Abstract
The anaerobic acetogenic bacterium Acetobacterium woodii carries out a unique type of Na+-motive, anaerobic respiration with caffeate as electron acceptor, termed “caffeate respiration.” Central, and so far the only identified membrane-bound reaction in this respiration pathway, is a ferredoxin:NAD+ oxidoreductase (Fno) activity. Here we show that inverted membrane vesicles of A. woodii couple electron transfer from reduced ferredoxin to NAD+ with the transport of Na+ from the outside into the lumen of the vesicles. Na+ transport was electrogenic, and accumulation was inhibited by sodium ionophores but not protonophores, demonstrating a direct coupling of Fno activity to Na+ transport. Results from inhibitor studies are consistent with the hypothesis that Fno activity coupled to Na+ translocation is catalyzed by the Rnf complex, a membrane-bound, iron–sulfur and flavin-containing electron transport complex encoded by many bacterial and some archaeal genomes. Fno is a unique type of primary Na+ pump and represents an early evolutionary mechanism of energy conservation that expands the redox range known to support life. In addition, it explains the lifestyle of many anaerobic bacteria and gives a mechanistic explanation for the enigma of the energetic driving force for the endergonic reduction of ferredoxin with NADH plus H+ as reductant in a number of aerobic bacteria.
Keywords: Acetobacterium, anaerobic respiration, electron transport, Na+ pump, Rnf
The mechanism(s) of energy conservation in a major group of strictly anaerobic bacteria, the acetate-forming, acetogenic bacteria, is still an enigma. Acetogens can grow autotrophically with hydrogen as electron donor. The electrons are channeled to the acceptor carbon dioxide that is reduced to acetate in the Wood–Ljungdahl pathway (1–3). How this pathway is coupled to energy conservation is obscure, but in the model acetogen Acetobacterium woodii at least one of the reactions is coupled to primary and electrogenic Na+ translocation across the membrane (4). It is known that the electrochemical sodium ion gradient drives the synthesis of ATP by a unique hybrid Na+ F1FO ATP synthase, but the enzyme generating the Na+ gradient remains to be identified (4–6).
In recent years it turned out that A. woodii can use the alternative electron acceptor caffeate [3-(3,4-dihydroxyphenyl)-2-propenoic acid] in a process called caffeate respiration (7, 8). The phenylacrylate caffeate is a major component of lignin and thus makes a considerable portion of plant-derived biomass in soils (9). Caffeate respiration in A. woodii is coupled to ATP synthesis by a chemiosmotic mechanism with Na+ as coupling ion (10). The pathway of caffeate respiration with hydrogen as reductant involves a ferredoxin-reducing hydrogenase (11), a ferredoxin:NAD+ oxidoreductase (Fno), an electron-transferring flavoprotein (Etf) that, in a complex with a caffeyl–CoA–dehydrogenase, reduces caffeyl–CoA to hydrocaffeyl–CoA with NADH as electron donor, and an activation of caffeate to caffeyl–CoA (12, 13). In the search for the Na+-translocating step of the pathway only the Fno was found to be membrane bound (13).
Here we present evidence that Fno is the Na+-translocating enzyme of the pathway. Furthermore, results from inhibitor studies are consistent with the hypothesis that the Fno activity is catalyzed by Rnf, a flavin- and FeS-containing membrane-bound electron transfer complex. Because the complex is widely distributed in anaerobic as well as aerobic species it is a unique general coupling site in bacteria and archaea (12, 14, 15).
Results
Fno Activity and 22Na+ Transport at Inverted Membrane Vesicles of A. woodii.
To determine ion transport, inverted membrane vesicles (IMV) were prepared as previously described (16), yielding almost exclusively inside-out vesicles. The vesicles were tightly closed and able to hold an artificial ΔpH, tested by creating and monitoring an artificial ammonium diffusion potential (17). The Fno activity at IMVs was measured as previously described for isolated membranes (13), with reduced ferredoxin [generated by reduction with titanium (III) citrate] as electron donor. The assay was carried out under strictly anaerobic conditions in 50 mM Mops buffer (pH 6.0) containing 20 mM MgSO4, 20 mM NaCl, 8 mM dithioerythritol (DTE), and 2.25 mg/L resazurin. Indeed, IMVs catalyzed Fno activity of 20–70 mU/mg, depending on the quality of the vesicle preparation (1 U corresponding to 1 μmol NAD+/min), and is in the same range as the ATPase activity (16). Fno activity was dependent on titanium (III) citrate or NAD+ in the assay. Ferredoxin requirement depended on the washing procedure. Vesicles washed only once did not require additional ferredoxin, whereas vesicles washed three times did. These data indicate a rather tight association of the ferredoxin with the membrane.
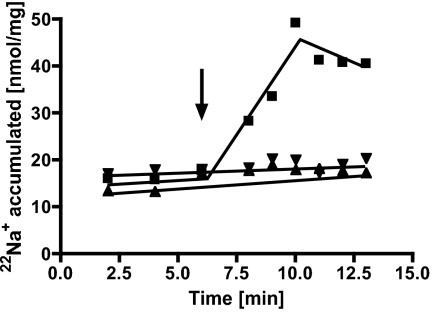
Transport of Na+ was measured at IMVs of A. woodii using the radioisotope 22Na+. Upon addition of reduced ferredoxin and NAD+, 22Na+ was accumulated in the lumen of IMVs (Fig. 1). 22Na+ transport was strictly dependent on the presence of titanium (III) citrate and NAD+. As expected, 22Na+ transport required addition of ferredoxin only in IMVs washed three times. 22Na+ translocation was observed at IMVs prepared from cells grown on fructose or on fructose plus caffeate, indicating that the Fno activity is produced under both growth conditions. When vesicles were prepared under aerobic conditions no NAD+ reduction was observed and subsequently transport was abolished, indicating that Fno contains oxygen-sensitive cofactors. Depending on the preparation of the IMVs, a stoichiometry of 2–35 electrons/22Na+ was obtained.
Fig. 1.
Fno catalyzed 22Na+ transport. Membrane vesicles (protein concentration 3.2 mg/mL) in 50 mM Mops buffer containing 20 mM MgSO4, 20 mM NaCl, 8 mM DTE, and 2.25 mg/L resazurin translocated 22Na+ upon addition of 7.5 μg ferredoxin, 5 mM titanium citrate, and 3.5 mM NAD+ (squares). In control experiments titanium citrate (downward-pointing triangles) or NAD+ (upward-pointing triangles) were omitted. Arrow indicates addition of NAD+.
Concentration Dependence of 22Na+ Transport.
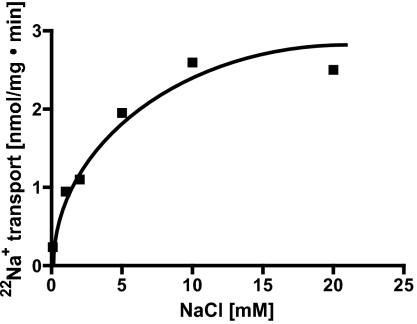
The rate of 22Na+ transport was dependent on the NaCl concentration of the buffer (Fig. 2). At 0.1 mM NaCl it was 0.24 nmol/min · mg protein but increased with increasing Na+ concentrations to 2.6 nmol/min · mg protein at 10 mM NaCl. The Km value for Na+ was determined to be 2.5 mM, and Vmax was 2.6 nmol/mg · min (Fig. 2). A 3.2-fold accumulation (0.48 nmol/mg) was observed at 0.1 mM NaCl, but it decreased to 2.1-fold (24 nmol/mg) at 20 mM NaCl. All subsequent experiments were performed in the presence of 20 mM NaCl.
Fig. 2.
Na+ dependence of transport activity. Membrane vesicles (protein concentration 4 mg/mL) were incubated in 50 mM Mops buffer, 20 mM MgSO4, 8 mM DTE, 2.25 mg/L resazurin, and varying NaCl concentrations. Transport rates were calculated from the initial slopes and plotted against the Na+ concentration.
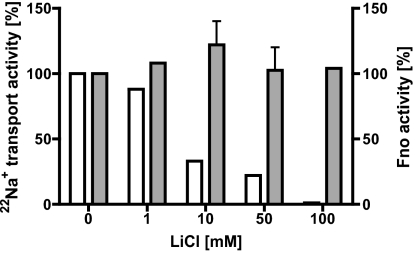
Increasing LiCl concentrations did not have an effect on NAD+ reduction, but with increasing LiCl concentrations 22Na+ transport decreased, indicating that Li+ can be transported instead of Na+ (Fig. 3).
Fig. 3.
Effect of LiCl on 22Na+ transport and Fno activity. White bars represent the rate of 22Na+ accumulation at IMVs; 100% corresponds to 0.9 nmol 22Na+/mg ⋅ min. Gray bars indicate Fno activity; 100% corresponds to 18 mU/mg.
22Na+ Transport Is Electrogenic and Primary.
If Na+ transport is vectorial and not accompanied by charge compensation, the build-up of an electrical field (ΔΨ, inside positive) in the IMVs should inhibit further accumulation of Na+. The K+ ionophore valinomycin used at high KCl concentrations should compensate for charge separation, eliminate the ΔΨ, and thus stimulate 22Na+ transport. Indeed, this was observed. Valinomycin (17 μM) in the presence of 150 mM KCl stimulated 22Na+ transport by 21%, indicating that 22Na+ transport is electrogenic and accompanied by the generation of an electrical field (ΔΨ) across the IMVs. This ΔΨ could then be used to drive the Na+ F1FO ATP synthase present in A. woodii. Indeed, inhibition of the Na+ F1FO ATP synthase by 1 mM N,N′-dicyclohexylcarbodiimide (DCCD) (16) increased Na+ accumulation by 47%.
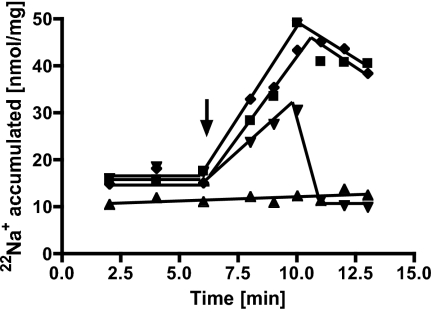
The protonophore 3,5-di-tert-butyl-hydroxybenzylidenemalonitrile (SF 6847; 100 μM) did not abolish 22Na+ translocation (Fig. 4), excluding the possibility that Fno activity is coupled to primary proton transport that then is used to energize secondary Na+ transport via Na+/H+ antiporter. The effectiveness of the protonophore was ensured by its ability to dissipate an artifical ΔpH created by an NH4+ diffusion potential in IMVs of A. woodii. There was no accumulation of 22Na+ in the presence of the Na+ ionophore N,N,N,N′-tetracyclohexyl-1,2-phenylendioxydiacetamide (ETH 2120; 100 μM); moreover, a previously established Na+ gradient was abolished by the Na+ ionophore (Fig. 4), indicating the generation of a transmembrane Na+ gradient (i.e., an uphill transport of 22Na+ upon oxidation of reduced ferredoxin). In summary, the ionophore studies present clear evidence that the Fno activity is coupled to a primary and electrogenic 22Na+ translocation.
Fig. 4.
22Na+ transport is a primary event. Membrane vesicles (protein concentration 3.2 mg/mL) in 50 mM Mops buffer (pH 6.0) containing 20 mM MgSO4, 20 mM NaCl, 8 mM DTE, and 2.25 mg/L resazurin showed 22Na+ transport upon addition of 7.5 μg ferredoxin, 5 mM titanium citrate, and 3.5 mM NAD+ (squares). ETH 2120 (100 μM) was added 6 min before (upward-pointing triangles) or 4 min after (downward-pointing triangles) addition of NAD+. SF 6847 (100 μM) (diamonds) was added 6 min before addition of NAD+. Arrow indicates addition of NAD+.
Inhibition Studies on Fno Activity.
So far, the data presented unequivocally demonstrate the presence of an Na+-translocating Fno activity but do not reveal the nature of this enzyme. Previously we had identified an operon in A. woodii that encodes a membrane-bound electron transfer complex with similartity to a gene cluster (rnf) (18) that was shown genetically to be involved in the production of reduced ferredoxin (required for nitrogen fixation) with NADH plus H+ as reductant in Rhodobacter capsulatus (19). The Rnf complexes of A. woodii and Clostridium tetanomorphum were partially purified and shown to catalyze Fno activity as well as NADH-dependent reduction of potassium ferricyanide (18, 20). Furthermore, it was shown that RnfG and RnfD of Vibrio cholerae contain covalently bound flavins (21). In addition, RnfC and RnfB have 4Fe4S clusters (22). To unravel whether this Rnf complex may catalyze the Na+-pumping Fno activity observed at IMVs, we first searched for inhibitors for the partially purified Rnf complex. Ag+ and Cu2+ turned out to be very efficient inhibitors; half-maximal inhibition of NAD+ reduction was observed at 3.75 ± 1 (SD) and 1.75 ± 0.35 μM, respectively. In addition, the inhibitors 1,10-phenantroline (I50 = 155 ± 21 μM), diphenyliodonium chloride (I50 = 1 ± 0.17 μM), and diphenyleniodonium chloride (I50 = 1 ± 0.035 μM) inhibited Fno activity. 1,10-Phenantroline is an iron chelating agent used to inhibit electron transport reactions (23, 24). Diphenyliodonium chloride and diphenyleniodonium chloride are used as flavin inhibitors (25, 26). The inhibitors inhibited not only Fno activity of the partially purified Rnf complex but also Fno activity catalyzed by IMVs. At the same time as Fno activity, 22Na+ transport catalyzed by the IMVs was inhibited by the different inhibitors of Rnf: AgNO3 inhibited transport completely at 100 μM, CuSO4 at 80 μM, 1,10-phenantroline at 500 μM, and diphenyliodonium chloride and diphenyleniodonium chloride at 40 μM (Table 1).
Table 1.
Inhibition of Fno activity
| Inhibitor | I50 (Fno activity) – partially purified Fno (μM) | I50 (Fno activity) – IMVs (μM) | Complete inhibition of 22Na+ transport (μM) |
| AgNO3 | 3.75 ± 1 | 5.4 ± 0.49 | 100 |
| CuSO4 | 1.75 ± 0.35 | 2 ± 0.07 | 80 |
| 1,10-Phenantroline | 155 ± 21 | 84 ± 19 | 500 |
| Diphenyliodonium chloride | 1 ± 0.17 | 4.1 ± 4 | 40 |
| Diphenyleniodonium chloride | 1 ± 0.035 | 2.5 ± 0.7 | 40 |
Inhibition of NAD+ reduction and 22Na+ transport at partially purified protein and IMVs. Errors are shown as SD.
Discussion
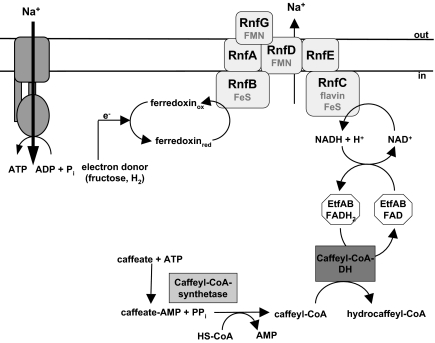
In summary, this works demonstrates a unique primary sodium ion pump that couples electron transfer from reduced ferredoxin to NAD+ with electrogenic movement of Na+ out of the cell. The energy stored in the electrochemical Na+ gradient may then drive ATP synthesis via the well-known Na+ F1FO ATP synthase present in A. woodii (6, 16). This Na+ pump is the only coupling site in caffeate respiration and thus explains how caffeate respiration is coupled to ATP synthesis with Na+ as coupling ion (Fig. 5).
Fig. 5.
Model of caffeate respiration in A. woodii. Flow of electrons from electron donors (fructose or hydrogen) to acceptor caffeate is shown. For explanations see text.
The free energy change of the Fno reaction is −19 kJ/mol, assuming a redox potential of ferredoxin of −420 mV and NAD+ of −320 mV. This would allow for the translocation of 1 mol Na+ across the membrane, assuming an electrochemical ion potential of −200 mV at the cytoplasmic membrane of A. woodii. Thus, during caffeate respiration 3 mol of ferredoxin have to be reduced to get the three ions required for synthesis of 1 mol ATP. However, it is conceivable that additional ferredoxin is reduced in the course of the caffeyl–CoA dehydrogenase reaction by electron bifurcation (see below) (27, 28).
What is the nature of the sodium-motive Fno in A. woodii? A membrane-bound protein complex with Fno activity was partially purified from A. woodii and shown to be encoded by the rnf operon (18). The partially purified Rnf complex is inhibited by AgNO3 and CuSO4, the flavin-directed inhibitors diphenyliodonium chloride and diphenyleniodonium chloride, and 1,10 phenantroline, which is described as an iron chelating agent. At the same time, these inhibitors inhibited ferredoxin-driven Na+ translocation in IMVs of A. woodii. This is consistent with the hypothesis that the Rnf complex catalyzes the observed Na+ translocation coupled to Fno.
Rnf is a proposed membrane-bound electron transport complex containing six subunits with flavins and iron–sulfur centers as electron carriers. It was originally described in mutants of R. capsulatus (Rhodobacter nitrogen fixation), but the genes are present in the genomes of many aerobic and anaerobic bacteria. NAD+ was regarded as the most electronegative electron donor in classic bioenergetics, but in recent years it turned out that ferredoxin is an electron carrier widely used by anaerobes (28, 29). Ferredoxin is used as primary electron acceptor by hydrogenases, the pyruvate:ferredoxin oxidoreductase, some pyruvate:formate lyases, formate dehydrogenases, and CO dehydrogenases (11, 30–33). In addition, some anaerobic bacteria couple the exergonic reduction of fermentation intermediates to the endergonic reduction of ferredoxin with NADH as a reductant in a process called “electron bifurcation” (27, 28). The reduced ferredoxin is then assumed to be oxidized by Rnf. In anaerobes it may often be the only way to generate an ion gradient for synthesis of ATP by a chemiosmotic mechanism. In aerobes, it may function to drive the endergonic reduction of ferredoxin or iron (sulfur) centers in other proteins. The use of Na+ as coupling ion for A. woodii Fno/Rnf was expected because it is known that A. woodii bases its bioenergetics on a sodium ion current across the cytoplasmic membrane (10, 15, 34). The same may be true in other anaerobes or aerobes. However, most organisms that harbor Rnf have no documented Na+ bioenergetics, and therefore it is likely that these species use H+ as coupling ion for Rnf (19).
Our work shows experimentally that electron transfer from reduced ferredoxin to NAD+ as acceptor drives the generation of a transmembrane ion gradient. It should be mentioned in this connection that other anaerobes, such as Methanosarcina or Pyrococcus species, have related systems that use Ech hydrogenase or Eha hydrogenase to use the redox difference between ferredoxin and H+/H2 to establish a transmembrane ion gradient (35–37).
Materials and Methods
Measurement of Fno Activity.
The measurement of Fno activity was conducted as previously described (13). The buffer used was 50 mM Mops (pH 6.0) containing in addition 20 mM MgSO4, 20 mM NaCl, 8 mM DTE, and 2.25 mg/L resazurin (to monitor redox state).
Growth of Cells and Preparation of Vesicles.
A. woodii (DSM 1030) was grown under anaerobic conditions using 20 mM fructose or 20 mM fructose and 5 mM caffeate as electron acceptor as previously described (16, 38). The preparation of vesicles was done under strictly anaerobic conditions in an anaerobic chamber (Coy Laboratory Products) as previously described (16). For preparation of vesicles the growth medium was supplemented with 420 mM sucrose and 8.1 mM MgSO4. Five liters of medium were inoculated (with 200 mL culture), and the optical density was followed at 600 nm. At OD600 of 0.7–0.9, 70 μg penicillin G/mg were added to the medium to induce protoplast formation. During further incubation A. woodii formed protoplasts as monitored by microscopic observations. After 20 h the culture consisted almost entirely of spherical forms that were highly sensitive to low osmolarity. These protoplasts were harvested anaerobically by centrifugation (6,250 × g, 20 min, 4 °C) and washed in cold vesicle buffer [50 mM Tris/HCl (pH 8.0) containing 25 mM MgSO4, 420 mM sucrose, 8 mM DTE, and 2.25 mg/L resazurin, flushed with N2 for 20 min]. After washing, the protoplasts were resuspended in a total volume of 300 mL vesicle buffer with lysozyme (1 mg/mL) and incubated for 30 min at room temperature. The protoplasts were centrifuged (6,250 × g, 20 min, 4 °C) and resuspended in 10–20 mL vesicle buffer. The protoplasts were passed through a French pressure cell at 41 MPa and centrifuged three times (4,500 × g, 35 min, 4 °C). The resulting supernatant (= crude vesicles) was centrifuged further by ultracentrifugation at 120,000 × g, 40 min, 4 °C. The pellet was washed in vesicle buffer and centrifuged again. The resulting pellet was solved in the same buffer in a volume of 3–5 mL.
Protein concentrations were determined by the method of Bradford (39).
Measurement of 22Na+-Translocation.
The experiments were performed under anaerobic conditions in 50 mM Mops buffer (pH 6.0) containing 20 mM MgSO4, 8 mM DTE, and 2.25 mg/L resazurin (to monitor redox state) at 30 °C in a shaking water bath as previously described (16). In 3.5-mL glass vials buffer, supplements (17 μM valinomycin, 1 mM DCCD, NaCl as indicated, 150 mM KCl, and 7.5 μg ferredoxin) and 22NaCl (final activity 0.5 μCi/mL) were combined and incubated for 120 min to ensure equilibration of 22Na+ before the reaction was started. Titanium citrate (5 mM), 100 μM ETH 2120 or 100 μM SF 6847 were added just before taking of samples started. After 6 min 3.5 mM NAD+ was added. When the effect of DCCD on transport was tested, sodium was omitted from the assay.
LiCl.
The sodium concentration in all experiments was 1 mM, and LiCl was increased from none to 100 mM. 22Na+ transport and Fno activity were measured in presence of different LiCl concentrations.
Inhibition Experiments.
AgNO3, CuSO4, 1,10-phenantroline, diphenyliodonium chloride, and diphenyleniodonium chloride were preincubated with IMVs for 10 min before starting the NAD+ reduction or transport. For experiments with AgNO3 and CuSO4, DTE was omitted from all buffers.
Partial Purification.
The protein was enriched as previously described (18).
Acknowledgments
This work was funded by a grant from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Ragsdale SW. Enzymology of the Wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood HG, Ragsdale SW, Pezacka E. The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol Rev. 1986;39:345–362. [Google Scholar]
- 3.Ljungdahl LG. In: Acetogenesis. Drake HL, editor. New York: Chapman & Hall; 1994. pp. 63–87. [Google Scholar]
- 4.Müller V. Energy conservation in acetogenic bacteria. Appl Environ Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fritz M, et al. An intermediate step in the evolution of ATPases: A hybrid F0V0 rotor in a bacterial Na+F1F0 ATP synthase. FEBS J. 2008;275:1999–2007. doi: 10.1111/j.1742-4658.2008.06354.x. [DOI] [PubMed] [Google Scholar]
- 6.Fritz M, Müller V. An intermediate step in the evolution of ATPases—the F1F0-ATPase from Acetobacterium woodii contains F-type and V-type rotor subunits and is capable of ATP synthesis. FEBS J. 2007;274:3421–3428. doi: 10.1111/j.1742-4658.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 7.Tschech A, Pfennig N. Growth yield increase linked to caffeate reduction in Acetobacterium woodii. Arch Microbiol. 1984;137:163–167. [Google Scholar]
- 8.Hansen B, Bokranz M, Schönheit P, Kröger A. ATP formation coupled to caffeate reduction by H2 in Acetobacterium woodii NZva16. Arch Microbiol. 1988;150:447–451. [Google Scholar]
- 9.Blum U, et al. Phenolic acid content of soils from wheat-no till, wheat-conventional till, and fallow-conventional till soybean cropping systems. J Chem Ecol. 1991;17:1045–1068. doi: 10.1007/BF01402933. [DOI] [PubMed] [Google Scholar]
- 10.Imkamp F, Müller V. Chemiosmotic energy conservation with Na+ as the coupling ion during hydrogen-dependent caffeate reduction by Acetobacterium woodii. J Bacteriol. 2002;184:1947–1951. doi: 10.1128/JB.184.7.1947-1951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragsdale SW, Ljungdahl LG. Hydrogenase from Acetobacterium woodii. Arch Microbiol. 1984;139:361–365. doi: 10.1007/BF00408380. [DOI] [PubMed] [Google Scholar]
- 12.Müller V, Imkamp F, Biegel E, Schmidt S, Dilling S. Discovery of a ferredoxin:NAD+-oxidoreductase (Rnf) in Acetobacterium woodii: A novel potential coupling site in acetogens. Ann N Y Acad Sci. 2008;1125:137–146. doi: 10.1196/annals.1419.011. [DOI] [PubMed] [Google Scholar]
- 13.Imkamp F, Biegel E, Jayamani E, Buckel W, Müller V. Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: Identification of an Rnf-type NADH dehydrogenase as a potential coupling site. J Bacteriol. 2007;189:8145–8153. doi: 10.1128/JB.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, et al. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J Bacteriol. 2006;188:702–710. doi: 10.1128/JB.188.2.702-710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt S, Biegel E, Müller V. The ins and outs of Na+ bioenergetics in Acetobacterium woodii. Biochim Biophys Acta. 2009;1787:691–696. doi: 10.1016/j.bbabio.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 16.Heise R, Müller V, Gottschalk G. Presence of a sodium-translocating ATPase in membrane vesicles of the homoacetogenic bacterium Acetobacterium woodii. Eur J Biochem. 1992;206:553–557. doi: 10.1111/j.1432-1033.1992.tb16959.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T, Hsu C, Rosen BP. Cation/proton antiport systems in Escherichia coli. Solubilization and reconstitution of delta pH-driven sodium/proton and calcium/proton antiporters. J Biol Chem. 1986;261:678–683. [PubMed] [Google Scholar]
- 18.Biegel E, Schmidt S, Müller V. Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii. Environ Microbiol. 2009;11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- 19.Schmehl M, et al. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: A putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 20.Boiangiu CD, et al. Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J Mol Microbiol Biotechnol. 2005;10:105–119. doi: 10.1159/000091558. [DOI] [PubMed] [Google Scholar]
- 21.Backiel J, et al. Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae. Biochemistry. 2008;47:11273–11284. doi: 10.1021/bi800920j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouanneau Y, Jeong HS, Hugo N, Meyer C, Willison JC. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus—characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 23.Harmon HJ, Crane FL. Inhibition of mitochondrial electron transport by hydrophilic metal chelators. Determination of dehydrogenase topography. Biochim Biophys Acta. 1976;440:45–58. doi: 10.1016/0005-2728(76)90112-2. [DOI] [PubMed] [Google Scholar]
- 24.Ponka P, Grady RW, Wilczynska A, Schulman HM. The effect of various chelating agents on the mobilization of iron from reticulocytes in the presence and absence of pyridoxal isonicotinoyl hydrazone. Biochim Biophys Acta. 1984;802:477–489. doi: 10.1016/0304-4165(84)90367-2. [DOI] [PubMed] [Google Scholar]
- 25.Majander A, Finel M, Wikström M. Diphenyleneiodonium inhibits reduction of iron-sulfur clusters in the mitochondrial NADH-ubiquinone oxidoreductase (Complex I) J Biol Chem. 1994;269:21037–21042. [PubMed] [Google Scholar]
- 26.Shiemke AK, Arp DJ, Sayavedra-Soto LA. Inhibition of membrane-bound methane monooxygenase and ammonia monooxygenase by diphenyliodonium: implications for electron transfer. J Bacteriol. 2004;186:928–937. doi: 10.1128/JB.186.4.928-937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–791. doi: 10.1128/JB.01422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jungermann K, Kirchniawy H, Thauer RK. Ferredoxin dependent CO2 reduction to formate in Clostridium pasteurianum. Biochem Biophys Res Commun. 1970;41:682–689. doi: 10.1016/0006-291x(70)90067-7. [DOI] [PubMed] [Google Scholar]
- 31.Thauer RK, Rupprecht E, Jungermann K. The synthesis of one-carbon units from CO2 via a new ferredoxin dependent monocarboxylic acid cycle. FEBS Lett. 1970;8:304–307. doi: 10.1016/0014-5793(70)80293-9. [DOI] [PubMed] [Google Scholar]
- 32.Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem. 2000;275:28494–28499. doi: 10.1074/jbc.M003291200. [DOI] [PubMed] [Google Scholar]
- 33.Shanmugasundaram T, Wood HG. Interaction of ferredoxin with carbon monoxide dehydrogenase from Clostridium thermoaceticum. J Biol Chem. 1992;267:897–900. [PubMed] [Google Scholar]
- 34.Heise R, Müller V, Gottschalk G. Acetogenesis and ATP synthesis in Acetobacterium woodii are coupled via a transmembrane primary sodium ion gradient. FEMS Microbiol Lett. 1993;112:261–268. [Google Scholar]
- 35.Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedderich R. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: Ancestors of complex I. J Bioenerg Biomembr. 2004;36:65–75. doi: 10.1023/b:jobb.0000019599.43969.33. [DOI] [PubMed] [Google Scholar]
- 37.Welte C, Krätzer C, Deppenmeier U. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 2010;277:3396–3403. doi: 10.1111/j.1742-4658.2010.07744.x. [DOI] [PubMed] [Google Scholar]
- 38.Heise R, Müller V, Gottschalk G. Sodium dependence of acetate formation by the acetogenic bacterium Acetobacterium woodii. J Bacteriol. 1989;171:5473–5478. doi: 10.1128/jb.171.10.5473-5478.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]