of RNA helicases: eIF4A or Ded1p, which are involved in eukaryotic translation. Both helicases lower the unfolding force of the hairpin during AFM pulling, suggesting that both helicases weaken the RNA hairpin stability. Yang et al. (Yang et al. 2007) applied smFRET techniques to study Ded1p. However, instead of studying the molecular motor properties of Ded1p, the authors focused on how Ded1p facilitates RNA conformational changes, which is another aspect of Ded1p function.
The nonstructural protein 3 (NS3) in hepatitis C virus (HCV) is a key component of the viral replication machinery for RNA directed RNA synthesis. It has both protease and helicase domains and can unwind both DNA and RNA duplexes. Dumont et. al (Dumont et al. 2006) used optical tweezers (Fig. 1) to study the translocation and unwinding mechanism of NS3 with an RNA hairpin substrate containing a short single-strand region for loading the NS3 monomer, and a hairpin region for the detection of the unwinding activity. It was found that NS3 unwinding of, and translocation on, RNA are coordinated by ATP in discrete steps of 11 ± 3 base pairs, and that each step is composed of three rapid substeps of 3.6 ± 1.3 base pairs, also triggered by ATP binding. Force does not affect the NS3 monomer unwinding step size, but force does increase NS3 processivity. The coupled duplex unwinding and translocation rate is much faster on A•U stretches than on G•C stretches, and the dependence on the base pair free energy suggests that NS3 actively destabilizes the RNA duplex to facilitate unwinding (Cheng et al. 2007). NS3 has a higher tendency to dissociate when encountering the barrier created by a G•C stretch following an A•U stretch, and NS3 processivity is affected up to six bases before the barrier. Using smFRET without applying force, Myong et al. observed that NS3 unwinds the DNA duplex with one base pair as the fundamental step, but several one base-pair steps accumulate tension on the NS3-DNA complex, which is relieved in a burst of three base pairs (Myong et al. 2009).
RIG-I is a cytosolic protein that detects pathogen-associated molecular patterns on viral RNA, and elicits an antiviral immune response. Wild-type RIG-I is composed of two amino-terminal tandem CARDs (caspase activation and recruitment domains), a central DExH box RNA helicase domain, and a carboxy-terminal regulatory domain (RD). Myong et al. (Myong et al. 2009) used protein-induced fluorescence enhancement (PIFE) (Fischer et al. 2004) to study RIG-I. In PIFE, the substrate is labeled with a dye, whose fluorescence emission is affected by the proximity of a protein (not fluorescently labeled). Two mutants were also studied: RIGh and svRIG, which have complete or partial deletions of the CARDs, respectively. Without ATP, all three forms of RIG-I bind steadily to the RNA substrate until dissociation. On a double-stranded RNA substrate, all three forms of RIG-I proteins translocate repetitively on the RNA substrate dependent on ATP concentration, temperature, and length of the RNA duplex region. The authors proposed that the repetitive shuttling of RIG-I on dsRNA regions of the viral genome might induce conformational changes in RIG-I important for antiviral immune response signaling.
4.3. Translation
Following movement of the ribosome along the mRNA using optical tweezers
Optical tweezers in the mode shown in Figure1A were used to monitor the motion of a single E. coli ribosome on a single messenger RNA during translation (Wen et al. 2008). A hairpin mRNA is used so that as the ribosome translates the 5′-side of the hairpin, double-stranded base pairs are converted to single strands leading to an increase in end-to-end distance of the RNA. A trajectory is seen in Figure 1C; it shows that a translating ribosome moves by a series of translocation-pause-translocation steps. Each translocation corresponds to translation of three nucleotides—one codon (with extension increase of ∼3 nm), with a mean time of approximately 25 ms (X. Qu, unpubl.). Each pause is on the order of seconds; during this time, all the biochemistry for the translation of one codon occurs.
The ability to see individual steps of translation and to separate the process into pauses and translocations, opens the door to learn how each component (either intrinsic or extrinsic) of the translation machinery affects each substep of the process of translation. Detailed mechanisms of the fidelity of translation can be obtained: amino acid misincorporation, frameshifting, read-through of stop codons, or premature termination. The effects of codon sequence, of mRNA structures such as hairpins and pseudoknots, of elongation factors and release factors, of antibiotics, etc., can all be assessed. Single-molecule methods will allow us to find answers that cannot be obtained by ensemble methods.
Transfer RNA and ribosome dynamics by smFRET
The Puglisi-Chu group pioneered the application of smFRET to observe the motion of fluorescently labeled tRNAs on the ribosome during translation (Blanchard et al. 2004a; Blanchard et al. 2004b; Kim et al. 2007a; Lee et al. 2007; Uemura et al. 2007). They found that tRNAs fluctuate between a classical state AA/PP (the anticodons are in the A-site and P-site of the 30S subunit, and the amino acid acceptor sites are in the A-site and P-site of the 50S subunit) and a hybrid state AP/PE (the anticodons remain in the A- and P-sites, but the amino acid acceptor sites move to the P- and E-sites of the 50S subunit). The hybrid state is favored by formation of the peptide bond. More recent results showed that there were two hybrid states and suggested that global conformational changes in the ribosome induced the different tRNA states (Munro et al. 2007).
smFRET measurements on fluorescently labeled ribosomes directly showed the conformational dynamics during the elongation phase of translation. The Noller and Ha groups (Cornish et al. 2008) labeled protein L9 of the 50S subunit with Cy3 (donor) and proteins S6 or S11 of the 30S subunit with Cy5 (acceptor). Spontaneous intersubunit fluctuations were seen consistent with the intersubunit rotation (racheting) seen by cryo-electron microscopy (Frank and Agrawal 2000), and the kinetics correspond to the tRNAs kinetics. The authors conclude that the intersubunit rotations correspond to the classical and hybrid states of the tRNAs. The fluctuations are thermally driven; they do not require EF-G, but the hybrid state is stabilized by EF-G binding. After translocation, the ribosome is left in the classical state.
Further characterization of ribosome and tRNA dynamics was obtained by measuring FRET between the L1 stalk of the 50S subunit and the tRNAs (Fei et al. 2008). Labeling two proteins of the large subunit, L1 and L33 (Cornish et al. 2009) or L1 and L9 (Fei et al. 2009), allows direct observation of the opening and closing of the L1 stalk, and its correlation with the classical-hybrid states of the tRNAs. The effects of the different tRNAs (initiator and elongator), the state of acylation of the tRNAs, and of EF-G on the dynamics have been measured. A detailed picture of the motions of the tRNAs, of the subunits, of individual proteins in the subunits, and of translocation dynamics along the mRNA is emerging.
It is clear that smFRET can provide useful information about relative motions, in the range of 1 to 10 nm, of the components of molecular machines. The kinetics of the motions can be accurately measured, with lifetimes in the range from milliseconds to minutes of each conformation. The long time limit is determined by the lifetime of the fluorophore; the short time limit depends on instrumental parameters.
5. CONCLUSIONS
Even though the ability to study individual molecules in solution or in biological cells has only recently become possible, its applications to biological questions are growing rapidly. Perhaps the detailed, step-by-step analysis of the mechanism of complex molecular motors, like the ribosome, will be an early success. In this article, we selected a very limited number of RNA-related studies as examples to show the capability of single-molecule techniques. Although we described only two specific single-molecule methods, optical tweezers and FRET, single-molecule techniques are actually very versatile and are evolving rapidly. We encourage the readers to explore further in single-molecule research; new methods are being discovered constantly, and some will surely help solve their problems.
ACKNOWLEDGMENTS
We thank Dr. Jin-Der Wen and Dr. Fei Liu for careful reading of the paper and for making helpful suggestions. Our work was supported by National Institute of Health Grant GM10840 and by the Human Frontiers Science Program.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X 2008. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature 453: 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard SC, Gonzalez RL Jr, Kim HD, Chu S, Puglisi JD 2004a. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol 11: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Blanchard SC, Kim HD, Gonzalez RL Jr, Puglisi JD, Chu S 2004b. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci 101: 12893–12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Zhuang X 2005. Single-molecule RNA folding. Acc Chem Res 38: 566–573 [DOI] [PubMed] [Google Scholar]
- Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM 1999. Mechanical and chemical unfolding of a single protein: A comparison. Proc Natl Acad Sci 96: 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S 2005. Direct observation of the three-state folding of a single protein molecule. Science 309: 2057–2060 [DOI] [PubMed] [Google Scholar]
- Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I Jr, 2009. Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc Natl Acad Sci 106: 12706–12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Wen JD, Tinoco I Jr, 2007. Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA 13: 2175–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Dumont S, Tinoco I Jr, Bustamante C 2007. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc Natl Acad Sci 104:, 13954–13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I Jr, Bustamante C 2005. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature 437: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T 2008. Spontaneous intersubunit rotation in single ribosomes. Mol Cell 30: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T 2009. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci 106: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE 1999. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys Rev E 60: 2721–2726 [DOI] [PubMed] [Google Scholar]
- Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG 2008. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res 36: 7088–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I Jr, Pyle AM, Bustamante C 2006. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Bronson J, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL Jr, 2009. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk direct tRNA movements during translation. Proc Natl Acad Sci 106: 15702–15707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr, 2008. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30: 348–359 [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM 2004. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol 344: 1287–1309 [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Green L, Kim CH, Bustamante C, Tinoco I Jr, 2008. Chacterization of the mechanical unfolding of RNA pseudoknots. J Mol Biol 375: 511–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319: 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T 2004. Structural dynamics and processing of nucleic acids revealed by single-molecule spectroscopy. Biochemistry 43: 4055–4063 [DOI] [PubMed] [Google Scholar]
- Hansen TM, Reihani SN, Oddershede LB, Sorensen MA 2007. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci 104: 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Joo C, Ha T 2004. Single-molecule three-color FRET. Biophys J 87: 1328–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DM, Ha T 2007. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the holliday junction. Science 318: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Michaelis J 2007. Mechanisms of nucleic acid translocases: Lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol 17: 87–95 [DOI] [PubMed] [Google Scholar]
- Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DM 2008. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci 105: 11176–11181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME 2007. RNA helicases - one fold for many functions. Curr Opin Struct Biol 17: 316–324 [DOI] [PubMed] [Google Scholar]
- Jarzynski C 1997. Nonequilibrium equality for free energy differences. Phys Rev Lett 78: 2690–2693 [Google Scholar]
- Kim HD, Puglisi JD, Chu S 2007a. Fluctuations of transfer RNAs between classical and hybrid states. Biophys J 93: 3575–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Blainey PC, Schroeder CM, Xie XS 2007b. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat Methods 4: 397–399 [DOI] [PubMed] [Google Scholar]
- Kim YG, Su L, Maas S, O’Neill A, Rich A 1999. Specific mutations in a viral RNA pseudoknot drastically change ribosomal frameshifting efficiency. Proc Natl Acad Sci 96: 14234–14239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S 2007. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci 104: 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PTX, Tinoco I Jr, 2009. Mechanical unfolding of two DIS RNA kissing complexes from HIV-1. J Mol Biol 386: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PTX, Bustamante C, Tinoco I Jr, 2007. Real-time control of the energy landscape by force directs the folding of RNA molecules. Proc Natl Acad Sci 104: 7039–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PTX, Vieregg J, Tinoco I Jr, 2008. How RNA Unfolds and Refolds. Ann Rev Biochem 77: 77–100 [DOI] [PubMed] [Google Scholar]
- Liphardt J, Dumont S, Smith SB, Tinoco I Jr, Bustamante C 2002. Equilibrium information from nonequilibrium measurements in an experimental test of Jarzynski’s equality. Science 296: 1832–1835 [DOI] [PubMed] [Google Scholar]
- Liphardt J, Napthine S, Kontos H, Brierley I 1999. Evidence for an RNA pseudoknot loop-helix interaction essential for efficient -1 ribosomal frameshifting. J Mol Biol 288: 321–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liphardt J, Onoa B, Smith SB, Tinoco I Jr, Bustamante C 2001. Reversible unfolding of single RNA molecules by mechanical force. Science 292: 733–737 [DOI] [PubMed] [Google Scholar]
- Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X 2008. Slide into action: Dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science 322: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bokinsky G, Walter NG, Zhuang X 2007. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic "fingerprinting". Proc Natl Acad Sci 104: 12634–12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Macosko J, Habel-Rodriguez D, Keller RW, Brozik JA, Keller DJ 2004. Closing of the fingers domain generates motor forces in the HIV reverse transcriptase. J Biol Chem 279: 54529–54532 [DOI] [PubMed] [Google Scholar]
- Marsden S, Nardelli M, Linder P, McCarthy JEG 2006. Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J Mol Biol 361: 327–335 [DOI] [PubMed] [Google Scholar]
- Munro JB, Altman RB, O’Connor N, Blanchard SC 2007. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell 25: 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, Ha T 2009. Cytosolic viral sensor RIG-I Is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323: 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DMJ, Ha T 2004. Observation of internal cleavage and ligation reactions of a ribozyme. Nat Struct Mol Biol 11: 1107–1113 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I Jr, Bustamante C 2003. Identifying kinetic barriers to mechanical unfolding of the T. thermophila ribozyme. Science 299: 1892–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz TP, Marshall JA, Meyer LA, Davis RW, Macosko JC, Hatch J, Keller DJ, Brozik JA 2005. Stepping statistics of single HIV-1 reverse transcriptase molecules during DNA polymerization. J Phys Chem B 109: 16127–16131 [DOI] [PubMed] [Google Scholar]
- Pereira MJ, Nikolova EN, Hiley SL, Jaikaran D, Collins RA, Walter NG 2008. Single VS ribozyme molecules reveal dynamic and hierarchical folding toward catalysis. J Mol Biol 382: 496–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM 2008. Translocation and unwinding mechanisms of RNA and DNA helicases. Ann Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF 2008. Single-molecule nonequilibrium periodic Mg2+-concentration jump experiments reveal details of the early folding pathways of a large RNA. Proc Natl Acad Sci 105: 6602–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, Myong S, Ha T 2006. Unraveling helicase mechanisms one molecule at a time. Nucleic Acids Res 34: 4225–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T 2008. A practical guide to single-molecule FRET. Nat Methods 5: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel R, Dekker C 2007. Single-molecule studies of nucleic acid motors. Curr Opin Struct Biol 17: 80–86 [DOI] [PubMed] [Google Scholar]
- Selvin PR, Ha T 2008. Single-molecule techniques, a laboratory manual. Cold Spring Harbor Laboratory [Google Scholar]
- Steiner M, Karunatilaka KS, Sigel RK, Rueda D 2008. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci 105: 13853–13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MD, Mihalusova M, O’Connor C M, Prathapam R, Collins K, Zhuang X 2007. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 446: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Chen L, Egli M, Berger JM, Rich A 1999. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat Struct Biol 6: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I Jr, Li PTX, Bustamante C 2006. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys 39: 325–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S 2007. Peptide bond formation destabilizes Shine-Dalgarno interaction on the ribosome. Nature 446: 454–457 [DOI] [PubMed] [Google Scholar]
- Walther KA, Grater F, Dougan L, Badilla CL, Berne BJ, Fernandez JM 2007. Signatures of hydrophobic collapse in extended proteins captured with force spectroscopy. Proc Natl Acad Sci 104: 7916–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I Jr, 2008. Following translation by single ribosomes one codon at a time. Nature 452: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Nahas M, Ha T, Lilley DM 2005. Folding and catalysis of the hairpin ribozyme. Biochem Soc Trans 33: 461–465 [DOI] [PubMed] [Google Scholar]
- Yang QS, Fairman ME, Jankowsky E 2007. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol 368: 1087–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]