Abstract
Translational repression mediated by RNA-binding proteins or micro RNAs has emerged as a major regulatory mechanism for fine-tuning important biological processes. In Caenorhabditis elegans, translational repression of the key sex-determination gene tra-2 (tra, transformer) is controlled by a 28-nucleotide repeat element, the TRA-2/GLI element (TGE), located in its 3′ untranslated region (UTR). Mutations that disrupt TGE or the germline-specific TGE-binding factor GLD-1 increase TRA-2 protein expression and inhibit sperm production in hermaphrodites. Here we report the characterization of the sup-26 gene, which regulates sex determination in the soma and encodes an RNA recognition motif (RRM)-containing protein. We show that SUP-26 regulates the level of the TRA-2 protein through TGE in vivo and binds directly to TGE in vitro through its RRM domain. Interestingly, SUP-26 associates with poly(A)-binding protein 1 (PAB-1) in vivo and may repress tra-2 expression by inhibiting the translation-stimulating activity of PAB-1. Taken together, our results provide further insight into how mRNA-binding factors repress translation and modulate sexual development in different tissues of C. elegans.
Translational repression through cis-acting elements in mRNAs is an important post-transcriptional regulatory mechanism in numerous biological systems (1). Analysis of the lengths of 5′ and 3′ untranslated regions (UTRs) shows that the average lengths of 5′ UTRs are relatively constant across phyla, whereas the lengths of 3′ UTRs increase with organism complexity (200 bp for yeast and 500 bp for humans), suggesting that they may be more highly regulated during animal development (2). Sequence elements or modifications in 3′ UTR are known to control the subcellular localization, stability, and translational efficiency of mRNAs. For example, the poly(A) sequence is important not only for the stability of mRNAs, but also for stimulating translation initiation by facilitating interaction of poly(A)-binding (PAB) protein with translation initiation factor eIF4G at the 5′ cap and formation of circularized mRNA (1). Moreover, numerous important developmental regulators, such as Bicoid and the cytoplasmic polyadenylation-element binding protein, act by binding 3′ UTRs and repressing translation (1). Finally, translational repression by microRNAs (miRNAs) is mediated primarily by formation of nonperfect duplexes between miRNAs and their mRNA targets at 3′ UTRs, which induces the formation of the translation repressive complex termed the RNA induced silencing complex (3). Therefore, cis elements in the 3′ UTR of mRNAs play critical roles in regulating the efficiency of translation.
Sex differentiation in Caenorhabditis elegans is determined by the X chromosome:autosome ratio: 1:2 results in XO males and 1:1 results in XX hermaphrodites (4, 5). Hermaphrodites are essentially females that produce sperm before oogenesis and are capable of self-fertilization and mating with other males. Male development is initiated by expression of a male-promoting secreted protein, HER-1 (hermaphrodization) (6, 7), which binds and inactivates the hermaphrodite-promoting transmembrane receptor TRA-2 (TRA, transformer), which is also important for sperm production (8, 9). TRA-2 interacts with and suppresses the male-promoting activity of an intracellular protein complex containing FEM-1 (feminization), FEM-2, and FEM-3 (10–13). How TRA-2 inhibits the activities of FEM proteins is poorly understood, but it may involve cleavage of the intracellular domain of TRA-2 by the TRA-3 calpain protease and subsequent translocation of the TRA-2 intracellular domain to the nucleus (14–16). The FEM-1/FEM-2/FEM-3 complex promotes male development by inhibiting the activity of the terminal sex-determination factor, TRA-1A, a zinc-finger transcription factor that promotes hermaphrodite development by repressing expression of genes required for sperm production and somatic male development (17, 18).
Translation repression plays an important role in regulating C. elegans sex differentiation. For example, the activities of fem-3 and tra-2 are regulated by translational repressors acting in specific tissues (19, 20). Several gain-of-function, feminizing mutations in the tra-2 gene were found to alter the tra-2 3′ UTR (20, 21). Further studies revealed that two different sequence elements in the tra-2 3′ UTR regulate TRA-2 translation. First, the TRA-2 retention element retains the tra-2 message in the nucleus and thus prevents translation (22). Second, the TRA-2/GLI element (TGE) is a conserved 28-nucleotide repeat element found in both C. elegans tra-2 and D. melanogaster GLI 3′ UTRs (20, 23). Mutations disrupting TGEs increase tra-2 poly(A) tail length (23, 24) and TRA-2 protein levels in both the germline and the soma (23, 25), suggesting that TGEs negatively regulate tra-2 expression. In the germline, repression of tra-2 translation is mediated by GLD-1 (germline development defective), a TGE-binding protein and a member of the STAR family of RNA-binding proteins (25), and by FOG-2 (feminization of germline), a GLD-1–interacting and F-box–containing protein (25, 26). FOG-2, GLD-1, and tra-2 3′ UTR form a ternary complex to repress tra-2 translation in the germline (26). However, GLD-1 and FOG-2 are expressed only in the germline, and it is unclear how TGEs mediate repression of tra-2 translation in somatic tissues (26, 27).
Here we report the molecular and biochemical characterization of sup-26 (suppressor). Loss-of-function (lf) mutations in sup-26 are semidominant suppressors of the masculinization defect in her-1(n695gf) XX animals and can suppress other masculinization defects in the absence of her-1, indicating that sup-26 likely acts downstream of her-1 to affect somatic sex determination (28). We find that sup-26 encodes an RNA recognition motif (RRM) containing protein that is expressed widely in somatic tissues, regulates the level of the tra-2 protein in the soma through the TGEs in the tra-2 3′ UTR, and binds directly to TGEs in vitro. Therefore, SUP-26 is a somatic TGE-binding factor that promotes male development by repressing tra-2 translation.
Results and Discussion
sup-26 Encodes a Protein with Two RRM Motifs.
sup-26(n1091) was isolated as a semidominant suppressor of the masculinized defect of her-1(n695gf) XX animals (Table S1) and mapped to a small genetic interval at approximately –3.2 genetic map units on linkage group (LG) III (28). To clone sup-26, fosmids covering this genetic interval were injected into sup-26(n1091); her-1(n695) animals and tested for restoration of the masculinized Tra phenotype (Fig. 1A). Two overlapping fosmids, WRM0627bE08 and WRM066dA01, each restored the Tra phenotype (Fig. 1A). The overlapping region of the two fosmids contains a single ORF, R10E4.2 (Fig. 1A). A translational GFP fusion that contains a 4-kb genomic fragment, including a 1.1-kb sequence 5′ of the R10E4.2 start codon, also restored the Tra phenotype in sup-26(n1091); her-1(n695) animals (Table S1; Methods), indicating that R10E4.2 is responsible for the rescuing activity. We determined R10E4.2 DNA sequences from two different sup-26 mutants, sup-26(ct49) and sup-26(n1091), and found a C-to-T transition in sup-26(ct49), which converts codon Q20 to an ochre stop codon, and a G-to-A transition in sup-26(n1091), which converts codon C215 to a tyrosine codon. Two independently isolated deletion mutations, gk403 (a 424-bp deletion) and gk426 (a 676-bp deletion), each of which removes the first two exons of R10E4.2 (Fig. 1A), also suppressed the her-1(n695) Tra phenotype, confirming that R10E4.2 is sup-26 (Table S1). Given the molecular nature of the two deletions and sup-26(ct49), they are likely strong lf or null mutations. However, sup-26 mutant males or hermaphrodites alone display no obvious defect in sex determination (Tables S1 and S2). Therefore, sup-26 appears to be a modulator of the sex-determination pathway, fine-tuning the pathway to ensure appropriate sexual development.
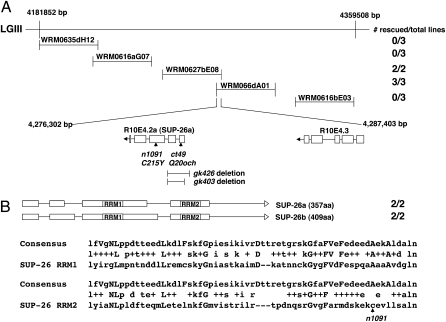
Fig. 1.
Cloning of sup-26. (A) Fosmids used in sup-26(n1091) rescue experiments and their relative base-pair positions on LGIII are shown. Transgenic sup-26(n1091); her-1(n695) animals carrying the indicated fosmid DNA as extrachromosomal arrays were generated and scored for restoration of the masculinized (Tra) phenotype as described in Methods. The number of rescued lines vs. total lines generated are indicated at the right. ORFs in the overlapping region of two rescuing fosmids (WRM0627bE08 and WRM066dA01) are indicated, with boxes representing exons and lines representing intronic sequences. The positions of ct40 and n1091 mutations are indicated by arrows. Two deletion alleles (gk403 and gk426) and the sup-26 regions removed by these mutations are represented below the sup-26 ORF. (B) A schematic of sup-26 transcripts and alignment of the consensus RRM (accession no. PF00076) with the two SUP-26 RRMs. Expression of these two transcripts under the control of the sup-26 promoter rescued the sup-26(n1091) phenotype. Uppercase letters indicate the most conserved residues of RRMs. The middle rows show residues that are identical (letters) or conservative changes (+). The RRM domains are identical in SUP-26a and SUP-26b. The residue affected by n1091 is indicated by an arrow.
We performed reverse transcription PCR amplification (RT-PCR) with primers corresponding to the predicted 5′ and 3′ ends of the sup-26 coding sequence (http://www.wormbase.org/) and identified two distinct transcripts, sup-26a and sup-26b, which encode 357 and 409 amino acid products, respectively (Fig. 1B). The predicted products of both transcripts contain two RRMs that share 77% and 74% sequence similarity to the consensus RRM sequence, respectively (Fig. 1B), suggesting that SUP-26 may bind RNA. When expressed under the control of the sup-26 promoter, each of the transcripts masculinized sup-26(n1091); her-1(n695) animals (Fig. 1B), indicating that both sup-26 isoforms are functional.
SUP-26 Is Broadly Expressed in Somatic Cells and Localizes to the Cytoplasm.
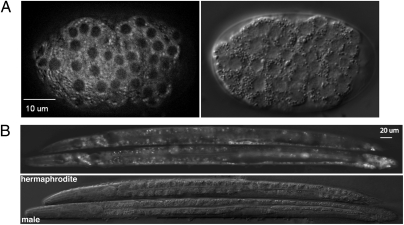
To determine where SUP-26 might function, we examined the expression pattern of the Psup-26sup-26::gfp translational fusion, which fully rescued the sup-26(n1091) phenotype (Table S1). We found that SUP-26::GFP was expressed in most, if not all, somatic cells, starting from the early gastrula through adulthood. SUP-26::GFP localized to the cytoplasm and was largely excluded from the nucleus (Fig. 2). There was no apparent difference in SUP-26::GFP expression patterns between male and hermaphrodite L4 larvae or adults (Fig. 2B). Based on data from the Nematode Expression Pattern DataBase (http://nematode.lab.nig.ac.jp), in situ hybridization experiments using either sup-26a or sup-26b cDNA as probes reveal that the sup-26 messages are absent from early meiotic-stage germ cells but are present in oocytes.
Fig. 2.
SUP-26 expression patterns in C. elegans embryos and larvae. (A) SUP-26::GFP observed in an early gastrula embryo carrying an integrated array containing Psup-26sup-26::gfp (Left) and the corresponding DIC image of the embryo (Right). (B) Expression of SUP-26::GFP in L4 stage hermaphrodite (Upper) and male (Lower) larvae, respectively. The corresponding DIC image is shown below.
TRA-2 Protein Expression Is Increased by sup-26 Loss-of-Function Mutations.
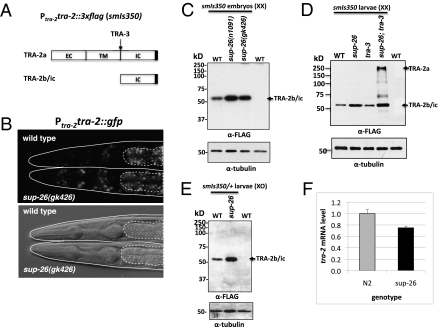
Previous genetic analysis indicates that sup-26 may regulate sexual development through tra-2 (28). We thus examined whether sup-26 mutations affect tra-2 gene expression. We generated a 15-kb transgene that contains the entire tra-2 operon (ppl-1 and tra-2), including an 816-bp promoter upstream of ppp-1, the first gene of the operon, the coding region of ppp-1, the tra-2–coding region fused at its carboxyl terminus with GFP or 3xFLAG epitope, and an 848-bp tra-2 3′ UTR. Stable integration lines were generated from these transgenes: smIs380 (Ptra-2tra-2::gfp) and smIs350 (Ptra-2tra-2::3xflag) (Methods). Both integrated lines fully rescued the tra-2(lf) defects (Fig. S1), suggesting that the TRA-2 fusion proteins are functional. Despite being a predicted transmembrane receptor (Fig. 3A) (8), TRA-2::GFP was observed exclusively in the nucleus as previously described (Fig. 3B) (15). Interestingly, TRA-2::GFP was expressed at higher levels in sup-26(gk426) animals than in wild-type animals (Fig. 3B). For example, only several cells in the head of wild-type animals displayed TRA-2::GFP, whereas many cells in sup-26(gk426) animals expressed TRA-2::GFP. In Western blot analysis of smIs350 hermaphrodites with different genetic backgrounds, we observed significantly increased levels of an ≈50-kDa TRA-2::3xFLAG polypeptide in sup-26 mutant embryos and L4 larvae compared with those in wild-type embryos and L4 larvae (Fig. 3 C and D). This TRA-2::3xFLAG polypeptide is similar in size to the TRA-2 product (TRA-2ic) generated by TRA-3 protease cleavage at the intracellular domain of TRA-2a (14) and to the predicted size of the TRA-2b isoform. It is also consistent with the size of the TRA-2 protein detected in immunoblot analysis using an antibody raised against the TRA-2 intracellular domain (15, 16). In sup-26(gk426); tra-3(e1107) smIs350 animals, we observed one additional high-molecular-weight form of TRA-2::3xFLAG consistent in size with full-length TRA-2 (Fig. 3D), indicating that TRA-2a is indeed cleaved by TRA-3 in C. elegans. In sup-26(gk426); smIs350/+ males, we observed a similar increase in the abundance of the 50-kDa TRA-2::3xFLAG polypeptide when compared with wild-type smIs350/+ males (Fig. 3E), which were mildly feminized due to TRA-2 overexpression from the smIs350 transgene (Table S2). The feminization phenotype of sup-26(gk426); smIs350/+ males was stronger than that of smIs350/+ males, which is consistent with more increased TRA-2 expression in sup-26(gk426); smIs350/+ males. These results suggest that in both males and hermaphrodites SUP-26 represses tra-2 protein expression. Moreover, sup-26 can inhibit translation from both tra-2 transcripts, which are transcribed from different promoters but share the same 3′ UTR (Fig. S1) (8). Real-time quantitative RT-PCR analysis revealed a slight decrease in tra-2 transcripts in sup-26(gk246) mixed-stage animals when compared with wild-type animals, indicating that sup-26 does not inhibit tra-2 transcription or reduce tra-2 mRNA stability (Fig. 3F). Therefore, our results are consistent with the model that sup-26 regulates tra-2 expression by inhibiting tra-2 translation.
Fig. 3.
Analysis of TRA-2 protein expression. (A) A schematic of tra-2 translation products (TRA-2a and TRA-2b) and the product (TRA-2ic) derived from processing of TRA-2a by the TRA-3 calpain protease (16). The predicted extracellular (EC), transmembrane (TM), and intracellular (IC) domains are indicated. The 3xFLAG is indicated by a solid box. (B) TRA-2::GFP expression in L4 stage wild-type (Upper) and sup-26(gk426) (Lower) hermaphrodites carrying an integrated array containing Ptra-2tra-2::gfp. Regions of intestinal auto-fluorescence are bounded by dashed lines. TRA-2::GFP was seen in nuclei of the head region. (Lower) Corresponding DIC image of the Upper panel. (C–E) Immunoblotting analysis of TRA-2 expression from an integrated transgene (smIs350; Ptra-2tra-2::3xflag) in different genetic backgrounds. (C) A total of 250 embryos of the indicated genotype carrying smIs350 or 250 nontransgenic wild-type (WT) embryos were solubilized with SDS sampling buffer, resolved on 10% SDS/PAGE, and then analyzed by immunoblotting using an anti-FLAG antibody or an anti-∂-tubulin antibody (as a loading control). (D) Twenty-five L4 larvae of the indicated genotype carrying smIs350 or a nontransgenic wild-type control were analyzed by immunoblotting as described above. The alleles used were sup-26(gk426) and tra-3(e1107). To resolve the high-molecular-weight TRA-2a transmembrane isoform that is prone to aggregate when boiled, the samples were sonicated in the SDS sampling buffer in a water bath and heated at 65 °C for 30 min before being resolved by a 8% SDS/PAGE. (E) Seventeen male L4 larvae of the indicated genotype, which were heterozygous for smIs350, were analyzed by 10% SDS/PAGE and immunoblotting as described above. smIs350 homozygous XO males are slightly feminized. Therefore, smIs350/+ XO males were used. (F) Abundance of the tra-2 transcripts in wild-type and sup-26(gk426) animals. Quantitative RT-PCR was performed on RNA samples from mixed-stage wild-type (N2) and sup-26(gk426) animals. rpl-26 was used as an internal reference. Mean value of tra-2 mRNAs is expressed as a ratio over rpl-26. Error bars are SDs.
SUP-26 Regulates tra-2 Expression Through the TGE Elements.
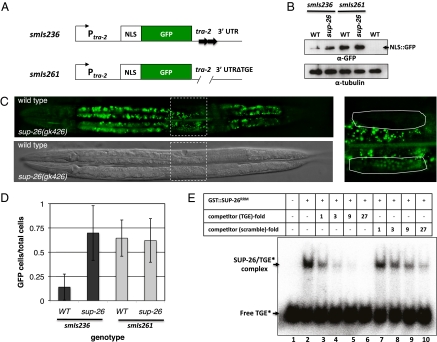
It was previously shown that translation of tra-2 in the germline is repressed by elements in its 3′ UTR (20). We thus tested whether sup-26 acts through the tra-2 3′ UTR. We generated GFP reporters that lack the TRA-2 coding sequence but contain the 816-bp tra-2 promoter, the coding region for nucleus-localized GFP (NLS::GFP), and an 848-bp tra-2 3′ UTR (Fig. 4A; Methods). An integrated transgene, smIs236 (Ptra-2NLS::GFP::3′UTRtra-2), had stronger GFP expression in sup-26(gk426) animals than in wild-type animals on the basis of the immunoblotting analysis (Fig. 4B) and the analysis of GFP fluorescence intensity (Fig. 4C). Increased NLS::GFP expression in sup-26(gk426) animals was apparent in most tissues and was particularly obvious in the uterus. For example, in smIs236 animals, an average of 17% uterine cells had visible NLS::GFP expression, compared with an average of 70% in sup-26(gk426); smIs236 animals (Fig. 4D). In contrast, a similar integrated transgene lacking both 28-bp TGEs, smIs261 [Ptra-2NLS::GFP::3′UTR(ΔTGE)tra-2], produced similar levels of NLS::GFP expression in sup-26(gk426) and wild-type animals (Fig. 4 B and D), suggesting that SUP-26 likely inhibits tra-2 translation through TGEs.
Fig. 4.
SUP-26 represses tra-2 translation by binding to the TGE elements in tra-2 3′ UTR. (A) A schematic showing two tra-2 transcriptional fusions used to examine the role of TGEs in regulating tra-2 expression. A GFP with four copies of the SV40 nucleus localization signal (NLS) is under the control of the tra-2 promoter and 3′ UTR with or without the two TGEs (Methods). (B–D) Expression levels of NLS::GFP in wild-type and sup-26(gk426) hermaphrodite larvae carrying two different integrated arrays, smIs236 (Ptra-2NLS::GFP::3′UTRtra-2) and smIs261 [Ptra-2NLS::GFP::3′UTR(ΔTGE)tra-2], which lacks two TGEs. (B) One hundred larvae of the indicated genotype were analyzed by 12% SDS/PAGE and then by immunoblotting using an anti-GFP antibody or an anti-∂-tubulin antibody as described in Fig. 3C. (C) GFP and DIC images of representative L4 stage wild-type (Upper) and sup-26(gk426) (Lower) hermaphrodites carrying smIs236. The region indicated by the dashed box is enlarged on the right, and the uterine cells bounded by the dashed box were scored for NLS::GFP expression. (D) Percentages of uterine cells that expressed NLS::GFP in wild-type or sup-26(gk426) hermaphrodites carrying smIs236 or smIs261. Images of 15 animals from each strain were captured and scored blind of the genotype for uterine cells with NLS::GFP, which are expressed as a ratio over the total number of uterine cells scored. Error bars are SDs. (E) SUP-26 binds specifically to the TGE element. A 32P-labeled 28-nt TGE RNA oligonucleotide (1.8 pmol) was incubated with or without 2.1 pmol of purified GST::SUP-26RRM (lanes 1 and 2) in the presence or absence of increasing concentrations of unlabeled TGE RNA oligonucleotide (lanes 3–6) or an RNA oligonucleotide with a scrambled TGE sequence (lanes 7–10), whose concentrations are presented as folds of the 32P-labeled TGE. The reactions were resolved by 5% nondenaturing polyacrylamide gel (Methods).
We then tested whether SUP-26 binds directly to the tra-2 3′ UTR in vitro. We found that a purified SUP-26 GST fusion (GST::SUP-26RRM), which contains the SUP-26a RRM domain (amino acids 81–259), formed a complex with a 32P-labeled TGE RNA oligonucleotide, displaying retarded mobility in a gel shift assay (Fig. 4E, lanes 1 and 2). Unlabeled TGE oligonucleotide competed effectively for binding to GST::SUP-26RRM in a concentration-dependent manner, blocking the complex formation (Fig. 4E, lanes 3–6). In contrast, an RNA oligonucleotide with the identical nucleotide composition but a scrambled sequence was much less effective in doing so, showing an approximately ninefold lower binding affinity (Fig. 4E, lanes 7–10). These results suggest that SUP-26 binds specifically to the 3′ UTR of the tra-2 mRNA through the 28-nt TGEs.
Polyadenylate-Binding Protein Associates with SUP-26 in Vivo.
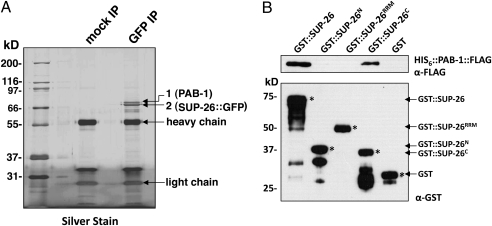
To identify factors that may act with SUP-26 to regulate tra-2 translation, we immunoprecipitated SUP-26::GFP from extracts of Psup-26sup-26::gfp transgenic animals (Methods). SDS polyacrylamide gel resolution of proteins coprecipitated with SUP-26::GFP revealed the presence of two major protein bands that were not observed in the mock immunoprecipitation (IP) sample (Fig. 5A). MALDI-TOF mass spectroscopy analysis determined that the lower band (Fig. 5A, band 2) corresponds to SUP-26::GFP and the upper band (Fig. 5A, band 1) corresponds to the poly(A)-binding protein PAB-1 (Table S3), which was confirmed by liquid chromatography-tandem mass spectrometry (LC MS/MS) analysis using LTQ Orbitrap (Fig. S2). To examine whether SUP-26 and PAB-1 directly interact, we performed a GST fusion protein pulldown assay. GST::SUP-26 and GST::SUP-26C [which contains the carboxyl terminal domain of SUP-26a (amino acids 260–357), but not GST], GST-SUP-26RRM, or GST::SUP-26N [which contains the amino terminal domain of SUP-26a (amino acids 1–80)] specifically pulled down His6::PAB-1::FLAG in the presence of RNase A (Fig. 5B). These results suggest that SUP-26 and PAB-1 can directly interact in vitro independently of RNA through the carboxyl terminal domain of SUP-26. Finally, we tested whether sup-26 may affect the length of the poly(A) tail of tra-2 mRNA using a PCR-based assay (Fig. S3) (23, 24). We were able to detect relatively short poly(A) tails on tra-2 mRNAs, similar to what has been reported previously (23, 24), but their lengths were not affected by mutations in sup-26 (Fig. S3B).
Fig. 5.
PAB-1 associates with SUP-26 both in vivo and in vitro. (A) Lysates from C. elegans animals expressing SUP-26::GFP (smIs259) were prepared as described in Methods, incubated with a mouse anti-GFP monoclonal antibody (GFP IP) or no antibody (mock IP), precipitated using Protein G Sepharose beads, resolved by 12% SDS/PAGE, and subjected to silver staining. Two major bands not observed in mock IP were excised from the gel, subjected to trypsin digestion, and analyzed by MALDI-TOF mass spectroscopy and LC-MS/MS. The upper band corresponds to PAB-1 and the lower band is SUP-26::GFP. (B) PAB-1 associates with SUP-26 in vitro through the carboxyl-terminal domain of SUP-26 in the presence of RNases. A total of 200 ng of purified GST, GST-SUP-26, GST-SUP-26N, GST-SUP-26RRM, and GST-SUP-26C were incubated with glutathione Sepharose beads and 100 ng of purified HIS6::PAB-1::FLAG. The bead-bound proteins were resolved by 12% SDS/PAGE and analyzed by immunoblotting with anti-GST and anti-FLAG antibodies, respectively. Asterisks indicate the corresponding GST fusion proteins.
In summary, we have identified an RRM-containing protein, SUP-26, that is ubiquitously expressed in C. elegans somatic cells, binds specifically to TGEs in the 3′ UTR of tra-2 mRNA, and modulates somatic sex determination by repressing tra-2 translation. Interestingly, GLD-1, a germ-cell–specific RNA-binding protein that shares no sequence similarity with SUP-26, also binds TGEs in the tra-2 3′ UTR to repress its translation in the germline and to promote spermatogenesis (25, 26). It appears that GLD-1 and SUP-26 use the same 3′ UTR cis-element (TGEs) but different cofactors or mechanisms to repress tra-2 translation. In the germline, FOG-2, a unique F-box protein and a germ-cell–specific factor, is proposed to act as a bridge to bring GLD-1–bound tra-2 mRNA into a translational repression complex (26). We find that PAB-1, a poly(A)-binding protein, associates with SUP-26 in vivo and interacts directly with SUP-26 in vitro independently of RNA. The PABPs have been shown to interact with translation initiation factors such as eIF4G to form a circular mRNA structure that facilitates active translation (1, 29). Their binding to the poly(A) sequences could also prevent deadenylation and thus stabilize mRNAs (1, 29), although loss of sup-26 does not appear to affect the lengths of tra-2 poly(A) tails. It seems more likely that the association of SUP-26 with PAB-1 at the tra-2 3′ UTR interferes with PAB-1’s function in stimulating tra-2 translation. If so, this would represent a different TGE-mediated translational repression mechanism from the one used in the germline and perhaps is similar to that used by inhibitory PABP-interacting proteins, which inhibit translation by antagonizing the translation-stimulating activity of PABPs in mammalian cells (30).
Methods
Strains.
Strains were maintained using standard procedures. Transgenic strains were generated by microinjection (31). Integration of extrachromosomal trangene arrays was performed by the γ-irradiation method (32). Mutations and integrated arrays used in this study were as follows: LGIII—sup-26(gk426, gk403, n1091, ct49); LGIV—tra-3(e1107), smIs350 (Ptra-2tra-2::3xflag), smIs261 [Ptra-2NLS::GFP::3′UTR(ΔTGE)tra-2]; LGV—unc-76(e911), her-1(n695); and LGX—smIs236 (Ptra-2NLS::GFP::3′UTRtra-2) and smIs259 (Psup-26sup-26::gfp). The chromosomal location of smIs380 (Ptra-2tra-2::gfp) has not been determined.
Molecular Biology and Transgenic Animals.
Sequences of all primers used in this study are listed in Table S4. Fosmids were injected into sup-26(n1091); her-1(n695) animals at 10 ng/μL using pRF4 as a co-injection marker (50 ng/μL). Psup-26sup-26::gfp was constructed by PCR amplification of the 4-kb sup-26 genomic fragment using the primers SUP-26pro and SUP-26cas and by subcloning the PCR fragment into a modified pPD117.01 vector using the standard Gateway cloning technique. Psup-26sup-26::gfp was injected at 5 ng/μL with pRF4 (50 ng/μL).
The sup-26 cDNAs were amplified from a cDNA library prepared from mixed-stage wild-type animals using primers complementary to the predicted 5′ and 3′ ends of the sup-26–coding sequence. The amplified cDNA fragments were cloned into the Gateway vector pDONR221. Of 20 cDNA clones analyzed by restriction enzyme digestion and DNA sequencing, 17 were sup-26a and 3 were sup-26b.
The Ptra-2NLS::GFP::3′UTRtra-2 reporter was generated by inserting an 816-bp tra-2 operon promoter fragment (XbaI-XmaI) and an 848-bp tra-2 3′ UTR fragment (EcoRI/SpeI) into pPD122.56. Ptra-2NLS::GFP::3′UTR(ΔTGE)tra-2, which lacks two TGEs, was generated by site-directed mutagenesis. These plasmids were injected individually into unc-76(e911) animals at 50 ng/μL with p76-16b (an unc-76 rescuing plasmid) at 25 ng/μL. To generate Ptra-2tra-2::3xflag, a 12,850-bp genomic fragment containing an 816-bp promoter upstream of ppp-1, the coding region of ppp-1, and the tra-2–coding region were fused to three tandem copies of the FLAG tag (DYKDHDGDYKDHDIDYKDDDDK). The 848-bp tra-2 3′ UTR was then fused to the 3′ end of the 3xFLAG tag. Ptra-2tra-2::gfp was made by replacing the 3xFLAG epitope sequence of Ptra-2tra-2::3xflag with a KpnI-EcoRI gfp fragment from pPD95.75. Ptra-2tra-2::3xflag or Ptra-2tra-2::gfp was injected into unc-76(e911) animals at 25 ng/μL with p76-16b (50 ng/μl).
Protein Purification and Gel Mobility Shift Assay.
Gel shift assays were performed as described previously (33). Briefly, GST::SUP-26RRM was purified from the BL21(DE3) Escherichia coli strain using glutathione Sepharose beads (GE Healthcare). An RNA oligonucleotide corresponding to the 28-nt TGE element was synthesized (Integrated DNA Technologies) and end-labeled with 32P using polynucleotide kinase (New England Biolabs). For the binding reaction, GST::SUP-26RRM was incubated at 25 °C with 32P-labeled RNA in a binding buffer containing 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, 100 mM KCl, 0.1 mM DTT, 5% glycerol, and 0.1 mg/mL BSA in the presence or absence of unlabeled RNA oligonucleotide competitors. After a 20-min incubation, the samples were resolved on a 5% nondenaturing polyacrylamide gel at 4 °C. The gel was then dried and exposed to a PhosphoImaging screen (Perkin-Elmer).
Microscopy Imaging.
Fluorescence and differential interference contrast (DIC) images were collected at 0.5-μm intervals with an Axioplan 2 microscope (Zeiss) and a cooled CCD camera (PCO SensiCam). Fluorescence images were subjected to deconvolution analysis using the Slidebook 5.0 software program (Intelligent Imaging Innovations).
Mass Spectroscopy Analysis.
Mixed-stage animals were harvested from nematode growth media (NGM) agar plates and lysed by sonication (3 × 10 s) in a buffer containing 250 mM NaCl, 100 mM Tris·HCl (pH 7.4), 1 mM EDTA, 5 mM DTT, 0.1% Triton X-100, 1% PMSF, and Roche Complete Protease Inhibitor Mixture. Lysates were clarified by centrifugation at 14,000 × g for 30 min, precleared with Protein G beads (GE Healthcare), and then incubated with an anti-GFP antibody and Protein G beads for 2 h at 4 °C with gentle rocking. After four extensive washes with the same buffer, the precipitated samples were resolved in 12% SDS/PAGE and silver-stained. In-gel tryptic digestion of silver-stained proteins and mass spectrometric analysis were carried out as described (SI Methods) (34).
GST Fusion Protein Pulldown Assay.
GST-SUP-26 fusion proteins were expressed and purified as described above. His6::PAB-1::FLAG expressed in BL21(DE3) was first purified using TALON Metal Affinity Resin (Clontech) and eluted from the resin with 200 mM imidazole. It was further affinity-purified using the anti-FLAG (M2) agarose beads (Sigma-Aldrich) and eluted with 100 μg/mL of the FLAG peptide. His6::PAB-1::FLAG was incubated with GST fusion proteins immobilized on glutathione Sepharose beads in the PBS buffer supplemented with 10% glycerol, 1 mM DTT, 0.01% Nonidet P-40, 0.5 mM EDTA, and 125 μg/mL RNase A at 4 °C for 12 h with gentle rotating. The Sepharose beads were washed four times with the binding buffer. The bound proteins were resolved by 12% SDS/PAGE and detected by immunoblotting.
Poly(A) Tail Length Assay.
The poly(A) tail length assay was carried out on tra-2 mRNAs using a protocol described previously with some modifications (23). mRNAs were isolated from wild-type and mutant strains and resuspended in 30 μL of H2O. cDNAs were synthesized from 3 μg of RNA using SuperScript III Reverse Transcriptase (Invitrogen) and 300 ng of oligo(dT)12. TRA-2 oligo 1 and oligo 2 are RT-PCR primers specific to the tra-2 3′ UTR and were end-labeled with 32P using polynucleotide kinase (New England Biolabs). A 25-cycle PCR was performed using 20 ng of oligo 1 or oligo 2, oligo(dT)12 remaining from the cDNA synthesis, and 3 μL of cDNA as templates. The PCR products were analyzed on a 2.5% agarose gel, which was dried and exposed to a PhosphoImaging screen (Perkin-Elmer).
Supplementary Material
Acknowledgments
We thank Tom Blumenthal, Tom Evans, and Bill Woods for comments on the manuscript. This work is supported by a Burroughs Wellcome Fund Award and by National Institutes of Health R01 Grants GM59083 and GM79097 (to D.X.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004513107/-/DCSupplemental.
References
- 1.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 2.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: The ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkin J. Primary sex determination in the nematode C. elegans. Development. 1987;101(Suppl):5–16. doi: 10.1242/dev.101.Supplement.5. [DOI] [PubMed] [Google Scholar]
- 5.Meyer BJ. Primary events in C. elegans sex determination and dosage compensation. Trends Genet. 1988;4:337–342. doi: 10.1016/0168-9525(88)90053-4. [DOI] [PubMed] [Google Scholar]
- 6.Trent C, et al. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech Dev. 1991;34:43–55. doi: 10.1016/0925-4773(91)90090-s. [DOI] [PubMed] [Google Scholar]
- 7.Perry MD, et al. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 1993;7:216–228. doi: 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- 8.Kuwabara PE, Okkema PG, Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol Biol Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamaoka BY, Dann CE, III, Geisbrecht BV, Leahy DJ. Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A. Proc Natl Acad Sci USA. 2004;101:11673–11678. doi: 10.1073/pnas.0402559101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimble J, Edgar L, Hirsh D. Specification of male development in Caenorhabditis elegans: The fem genes. Dev Biol. 1984;105:234–239. doi: 10.1016/0012-1606(84)90279-3. [DOI] [PubMed] [Google Scholar]
- 11.Doniach T, Hodgkin J. A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev Biol. 1984;106:223–235. doi: 10.1016/0012-1606(84)90077-0. [DOI] [PubMed] [Google Scholar]
- 12.Chin-Sang ID, Spence AM. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- 13.Mehra A, Gaudet J, Heck L, Kuwabara PE, Spence AM. Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 1999;13:1453–1463. doi: 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol SB, Kuwabara PE. Proteolysis in Caenorhabditis elegans sex determination: Cleavage of TRA-2A by TRA-3. Genes Dev. 2000;14:901–906. [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada M, Kanematsu K, Tanaka K, Yokosawa H, Kawahara H. Proteasomal ubiquitin receptor RPN-10 controls sex determination in Caenorhabditis elegans. Mol Biol Cell. 2006;17:5356–5371. doi: 10.1091/mbc.E06-05-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubert A, Anderson P. The C. elegans sex determination gene laf-1 encodes a putative DEAD-box RNA helicase. Dev Biol. 2009;330:358–367. doi: 10.1016/j.ydbio.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: A gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- 18.Zarkower D, Hodgkin J. Zinc fingers in sex determination: Only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993;21:3691–3698. doi: 10.1093/nar/21.16.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin EB, Okkema PG, Evans TC, Kimble J. Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C. elegans. Cell. 1993;75:329–339. doi: 10.1016/0092-8674(93)80074-o. [DOI] [PubMed] [Google Scholar]
- 21.Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986;114:53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuersten S, Segal SP, Verheyden J, LaMartina SM, Goodwin EB. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol Cell. 2004;14:599–610. doi: 10.1016/j.molcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Jan E, Yoon JW, Walterhouse D, Iannaccone P, Goodwin EB. Conservation of the C. elegans tra-2 3′UTR translational control. EMBO J. 1997;16:6301–6313. doi: 10.1093/emboj/16.20.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson SR, Goodwin EB, Wickens M. Rapid deadenylation and poly(A)-dependent translational repression mediated by the Caenorhabditis elegans tra-2 3′ untranslated region in Xenopus embryos. Mol Cell Biol. 2000;20:2129–2137. doi: 10.1128/mcb.20.6.2129-2137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clifford R, et al. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- 27.Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- 28.Manser J, Wood WB, Perry MD. Extragenic suppressors of a dominant masculinizing her-1 mutation in C. elegans identify two new genes that affect sex determination in different ways. Genesis. 2002;34:184–195. doi: 10.1002/gene.10118. [DOI] [PubMed] [Google Scholar]
- 29.Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: A developmental perspective. Brief Funct Genomics Proteomics. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 30.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 31.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 33.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2:1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, Yu JS. Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Mol Biol Cell. 2010;21:287–301. doi: 10.1091/mbc.E09-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.