Abstract
A set of currently known alleles increasing the risk for coronary artery disease, cancer, and type 2 diabetes as identified by genome-wide association studies was tested for compatibility with human longevity. Here, we show that nonagenarian siblings from long-lived families and singletons older than 85 y of age from the general population carry the same number of disease risk alleles as young controls. Longevity in this study population is not compromised by the cumulative effect of this set of risk alleles for common disease.
Keywords: association, aging SNP
Members of long-lived families have a lifelong survival advantage (1, 2) that can be attributed to a lower risk for coronary artery disease, cancer, and type 2 diabetes (3–5). These observations may be explained by the presence of alleles protecting against diseases that contribute to population mortality or the absence of alleles promoting such diseases. The latter hypothesis can be tested now that genome-wide association studies (GWASs) have produced a series of SNPs showing robust associations with common diseases that are the main causes of death.
We tested whether nonagenarians from the Leiden Longevity Study (LLS) and octogenarians from the Leiden 85 Plus Study carried lower total numbers of 30 major GWAS-identified disease susceptibility alleles than younger controls.
Results
Reviewing the catalog of published GWASs for coronary artery disease, heart failure, cancer, and type 2 diabetes up to February 2009 revealed 30 SNPs in 22 disease-associated loci (Table S1). These SNPs were investigated for their deleterious role in survival beyond 85 y of age by comparing 723 nonagenarian siblings (mean age of 94 y) from the LLS (1), representing familial long-lived cases, and 721 unrelated younger controls (mean age of 52 y). We also compared 979 singleton long-lived individuals older than 85 y (mean age of 87 y) of age from the population-based Leiden 85 Plus Study (6, 7) and 1,167 younger controls (mean age of 41 y) from the Netherlands Twin Register (NTR) (8).
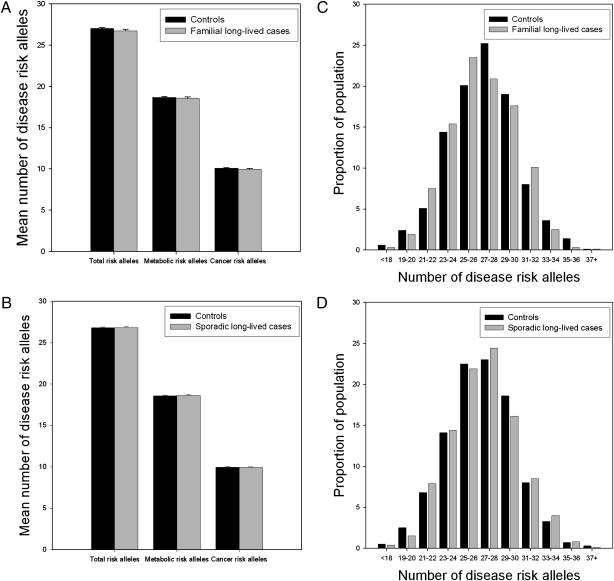
We tested whether the number of disease alleles was different between long-lived individuals and younger controls, because the cumulative effect of disease alleles is expected to be more pronounced than that of individual SNPs, with a modest effect on disease risk (9, 10). On average the nonagenarian siblings carried 26.7 ± 0.19 disease risk alleles, which was virtually identical to the number carried by younger controls (27.0 ± 0.12; P = 0.127; Fig. 1A), and the same was observed for the number carried by sporadic long-lived subjects (26.8 ± 0.11) and younger controls (26.8 ± 0.10; P = 0.847; Fig. 1B). The distribution of the number of risk alleles was the same in highly aged and young subjects, even in the tails of the distribution (Fig. 1 C and D). Similar results were obtained when the 19 alleles associated with metabolic disease or the 11 cancer-associated alleles were considered separately. We compared 391 long-lived subjects and 409 younger controls in the 10% tails of the sum of risk allele distribution. The carriers of the 10% upper tail of the sum of risk allele distributions did not have a decreased risk of surviving beyond 85 y of age, because the odds ratio (OR) estimates are close to unity (Tables S2–S4). The 95% confidence interval (CI) of the OR per extra risk allele ranges from 0.97 to 1.01. Further exploration for different effect sizes and interacting alleles did not reveal any evidence for a subset of alleles associating with longevity via main effects or as interactions.
Fig. 1.
GWAS-identified disease risk alleles in nonagenarians from long-lived families and sporadic long-lived individuals. (A) Average number of risk alleles among nonagenarian siblings and controls of the LLS. (B) Average number of risk alleles among sporadic long-lived individuals of the Leiden 85 Plus Study and NTR controls. (C) Distribution of the number of disease risk alleles among nonagenarian siblings and controls of the LLS. (D) Distribution of the number of disease risk alleles among sporadic long-lived participants of the Leiden 85 Plus Study and NTR controls.
In a meta-analysis of the two cohorts, nominal significant associations with survival into old age were observed for seven individual SNPs (0.023 < P < 0.046; Table S5), but none was significant when accounting for multiple testing (P < 0.001).
The effect on fatal disease can also be analyzed by a cross-sectional comparison of healthy groups of different age categories (11–13). Because disease risk alleles contribute to morbidity and mortality, the frequencies of those risk alleles are expected to decrease in the healthy elderly individuals of the population at large. However, the frequency of deleterious genotypes might increase among long-lived individuals because their protective genotype allows disease-related genes to accumulate (12). To investigate the presence of such a buffering mechanism, we divided all subjects and controls (n = 3,590) into 10 age groups (Table S6) and determined the mean number of risk alleles and the allele frequencies of the 30 SNPs in each age group. We did not observe differences in the mean number of risk alleles (Fig. S1) or in SNP-specific allele frequencies (Dataset S1).
Discussion
A lower genetic predisposition for disease in long-lived individuals may explain why long-lived families have a delayed age of onset and lower mortality attributable to common age-related diseases (3–5). Therefore, our aim was to investigate whether the absence of currently known genetic susceptibility alleles is essential for survival into old age. Hence, we compared the cumulative effect of alleles that have convincingly been identified as risk factors for cardiovascular disease, type 2 diabetes, and cancer between long-lived individuals (aged 85 y and older) and the middle-aged general population, in whom the disease risk alleles are generally discovered.
Despite the fact that cardiovascular disease, cancer, and type 2 diabetes contribute to the majority of deaths in modern societies, the cumulative effects of this set of 30 risk alleles do not restrain the study group from surviving into old age. This observation, together with the observations that the offspring of the nonagenarian siblings and centenarians have a decreased prevalence of cardiovascular disease, hypertension, and type 2 diabetes as compared with controls (5, 14), suggests that the absence of these disease loci does not explain the lower morbidity in the offspring. Also, offspring of centenarians have lower mortality from cancer despite the same cancer prevalence as compared with controls (4). It may be possible that other factors, such as unidentified (rare) genetic variants that associate with fatal disease, are lacking in the study group. Alternatively, alleles protecting individuals from cardiovascular disease, cancer, and type 2 diabetes by delaying the onset or decreasing the severity may be prevalent in this study group. To identify loci essential for delay of disease and survival, a GWAS for survival into old age may be an approach by which protective alleles could also be identified.
The sum of risk alleles has commonly been used to estimate a cumulative effect of the risk alleles to the disease prevalence of type 2 diabetes and cancer (9, 15–21). Previous studies reported substantially increased risk among the upper 10% of the population carrying the highest number of risk alleles as compared with the 10% with the lowest number of risk alleles [e.g., type 2 diabetes, OR= 8.7 (9); prostate cancer, OR = 9.5 (19)]. Of 20 known variants associated with type 2 diabetes, it has been estimated that they would account for 5–10% of an inherited predisposition for type 2 diabetes (22). The SNP sets that have been investigated for their cumulative effect on the risk for specific diseases in these studies overlap with the SNP set investigated in the current study. No effect of a subset of SNPs or interactions among SNPs was detected in the current data, which are in agreement with previous observations that additive variance often accounts for almost all genetic variance (23). Putative associations are thus best described with a multiplicative model in line with previous disease-focused studies on GWAS SNP sets (9, 15,15–21). An accurate estimation of a predicted cumulative effect of these SNPs on longevity was hardly possible because the genetic associations used for this estimation derive mostly from studies focused on disease risk and not on the effect of disease-specific mortality.
In our study, we detected the odds of the carriers with the highest number of risk alleles, with an OR of 0.99 (95% CI: 0.97–1.03), to survive into old age. The boundaries of the 95% CI in this analysis are thus close to unity, which was also the case for the boundaries of the OR per extra risk allele. Although, in general, the association of disease risk alleles with longevity remains to be investigated (if all common and uncommon disease risk alleles are discovered and tested for their association with human longevity accounting for their interaction with environmental factors), our data suggest that longevity is not likely determined by the absence of GWAS-identified risk alleles for cardiovascular disease, cancer, and type 2 diabetes.
Because the current GWAS-identified SNPs have a relatively low predictive value for disease risk (24–26), it was expected that such risk alleles only marginally affect population-wide survival. However, the cumulative effect of cardiovascular disease, cancer, and type 2 diabetes risk alleles, which was expected to be more pronounced (9, 10), did not affect population survival. The extent to which disease risk attributable to the cumulative effects of susceptibility alleles is associated with population-based mortality risk should be tested in patient-based studies. Alternatively, other genetic and environmental factors present in individuals surviving into old age may counteract the detrimental effects of disease susceptibility alleles.
Materials and Methods
Study Populations.
All participants of the LLS, Leiden 85 Plus Study, and NTR provided written informed consent after receiving an explanation of the nature and consequences of the study.
LLS and Control Samples.
For the LLS, nonagenarian siblings of European descent were recruited together with their offspring and the partners of the offspring, who served as population controls. Families were recruited if at least two long-lived siblings were alive and fulfilled the age criterion of 89 y or older for men and 91 y or older for women, representing less than 0.5% of the Dutch population in 2001 (1). In total, 944 long-lived proband siblings with a mean age of 94 y (range: 89–104 y), 1,671 offspring with a mean age of 60 y (range: 39–81 y), and 744 partners with a mean age of 60 y (range: 36–79 y) were included.
Because we recently obtained Illumina Human660W-Quad and HumanOmniExpress GWAS data for participants in the LLS, we were able to investigate potential substructure. First, in PLINK version 1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (27) for 10,000 random SNPs, IBS (identity-by-state) estimates for all pairs of subjects in the samples were computed. Next, we performed a multidimensional scaling (mds) analysis on the N × N matrix of genome-wide IBS pairwise distances, using the mds-plot option in conjunction with clustering. The C1 values were plotted against C2 values to identify clustering among the subjects and controls (Fig. S2). We conclude that there is no substructure among the participants of the LLS to an extent that would affect our conclusions.
Leiden 85 Plus Study.
In the Leiden 85 Plus Study, two prospective population-based cohorts consisting of inhabitants of Leiden aged 85 y and older were followed. Between 1987 and 1989 (cohort 1), 673 subjects aged 85 y and older of Dutch ancestry and white ethnicity were enrolled and followed up for survival for 17 y; during this time, 672 subjects (99.9%) died. Between 1997 and 1999, 563 subjects were enrolled in the month of their 85th birthday (cohort 2) and followed up for survival for 10 y; during this time, 453 subjects (81%) died. Subjects were visited at their home, and there were no exclusion criteria related to health. DNA was available from the combined cohorts for 1,245 subjects aged 85 y and older.
NTR as Population Controls.
From the NTR, 1,203 unrelated participants of European descent with mean age of 41 y (range: 18–80 y) were selected as young population controls for whom DNA was available (8). The substructure in the NTR control group has already been reported (28), and in our study, we included the samples of Dutch descent without known family relations (i.e., those without any substructure).
SNP Selection.
Using the catalog of published GWASs published up to February 2009 (http://www.genome.gov/26525384), we reviewed 266 GWASs, 69 of which reported on associations with coronary artery disease, heart failure, cancer, and type 2 diabetes. These 69 GWASs reported 23 disease-associated loci resulting from at least 2 independent GWASs, harboring 81 associated SNPs with P < 10−4 (Table S1). For each locus, the most replicated SNP was selected from the 81 SNPs, and, subsequently, in case of an equal number of replications, the SNP with the lowest reported P value was selected. For 5 loci, 9 further disease-associated SNPs were selected that were in low to moderate linkage disequilibrium (r2 < 0.80) with the most replicated SNP. Thus, 32 SNPs were selected to be genotyped with the Sequenom iPLEX. Because 2 SNPs failed in genotyping, 30 SNPs covering 22 loci were studied. Seventeen loci are represented by 1 SNP, 3 loci are represented by 2 SNPs, 1 locus is represented by 3 SNPs (8q24.21), and 1 locus is represented by 4 SNPs (9p21.3).
Genotyping.
Genotyping of the selected SNPs was performed using the Sequenom MassARRAY iPLEXGold. The average genotype call rate for genotyped SNPs was 96.3%, and the average concordance rate was 99.7% among 4% duplicated control samples. All 30 SNPs were in Hardy–Weinberg equilibrium (P ≥ 0.002) among the controls.
Statistical Analysis.
Single SNP analysis meta-analysis.
We applied a variance-modified Cochrane–Armitage test to take into account the relatedness between the highly aged sibling cases when computing the variances of the scores (29). In the meta-analysis of both the LLS and Leiden 85 Plus Study/NTR controls, the score statistics of the two studies were combined using a fixed-effects approach.
Cumulative SNP analysis.
For each individual who was genotyped for all 30 SNPs, the total number of disease risk alleles was counted. The difference in the number of disease risk alleles between long-lived subjects and population controls was assessed by linear regression. In these analyses, the number of risk alleles was the dependent variable and the case status was included in the model as a categorical variable. To take into account dependencies within the long-lived sibships of the LLS, robust SEs were used [i.e., the variance was computed from the between-family variation (30)]. P values were also based on these robust SEs.
Logistical regression was performed with control/long-lived status as the outcome, study and gender as covariates, and 10% upper or lower tail as a variable. For the estimation of the OR per extra risk allele, logistical regression was performed with control/long-lived status as the outcome, study and gender as covariates, and the sum of risk alleles as a variable. Analyses were performed using the software package STATA/SE 11.0 (DPC Software).
Interaction analysis.
Polychotomous regression (31) and logic regression (32) were applied to the datasets of the LLS and Leiden 85 Plus/NTR controls to identify subsets of SNPs that might have a joint effect on longevity. Cross-validation was used for selecting the best subset of SNPs and the best functional forms of those subsets. The two approaches are different in that the polychotomous regression is flexible in terms of inheritance modes of the SNPs, although only pairwise interactions are considered, whereas the logic regression is used to identify multiple interacting dominant and recessive effects of SNPs.
Supplementary Material
Acknowledgments
We thank all participants of the Leiden Longevity Study, Leiden 85 Plus Study, and Netherlands Twin Register. This study was supported by a grant from the Innovation Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the National Institute for Healthy Ageing (Grant 05060810), all in the framework of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The data collection of the Netherlands Twin Register is supported by the Netherlands Organization for Scientific Research (Grants NWO904-61-090, NWO904-61-193, NWO480-04-004, and NWO400-05-717), Center for Medical Systems Biology (NWO Genomics), and Spinozapremie (Grant NWO-SPI 56-464-14192).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003540107/-/DCSupplemental.
References
- 1.Schoenmaker M, et al. Evidence of genetic enrichment for exceptional survival using a family approach: The Leiden Longevity Study. Eur J Hum Genet. 2006;14:79–84. doi: 10.1038/sj.ejhg.5201508. [DOI] [PubMed] [Google Scholar]
- 2.Perls TT, et al. Life-long sustained mortality advantage of siblings of centenarians. Proc Natl Acad Sci USA. 2002;99:8442–8447. doi: 10.1073/pnas.122587599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terry DF, Wilcox M, McCormick MA, Lawler E, Perls TT. Cardiovascular advantages among the offspring of centenarians. J Gerontol A Biol Sci Med Sci. 2003;58:M425–M431. doi: 10.1093/gerona/58.5.m425. [DOI] [PubMed] [Google Scholar]
- 4.Terry DF, et al. Lower all-cause, cardiovascular, and cancer mortality in centenarians’ offspring. J Am Geriatr Soc. 2004;52:2074–2076. doi: 10.1111/j.1532-5415.2004.52561.x. [DOI] [PubMed] [Google Scholar]
- 5.Westendorp RG, et al. Leiden Longevity Study Group Nonagenarian siblings and their offspring display lower risk of mortality and morbidity than sporadic nonagenarians: The Leiden Longevity Study. J Am Geriatr Soc. 2009;57:1634–1637. doi: 10.1111/j.1532-5415.2009.02381.x. [DOI] [PubMed] [Google Scholar]
- 6.Bootsma-van der Wiel A, et al. Disability in the oldest old: “Can do” or “do do”? J Am Geriatr Soc. 2001;49:909–914. doi: 10.1046/j.1532-5415.2001.49181.x. [DOI] [PubMed] [Google Scholar]
- 7.Kuningas M, et al. Haplotypes in the human Foxo1a and Foxo3a genes; Impact on disease and mortality at old age. Eur J Hum Genet. 2007;15:294–301. doi: 10.1038/sj.ejhg.5201766. [DOI] [PubMed] [Google Scholar]
- 8.Boomsma DI, et al. Genome-wide association of major depression: Description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 9.Cauchi S, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS ONE. 2008;3:e2031. doi: 10.1371/journal.pone.0002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lango H, et al. UK Type 2 Diabetes Genetics Consortium Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes. 2008;57:3129–3135. doi: 10.2337/db08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heijmans BT, Westendorp RG, Slagboom PE. Common gene variants, mortality and extreme longevity in humans. Exp Gerontol. 2000;35:865–877. doi: 10.1016/s0531-5565(00)00171-6. [DOI] [PubMed] [Google Scholar]
- 12.Bergman A, Atzmon G, Ye K, MacCarthy T, Barzilai N. Buffering mechanisms in aging: A systems approach toward uncovering the genetic component of aging. PLOS Comput Biol. 2007;3:e170. doi: 10.1371/journal.pcbi.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kammerer S, et al. Amino acid variant in the kinase binding domain of dual-specific A kinase-anchoring protein 2: A disease susceptibility polymorphism. Proc Natl Acad Sci USA. 2003;100:4066–4071. doi: 10.1073/pnas.2628028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terry DF, Wilcox MA, McCormick MA, Perls TT. Cardiovascular disease delay in centenarian offspring. J Gerontol A Biol Sci Med Sci. 2004;59:385–389. doi: 10.1093/gerona/59.4.m385. [DOI] [PubMed] [Google Scholar]
- 15.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358:2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- 16.Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100:1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, et al. Cumulative effect of five genetic variants on prostate cancer risk in multiple study populations. Prostate. 2008;68:1257–1262. doi: 10.1002/pros.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 19.Yamada H, et al. Replication of prostate cancer risk loci in a Japanese case-control association study. J Natl Cancer Inst. 2009;101:1330–1336. doi: 10.1093/jnci/djp287. [DOI] [PubMed] [Google Scholar]
- 20.Wacholder S, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362:986–993. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paynter NP, et al. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA. 2010;303:631–637. doi: 10.1001/jama.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9:164–171. doi: 10.1007/s11892-009-0027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 25.Hirschhorn JN. Genomewide association studies—Illuminating biologic pathways. N Engl J Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 26.Kraft P, Hunter DJ. Genetic risk prediction—Are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 27.Purcell S, et al. PLINK: A toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan PF, et al. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slager SL, Schaid DJ. Evaluation of candidate genes in case-control studies: A statistical method to account for related subjects. Am J Hum Genet. 2001;68:1457–1462. doi: 10.1086/320608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diggle PJ, Liang KY, Zegers SL. Analysis of Longitudinal Data. Oxford: Oxford Univ Press; 1994. [Google Scholar]
- 31.Kooperberg C, Ruczinski I. Identifying interacting SNPs using Monte Carlo logic regression. Genet Epidemiol. 2005;28:157–170. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- 32.Ruczinski I, Kooperberg C, LeBlanc M. Logic regression. J Comput Graph Statist. 2003;12:475–511. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.