Abstract
Diatoms are prominent phytoplanktonic organisms that contribute around 40% of carbon assimilation in the oceans. They grow and perform optimally in variable environments, being able to cope with unpredictable changes in the amount and quality of light. The molecular mechanisms regulating diatom light responses are, however, still obscure. Using knockdown Phaeodactylum tricornutum transgenic lines, we reveal the key function of a member of the light-harvesting complex stress-related (LHCSR) protein family, denoted LHCX1, in modulation of excess light energy dissipation. In contrast to green algae, this gene is already maximally expressed in nonstressful light conditions and encodes a protein required for efficient light responses and growth. LHCX1 also influences natural variability in photoresponse, as evidenced in ecotypes isolated from different latitudes that display different LHCX1 protein levels. We conclude, therefore, that this gene plays a pivotal role in managing light responses in diatoms.
Keywords: diatoms, light acclimation, nonphotochemical quenching, RNA interference, ecotypes
Marine diatoms are believed to contribute around 40% of oceanic organic carbon production and to constitute an important component of the biological carbon pump (1, 2). They are widespread throughout the oceans and show optimal photosynthetic activity over a wide range of environments (3–5). Their extreme flexibility to changing conditions is thought to be a key element that has driven their rise to dominance in contemporary oceans (6). For example, they show an outstanding capacity to cope with light stress. Their ability to dissipate excess energy in high light can surpass that of plants, as witnessed by the impressive levels of nonphotochemical quenching (NPQ) of chlorophyll fluorescence observed in the model diatom Phaeodactylum tricornutum in some conditions (7). The term NPQ describes the enhancement of thermal dissipation of absorbed energy that occurs in the pigment-containing proteins of photosystem (PS) II, whenever light absorption exceeds the maximum rate of CO2 assimilation. In plants, NPQ relies on acidification of the luminal pH, which affects zeaxanthin-violaxanthin pigment composition via the xanthophyll cycle (XC) and the activity of the PSII subunit PsbS (8, 9). State transitions of the light-harvesting complexes between PSII and PSI add an additional layer of photoprotection (8). In diatoms, state transitions have not been found (10) and the PsbS gene is absent from the nuclear genomes sequenced to date (2). Instead, the superior capacity of diatoms for NPQ has been attributed to the existence of an alternative XC, also observed in other chromophytes (11), which catalyzes the de-epoxidation of diadinoxanthin (DD) to diatoxanthin (DT), the better studied cycle present in plants being observed only in high light grown diatom cells (12).
To test the existence of additional NPQ effectors in diatoms, expression of light stress response proteins was examined in P. tricornutum. In contrast with green algae, we found that LHCX1, one of the putative high light response genes of the LHCSR family of diatoms, is already maximally expressed in low light and that its gene product modulates NPQ capacity during a typical light/dark cycle. Transgenic lines with reduced expression of the gene have significantly reduced NPQ and growth capacities. Analysis of natural NPQ variants of P. tricornutum indicates a key role of LHCX1 not only in the transient response to light, but also in constitutive adaptation to environmental constraints.
Results
LHCX1 Is Required for Light Responses in Both Stress and Nonstressful Conditions.
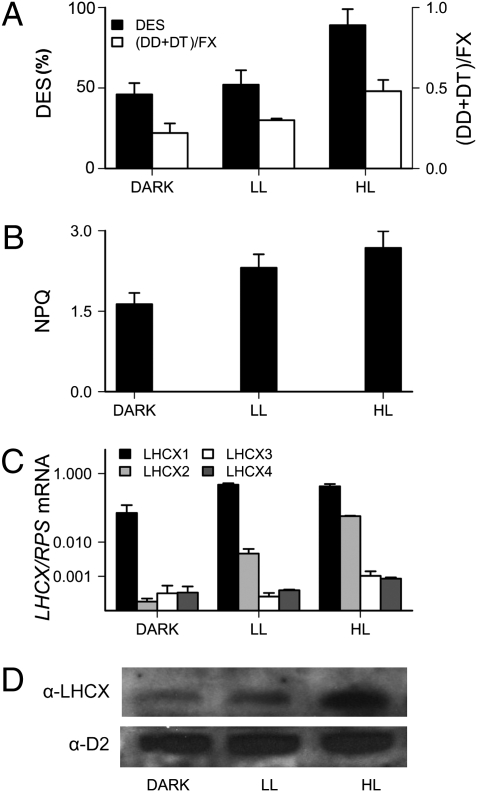
Genomic and phylogenetic analyses reveal in diatoms the presence of multiple members of the light-harvesting complex stress-related proteins LHCX (also known as LHCSR/LI818 in green algae, refs. 13, 14), and hereafter named LHCX in this study (Fig. S1). Genes encoding these proteins are absent in higher plants but are induced by photooxidative stress in some unicellular photosynthetic organisms (Fig. S1 and refs. 13, 14). Their accumulation upon light stress has been associated with enhanced photoprotection in the green freshwater alga Chlamydomonas reinhardtii (15). A similar role was recently proposed for diatom orthologs that are upregulated in high light (16–18). On the other hand, gene expression analyses revealed that one of the four LHCX genes of P. tricornutum (LHCX1) is not light-stress responsive (Fig. 1). In cells grown under 12:12 h light:dark cycles, exposure to low irradiance (30–70 μmol photons m−2·s−1) already induced maximal LHCX1 induction, whereas expression of the other isoforms remained almost undetectable. LHCX1 mRNA levels remain stable even with increasing light intensities (Fig. 1C and ref. 17). Recent data have suggested that this isoform would not be responsible for NPQ responses, but would play instead a structural role within the PSII–flucoxanthin chlorophyll protein (FCP) supercomplex (18). However, induction of this gene upon a dark-to-low light transition paralleled with an enhancement of the NPQ capacity (Fig. 1B), i.e., the maximum extent of energy quenching (see Materials and Methods) for a constant XC activity (Fig. 1A). Further exposure to high light promoted the expression of additional LHCX isoforms, mainly LHCX2 (Fig. 1C and ref. 17). This paralleled a significant increase in the accumulation of LHCX proteins in the cell (Fig. 1D). Because LHCX1 always remained the most abundant mRNA, this finding suggests possible differences in translation efficiency and/or protein stability between the different isoforms. High light exposure induced other typical photoprotective responses (increased activity of the XC and enhanced xanthophyll accumulation in the cells) (7, 12), ultimately leading to an additional (albeit small) increase in NPQ (Fig. 1B), in agreement with previous studies (19). Altogether, these data suggest that induction of LHCX proteins in high light increases energy quenching as part of a more general photoprotective response. Conversely, expression of the LHCX1 gene in low light seems to provide P. tricornutum cells with nearly maximum NPQ capacity, pinpointing the LHCX1 isoform as a likely NPQ effector in diatoms.
Fig. 1.
Diatom LHCX expression and NPQ characteristics in dark, low light and high light conditions. (A) De-epoxidation state (DES) determined as DT/(DD + DT), and relative level of the pool of xanthophyll (DT + DD), normalized to the fucoxanthin (Fx) content, obtained from HPLC analysis. (B) NPQ response from cells grown as described above. NPQ was calculated as (Fm-Fm′)/Fm′ (34), where Fm and Fm′ are the maximum fluorescence emission measured in dark (30-min adaptation before exposure) and cells exposed for 5 min to 700 μmol photons m−2·s−1, respectively. (C) Accumulation of the four P. tricornutum LHCX transcripts determined by qRT-PCR from cells grown in a 12:12 h light:dark cycle and collected in the dark (dark), after 2 h of low light treatment (70 μmol photons m−2·s−1, LL), and after 1 h of a subsequent low light to high light shift (600 μmol photons m−2·s−1, HL). RPS was used as reference gene. Error bars are relative to three independent experiments. (D) Western blotting showing LHCX protein accumulation. Proteins were detected with antibodies against the LHCSR/LHCX and the PSII subunit D2 was used as loading control.
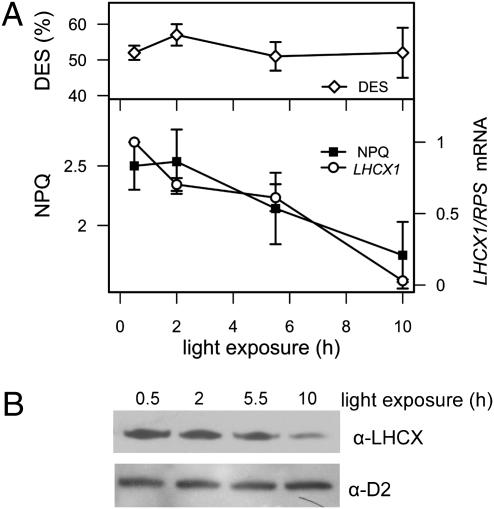
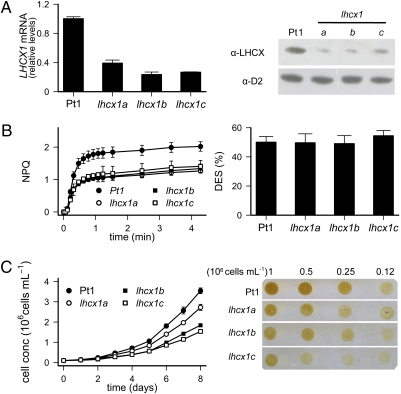
Analysis of NPQ changes in diatoms exposed to a 12:12 h light:dark cycle at low light (70 μmol photons m−2·s−1) further confirmed this observation. We observed that in low light, expression of the LHCX1 gene slowly declined after its induction following a dark-to-light shift. We found a tight correlation between NPQ capacity, LHCX1 mRNA expression levels, and LHCX protein abundance, contrasting with the stable XC activity (Fig. 2). This suggests that LHCX1 actively participates in modulating the extent of energy dissipation in P. tricornutum, for a given de-epoxidation state (DES). This possibility was further investigated by generating knockdown transgenic lines by RNA interference (RNAi). Three clones were identified, which showed reduced expression of the LHCX1 gene and a consequently lower protein accumulation (Fig. 3A). The transgenic lines showed a ∼50% reduction in NPQ capacity (Fig. 3B), although their XC capacity (Fig. 3B), PSII efficiency, maximal photosynthetic electron flow, PSII absorption cross-section (Table 1), and pigment composition (Table S1) were the same as in wild-type cells, when measured in unstressed cells. This effect was specific, as no reduction in NPQ capacity could be detected in clones in which other isoforms were targeted for silencing. Silencing specificity was confirmed for the lhcx1a line (Fig. S2B), for which a reduced NPQ capacity throughout the day correlated with the reduced expression of LHCX1 and an unchanged DES (Fig. S2A). Whereas photosynthesis showed a similar light saturation profile in the Pt1 and lhcx1a strains (Fig. S3A), the NPQ response was reduced throughout the whole light intensity range in the silenced line. In particular, the finding that NPQ was smaller in the lhcx1a even in conditions where LHCX isoforms other than LHCX1 were induced (Fig. 1 and Fig S2) strongly suggests that the expression of these genes cannot compensate for the lower LHCX1 accumulation in this strain. Reduced NPQ capacity paralleled with enhanced photoinhibition in lhcx1a cells, as evidenced by the declined oxygen evolution capacity measured upon prolonged exposure to strong light (Fig. S3B).
Fig. 2.
Diurnal changes in DES, NPQ, and LHCX1 transcript (A), and LHCX protein levels (B). Cells were exposed to a 12:12 h light:dark regime at 70 μmol photons m−2·s−1, and samples were collected at the indicated times during the light phase for the different measurements. Error bars refer to duplicate measurements in three biological samples. NPQ, DES, and blot analyses were performed as specified in Fig. 1.
Fig. 3.
LHCX1 regulates NPQ and growth in P. tricornutum. (A) Analyses of LHCX1 mRNA by qRT-PCR (Left) and protein levels (Right), and (B) NPQ (Left) and DES (Right) values, in wild-type (Pt1) and three RNAi silenced lines, sampled 8 h after the onset of illumination. Analyses were performed as described in Fig. 1. Relative transcript levels were determined using RPS as a reference gene and values normalized to gene expression levels of the wild type (Pt1). (C) Growth curve of the Pt1 and lhcx1 cells (Left) under 12 h:12 h light:dark regime at 30 μmol photons m−2·s−1 (SE refers to duplicate measurements from three biological samples), and growth tests on cells spotted on solid media (Right). A total of 5 μL of different cell dilutions (1, 0.5, 0.25, and 0.12·106·mL cells) were spotted and pictures were taken after 5 d.
Table 1.
Photosynthetic parameters in wild types and transgenic lines with a modulated LHCX1 content
| Strain | PSII efficiency (Fv/Fm) | Photosynthetic electron flow (ΦPSII) | PSII absorption cross section (photons PSII−1·s−1) |
| Pt1 | 0.57 ± 0.01 | 0.32 ± 0.02 | 185 ± 7 |
| lhcx1a | 0.59 ± 0.02 | 0.32 ± 0.03 | 194 ± 6 |
| lhcx1b | 0.61 ± 0.02 | 0.31 ± 0.01 | 186 ± 2 |
| lhcx1c | 0.56 ± 0.01 | 0.27 ± 0.01 | 173 ± 9 |
| Pt4 | 0.63 ± 0.01 | 0.35 ± 0.02 | 155 ± 6 |
| Pt4OE1 | 0.59 ± 0.01 | 0.28 ± 0.01 | 184 ± 8 |
| Pt4OE2 | 0.61 ± 0.01 | 0.32 ± 0.01 | 166 ± 12 |
PSII efficiency (Fv/Fm = (Fm-Fo)/Fm) and photosynthetic electron flow (Fm′–Fs)/Fm′, (36) were calculated from minimum (Fo), steady state (Fs) and maximum fluorescence emission (Fm and Fm′, measured in dark and light exposed cells, respectively). PSII absorption cross-section is the number of photons absorbed by a single PSII per unit of time. This parameter is evaluated from the rate of fluorescence induction upon inhibition of PSII with DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea) as 1/t, where t is the time at which variable fluorescence (F-Fo) has reached 2/3 of its maximum value. SE is relative to three independent measurements.
The notion of a central role of LHCX1 in light acclimation in diatoms was further reinforced by measuring the growth capacity in wild-type and RNAi lines. Reduced growth was seen in the three strains under nonstressful light conditions (Fig. 3C and Fig. S4A), in high light (Fig. S4B), and also upon exposure to intermittent light (Fig. S4C), a condition particularly favorable for the development of NPQ in P. tricornutum (7). Because reduced growth could not be accounted for by a modification of the overall photosynthetic performance in unstressed cells (Table 1), we exclude possible pleiotropic effects of LHCX1 misregulation on photosynthesis. Conversely, the reduced fitness of lhcx1 strains suggests that LHCX1 could be important for optimum light utilization in diatoms in a wide range of photon fluxes.
Natural NPQ Variants of P. tricornutum with Altered LHCX1 Expression.
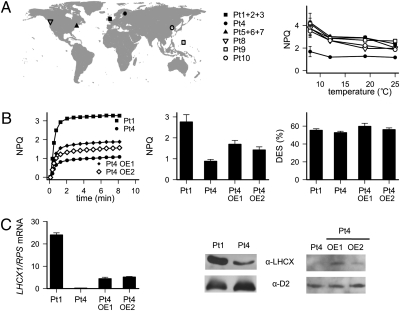
Although P. tricornutum is generally considered to have only limited ecological relevance, it has been found at several locations worldwide (Fig. 4A and ref. 20). We used the available strains to screen for natural NPQ variants. Only minor differences were observed in most of them, with the significant exception of Pt4, the strain isolated from the highest latitude and therefore adapted to the lowest ambient light intensities (Fig. 4A and ref. 20). This ecotype displayed systematically reduced NPQ levels with respect to other strains in all conditions tested, including the low temperatures (Fig. 4A and Fig. S5) that characterize its natural environment (21). This suggests that Pt4 is a natural NPQ variant, most probably reflecting a constitutive adaptation to its environment.
Fig. 4.
Pt4 is a natural NPQ variant displaying altered LHCX expression. (A) Analysis of 10 P. tricornutum ecotypes isolated from different locations (Left). Maximum NPQ as a function of growth temperature (Right). Cells were grown at the indicated temperatures for at least 3 wk. Samples were collected 2 h after the onset of illumination and NPQ was measured as in Fig. 1. (B) NPQ (Left and Center) in Pt1, Pt4, and two Pt4 transgenic lines overexpressing LHCX1. Right shows their DES values. (C) LHCX1 mRNA levels by qRT-PCR using RPS as reference gene (Left) and protein accumulation (Right). A total of 50 μg of total protein extracts was used in the Western blot analysis for Pt4 and Pt1. To better highlight differences, 20 μg has been loaded for the Pt4 and Pt4 overexpressing lines (OE1 and OE2). Proteins were detected with antibodies against the LHCSR/LHCX, and the photosystem II subunit D2 was used as a loading control.
In agreement with the results obtained from the lhcx1 RNAi lines, the reduced NPQ capacity in Pt4 did not correlate with a diminished XC capacity (Fig. 4B) or a reduced photosynthetic performance (Table 1), but could be linked to a specific decrease in the expression of the LHCX1 gene (Fig. 4C). This was confirmed by generating transgenic Pt4 lines in which the LHCX1 gene was overexpressed (OE). Several lines were isolated, and it was found that enhanced levels of LHCX1 expression could partially rescue the NPQ capacity, in the absence of any alteration in XC pigments with respect to the wild-type Pt4 (Fig. 4 B and C). These studies provide a definitive confirmation for the link between the abundance of the LHCX1 gene product and the capacity for NPQ in diatoms.
Discussion
Unlike Chlamydomonas and Ostreococcus (15), P. tricornutum possesses a LHCSR gene family member, LHCX1, which is highly expressed in low light-acclimated cells and is not further induced by high light stress. Its product rapidly accumulates upon a dark-to-light shift and then slowly decreases during a 12:12 h light:dark cycle, correlating with a decrease in NPQ capacity. Changes in LHCX1 levels are directly related to the ability of cells to quench excess energy (for a given amount of DT synthesized), suggesting that the function of this gene closely resembles that of the plant PSII subunit PsbS. In higher plants, PsbS has a dual role: it triggers the onset of NPQ by sensing luminal pH changes (via protonation of two conserved glutamic acid residues), and it amplifies fluorescence quenching in a concentration-dependent manner (9). In diatoms, the LHCX proteins do not share these acidic amino acids (Fig. S1 B and C), questioning their role as pH sensors. In agreement with a different modulation of NPQ by the proton gradient in diatoms, previous data have shown that pH changes modulate fluorescence quenching in P. tricornutum only through their control on the turnover of the XC enzymes (22). On the other hand, it is clear from our data and previous evidence that both PsbS and LHCX/LHCSR proteins share the capacity to amplify quenching. It is tempting, therefore, to propose that this function could have been the ancient property of these proteins. Consistent with this, NPQ in Chlamydomonas shows the same pH modulation as in plants, but relies on LHCSR proteins (15) that are extremely rich in conserved acidic amino acids when compared with the P. tricornutum counterparts (Fig. S1 B and C). Sequence analysis reveals that these amino acids are confined to an additional protein domain in Chlamydomonas, which is absent in the diatom orthologs. Assuming a similar topology for LHCSR and LHCII (23) proteins, the “extra” domain of Chlamydomonas LHCSR should be localized in the thylakoid lumen, where it could confer the observed pH sensitivity to NPQ.
Previous studies have revealed the existence of different NPQ effectors (PsbS and LHCSRs) and different xanthophyll cycles (violaxanthin/zeaxanthin or diatoxanthin/diadinoxanthin), having different efficiencies and ΔpH requirements (8, 9). This study, as well as recent findings in Chlamydomonas and moss (15, 24), allow pinpointing the key role of the molecular NPQ effectors, showing that efficient photoprotection in different environments is achieved using diverse NPQ machineries: PsbS (9), LHCSR–LHCX (ref. 15 and this work), or both (24). Several studies have shown changes in the expression pattern of LHCX genes depending on light conditions (refs. 16–18 and this work). However, our work underlines that the presence of the constitutive LHCX1 gene product is essential for proper light acclimation in P. tricornutum, its decrease resulting in reduced growth capacity in low light, high light, and intermittent light (Fig. S4). We believe that besides preventing photoinhibition at high light, LHCSX1 could also influence the ability of P. tricornutum to acclimate to nonstressful light regimes by modulating light acclimation during repeated diurnal dark-to-light shifts during exponential growth. This would mainly stem from changes in the transient NPQ response, which is observed at the onset of illumination at moderate light intensities (Fig. S6).
In principle, the recent hypothesis that in Thalassiosira pseudonana LHCX1 may play a structural role in the PSII–FCP complex (18) could also be consistent with the reduced growth observed in the P. tricornutum lhcx1 knock-down lines in nonstressful light conditions. However, this hypothesis is difficult to reconcile with our findings that both the antenna size and the photosynthetic activity are unmodified in the lhcx1 cells when compared with their wild-type counterpart (Table 1 and Fig. S3A).
The identification of a natural NPQ variant that displays reduced LHCX1 expression (Pt4) indicates that NPQ effectors have also been targeted for adaptive evolution to specific environments. Genetic analyses indicate that Pt4 is the most diverse P. tricornutum strain (20), and the lower NPQ capacity may reflect its adaptation to high latitudes, with exposure to lower light intensities and to less drastic diurnal light variations. It is possible, therefore, that this ecotype may represent the result of a favorable mutation event in the Baltic environment. Because of the very recent flooding of this basin after the last glaciation and its strong isolation due to topographic and thermohaline constraints (25), this mutation could have been confined within this area. Interestingly, although similar variation in NPQ has also been observed in Arabidopsis ecotypes (26), no correlation with PsbS levels has been found.
In conclusion, our data challenge the long-standing dogma that the peculiar characteristics of NPQ in diatoms are due solely to the presence of a novel xanthophyll cycle. Our molecular study in P. tricornutum unveils the key role of LHCX1 as a molecular gauge controlling quantitatively the level of NPQ. The constitutive presence of this protein in cells acclimated to nonstressful light conditions could provide diatoms with a machinery capable of anticipating sudden changes in the underwater light field. This property could have offered a selective growth advantage in turbulent waters. Diatom LHCX genes appear to be widely dispersed in the ocean (http://camera.calit2.net/), further reinforcing our tenet that they have contributed to the ecological success of diatoms in ocean environments.
Materials and Methods
Cell Cultures.
Axenic P. tricornutum cells were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton and grown in f/2 medium (27) at 19 °C in a 12-h photoperiod. Cells were grown under low light (30 or 70 μmol photons m−2·s−1) and collected during the exponential growth phase. In the experiments shown in Fig. 1, cells were first acclimated to 70 μmol photons m−2·s−1 for 2 h and then shifted to 600 μmol photons m−2·s−1 white light for 1 h. Cells used for data presented in Fig. 3A and Fig. S6 were grown at the indicated temperatures, light intensities, and salinities for at least 3 wk before measurements. These parameters were varied within a range consistent with possible variation at the ocean surface (21, 28).
Construction of Vectors for Gene Silencing and Overexpression.
Vectors for antisense constructs were generated using standard molecular cloning procedures (29). Vectors containing antisense fragments from the different LHCX genes were generated in a vector bearing a phleomycin resistance cassette, as described in SI Materials and Methods. Vectors were introduced into wild-type P. tricornutum strains (Pt1 and Pt4 ecotypes) by microparticle bombardment using a BiolisticPDS-1000/He Particle Delivery System (Bio-Rad) (30). BLAST searches in the Phaedactylum genome were performed to exclude that the selected fragment recognizes the other LHCX transcripts. Putative silenced clones in the Pt1 ecotype (50 clones for each transformation series with the different silencing vectors) were first selected on 1% agar plates (50% f/2 medium) containing 100 μg/mL phleomycin (Invivogen) in low light (30 μmol photons m−2·s−1) and then further screened for NPQ capacity (see below).
Gene Expression and Protein Analysis.
RNA extraction and quantitative PCR (qRT-PCR) were performed as described in ref. 31. The different LHCX transcripts were amplified as described in SI Materials and Methods. Proteins were extracted as described in ref. 32. The diatom LHCX proteins (around 22 kDa) were detected using a rabbit polyclonal anti-LHCSR antibody from Chlamydomonas (gift of Graham Peers, University of California, Berkeley, CA), used at a dilution of 1:5,000 (referred to as α-LHCX in the figures). The anti-D2 antibody (gift of Jean-David Rochaix, University of Geneva, Geneva, Switzerland) was used as a loading control (1:10,000 dilution).
Oxygen Evolution and Chlorophyll Fluorescence.
Light-induced fluorescence kinetics was measured using a home-built fluorescence CCD camera recorder, as described in ref. 33. A crown of green (532 nm) LEDS provided actinic light (50–1,100 μmol photons m−2·s−1). NPQ was calculated as (Fm-Fm′)/Fm′ (34), where Fm and Fm′ are the maximum fluorescence emission level in the dark and light, measured with the saturating pulse of light. For all of the measurements of NPQ capacity (i.e., the maximal NPQ response measured upon a short exposure to a saturating green light of 700 μmol photons m−2·s−1, ∼2,000 photons PSII−1·s−1), samples were dark adapted for 30 min at 20 °C. They were then exposed for 5 min to 700 μmol photons m−2·s−1 of green light. We checked that increasing the actinic light intensity and/or the time of illumination did not enhance NPQ amplitude. Oxygen evolution was measured with a Clark electrode (Hansatech), coupled to a PAM fluorometer (Walz) to estimate NPQ at the same time (Fig. S3).
Pigment Analysis.
For pigment analysis 100 μL of either 1-h dark-adapted or illuminated (directly sampled from the CCD camera recorder) cells were added to 900 μL methanol. Debris was removed by centrifugation at 10,000 × g for 15 min and the supernatant was frozen in liquid nitrogen. A total of 15 μL of pigment extract was subjected to reverse-phase HPLC analysis (Waters), and pigments were estimated according to Lichtenthaler (35).
Phylogenetic Analysis.
Phylogenetic analysis was performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank F. Rappaport and F. A. Wollman for input during the initial phase of this study; M. Ribera d'Alcalà, D. Iudicone, C. Brunet, A. R. Grossman, and B. R. Green for critical suggestions; and G. Peers and J.-D. Rochaix for antibodies. Help from A. Meichenin for cell cultures, from Marcel Kuntz for HPLC measurements, and from Alix Boulouis for sampling during time-course experiments is also acknowledged. The project was supported by grants from the JST-Centre National de la Recherche Scientifique (CNRS) cooperative project on Marine Genomics and Marine Biology, the Agence National de la Recherche (ANR) Phytadapt project (to G.F. and C.B.), the Human Frontier Science Program (HFSP) Career Development Award (0014/2006), the FP7 Marie Curie Initial Training Network (ITN) (COSI; 215174) and the Action Thématique et Incitative sur Programme (ATIP) award (2009) from CNRS (to A.F.). P.C. is an Fond de la Recherche Scientifique; Fond National de la Recherche (FRS-FNRS) research associate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007703107/-/DCSupplemental.
References
- 1.Smetacek V. Diatoms and the ocean carbon cycle. Protist. 1999;150:25–32. doi: 10.1016/S1434-4610(99)70006-4. [DOI] [PubMed] [Google Scholar]
- 2.Bowler C, Vardi A, Allen AE. Oceanographic and biogeochemical insights from diatom genomes. Annu Rev Mar Sci. 2010;2:333–365. doi: 10.1146/annurev-marine-120308-081051. [DOI] [PubMed] [Google Scholar]
- 3.Kirk JYO. Light and Photosynthesis in the Aquatic Ecosystems. Cambridge, UK: Cambridge Univ Press; 1994. [Google Scholar]
- 4.Harris G. Phytoplankton Ecology. Structure, Function and Fluctuation. London, UK: Chapman and Hall; 1986. [Google Scholar]
- 5.Litchman E, Klausmeier CA, Yoshiyama K. Contrasting size evolution in marine and freshwater diatoms. Proc Natl Acad Sci USA. 2009;106:2665–2670. doi: 10.1073/pnas.0810891106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 7.Lavaud J, Rousseau B, van Gorkom HJ, Etienne AL. Influence of the diadinoxanthin pool size on photoprotection in the marine planktonic diatom Phaeodactylum tricornutum. Plant Physiol. 2002;129:1398–1406. doi: 10.1104/pp.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard S, Finazzi G, Wollman FA. The dynamics of photosynthesis. Annu Rev Genet. 2008;42:463–515. doi: 10.1146/annurev.genet.42.110807.091452. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 10.Owens TG. Light-harvesting function in the diatom Phaeodactylum tricornutum. II. Distribution of excitation energy between the photosystems. Plant Physiol. 1986;80:739–746. doi: 10.1104/pp.80.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hager A, Stransky H. The carotenoid pattern and the occurrence of the light-induced xanthophyll cycle in various classes of algae. V. A few members of Cryptophyceae, Euglenophyceae, Bacillariophyceae, Chrysophyceae and Phaeophyceae (Translated from German) Arch Mikrobiol. 1970;73:77–89. [PubMed] [Google Scholar]
- 12.Lohr M, Wilhelm C. Algae displaying the diadinoxanthin cycle also possess the violaxanthin cycle. Proc Natl Acad Sci USA. 1999;96:8784–8789. doi: 10.1073/pnas.96.15.8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savard F, Richard C, Guertin M. The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the cabI/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol Biol. 1996;32:461–473. doi: 10.1007/BF00019098. [DOI] [PubMed] [Google Scholar]
- 14.Neilson JA, Durnford DG. Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth Res. 2010 doi: 10.1007/s11120-010-9576-2. 10.1007/s11120-010-9576-2. [DOI] [PubMed] [Google Scholar]
- 15.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 16.Park S, Jung G, Hwang YS, Jin E. Dynamic response of the transcriptome of a psychrophilic diatom, Chaetoceros neogracile, to high irradiance. Planta. 2010;231:349–360. doi: 10.1007/s00425-009-1044-x. [DOI] [PubMed] [Google Scholar]
- 17.Nymark M, et al. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE. 2009;4:e7743. doi: 10.1371/journal.pone.0007743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu SH, Green BR. Photoprotection in the diatom Thalassiosira pseudonana: Role of LI818-like proteins in response to high light stress. Biochim Biophys Acta. 2010;1797:1449–1457. doi: 10.1016/j.bbabio.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Olaizola M, et al. Non-photochemical fluorescence quenching and the diadinoxanthin cycle in a marine diatom. Photosynth Res. 1994;41:357–370. doi: 10.1007/BF00019413. [DOI] [PubMed] [Google Scholar]
- 20.De Martino A, Meichenin A, Shi X, Pan K, Bowler C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J Phycol. 2007;43:992–1009. [Google Scholar]
- 21.Locarnini RA, Mishonov AV, Antonov JI, Boyer TP, Garcia HE. In: World Ocean Atlas 2005, Volume 1: Temperature. Levitus S, editor. Washington, DC: NOAA Atlas NESDIS 61, US Government Printing Office; 2006. p. 182. [Google Scholar]
- 22.Goss R, Ann Pinto E, Wilhelm C, Richter M. The importance of a highly active and DeltapH-regulated diatoxanthin epoxidase for the regulation of the PS II antenna function in diadinoxanthin cycle containing algae. J Plant Physiol. 2006;163:1008–1021. doi: 10.1016/j.jplph.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 24.Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA. 2010;107:11128–11133. doi: 10.1073/pnas.1002873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannesson K, André C. Life on the margin: Genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol. 2006;15:2013–2029. doi: 10.1111/j.1365-294X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- 26.Jung HS, Niyogi KK. Quantitative genetic analysis of thermal dissipation in Arabidopsis. Plant Physiol. 2009;150:977–986. doi: 10.1104/pp.109.137828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillard RL. In: Culture of Phytoplankton for Feeding Marine Invertebrates, Culture of Marine Invertebrates Animals. Smith WL, Chanley MH, editors. New York: Plenum; 1975. pp. 29–60. [Google Scholar]
- 28.Antonov JI, Locarnini RA, Boyer TP, Mishonov AV, Garcia HE. In: World Ocean Atlas 2005, Volume 2: Salinity. Levitus S, editor. Washington, DC: NOAA Atlas NESDIS 62, US Government Printing Office; 2006. p. 182. [Google Scholar]
- 29.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY) 1999;1:239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- 31.De Riso V, et al. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009;37:e96. doi: 10.1093/nar/gkp448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coesel S, et al. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 2009;10:655–661. doi: 10.1038/embor.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson X, et al. A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynth Res. 2009;102:85–93. doi: 10.1007/s11120-009-9487-2. [DOI] [PubMed] [Google Scholar]
- 34.Bilger W, Björkman O. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res. 1990;25:173–186. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- 35.Lichtenthaler HK. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- 36.Genty B, Harbinson J, Briantais J-M, Baker NR. The relationship between non-photochemical quenching of chlorophyll fluorescence and the rate of photosystem 2 photochemistry in leaves. Photosynth Res. 1990;25:249–257. doi: 10.1007/BF00033166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.