Abstract
Elicitation of antibodies against targets that are immunorecessive, cryptic, or transient in their native context has been a challenge for vaccine design. Here we demonstrate the elicitation of structure-specific antibodies against the HIV-1 gp41 epitope of the broadly neutralizing antibody 2F5. This conformationally flexible region of gp41 assumes mostly helical conformations but adopts a kinked, extended structure when bound by antibody 2F5. Computational techniques were employed to transplant the 2F5 epitope into select acceptor scaffolds. The resultant “2F5-epitope scaffolds” possessed nanomolar affinity for antibody 2F5 and a range of epitope flexibilities and antigenic specificities. Crystallographic characterization of the epitope scaffold with highest affinity and antigenic discrimination confirmed good to near perfect attainment of the target conformation for the gp41 molecular graft in free and 2F5-bound states, respectively. Animals immunized with 2F5-epitope scaffolds showed levels of graft-specific immune responses that correlated with graft flexibility (p < 0.04), while antibody responses against the graft—as dissected residue-by-residue with alanine substitutions—resembled more closely those of 2F5 than sera elicited with flexible or cyclized peptides, a resemblance heightened by heterologous prime-boost. Lastly, crystal structures of a gp41 peptide in complex with monoclonal antibodies elicited by the 2F5-epitope scaffolds revealed that the elicited antibodies induce gp41 to assume its 2F5-recognized shape. Epitope scaffolds thus provide a means to elicit antibodies that recognize a predetermined target shape and sequence, even if that shape is transient in nature, and a means by which to dissect factors influencing such elicitation.
Keywords: computational design, epitope transplantation, structural mimicry
Monoclonal antibodies of enormous utility have been identified, revolutionizing treatments for autoimmune disorders, infectious disease, and different types of cancers (reviewed in ref. 1). Requirements for nonoral means of delivery and in some contexts prolonged treatment regimens, however, have limited their use. While vaccine modalities have potential for improvements, no clear path exists from a clinically useful monoclonal antibody to elicitation of similar antibodies in a vaccine context. One potential solution is precise immunogen design. The ability of structural biology to provide atomic-level definition of antibody–antigen interactions and of computational biology to manipulate protein structure has raised the possibility—at least for protein antigens—of precisely replicating the antigenic surface recognized by a target antibody. We hypothesized that appropriate immunization with such an antigenic mimic might succeed in eliciting replicas of the original target antibody.
As a first step toward solving the vaccine problem of “reelicitation,” we undertook the challenge of structure-specific elicitation—the elicitation of antibodies capable of binding the sequence and of inducing the structure of a predetermined target epitope. Various protein–scaffold platforms have been described in which structural elements of scaffold proteins act as acceptors of functional or antigenic regions from other proteins (2–4). Here we describe a platform for the elicitation of structure-specific antibodies—the epitope-scaffold platform—in which structural mimics of viral neutralizing determinants are grafted into heterologous protein scaffolds using techniques of computational protein design. As a test system, we chose the 2F5 antibody (5, 6), which recognizes an epitope in the membrane-proximal external region (MPER) of the HIV-1 gp41 transmembrane glycoprotein, for which we and others have determined a number of atomic-level structures (7–14). Although recognition by 2F5 involves not only the structure-specific binding of a gp41 epitope but also nonspecific interactions with membrane (13, 15–17), the system was nonetheless attractive because of the conformational diversity of the MPER, its extensive structural characterization, and the linear nature of the epitope. We show that immunization of animals with epitope–scaffold mimics of the target 2F5 epitope leads to the elicitation of polyclonal serum responses that mimic those of antibody 2F5. Moreover, we confirm crystallographically that monoclonal antibodies elicited by 2F5-epitope scaffolds are capable of binding the sequence and of inducing the conformation of the 2F5 epitope in a flexible gp41 peptide, a conformation that would otherwise only rarely be assumed.
Results
Computational Design of Epitope Scaffolds.
To translate structural information into immunogen design, we devised a semiautomated procedure involving the following steps: First, the entire Protein Data Bank was searched for appropriate acceptor proteins (scaffolds) with backbone structural similarity to segments of the 2F5-bound epitope on gp41. Second, a filtering step was applied in which initial structural matches were only retained if the scaffolds could be bound by antibody without significant clashes. Third, epitope side chains were transplanted at appropriate positions. Fourth, additional mutations were introduced into each of the scaffolds to optimize stability, to enhance epitope exposure, and to minimize nonepitope interactions with antibody (Fig. 1).
Fig. 1.
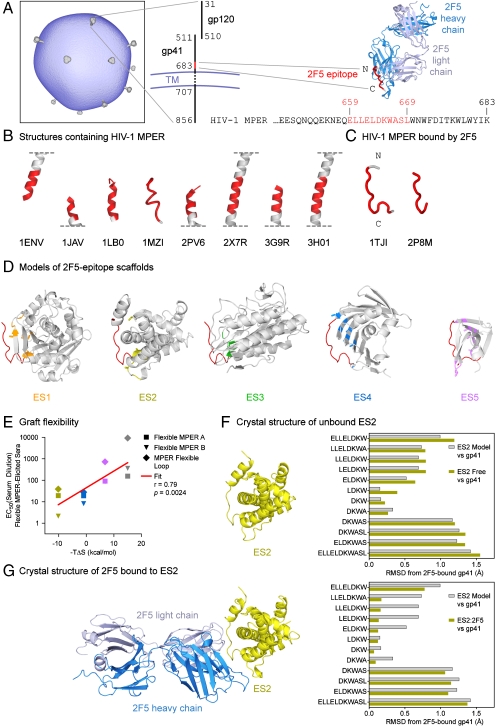
Target epitope and transplantation to select acceptor scaffolds. (A) Epitope for antibody 2F5. The HIV-1 virion (schematic based on ref. 40) employs numerous mechanisms of immune evasion to avoid recognition by neutralizing antibody. A potential site of vulnerability is recognized by antibody 2F5, depicted as an antigen-binding fragment (Fab) with heavy chain in blue and light chain in gray, which binds to residues 659–669 of gp41, depicted in red. The sequence of the HIV-1 MPER is from strain HxB2. (B) Shown are diverse structures of gp41 that have been previously determined (7–11, 14, 41), with residues 659–669 highlighted in red. (C) The 2F5-recognized conformation of the epitope shows essentially the same conformation in two different crystal lattices (12, 13). (D) Computational methods were used to transplant the 2F5 epitope into acceptor scaffolds, which were selected and further modified to present the gp41 epitope (red) in its 2F5-recognized conformation. Epitope scaffolds are drawn in gray as Cα-ribbons, with scaffold residues altered to accept transplantation highlighted in stick representation, and colored orange for ES1, yellow for ES2, green for ES3, blue for ES4, and purple for ES5. (E) Two potential measures of graft flexibility, recognition by sera generated by flexible representations of the epitope (vertical axis) and entropy of 2F5 recognition (horizontal axis), were found to correlate. Data points are colored according to epitope scaffold as in D, with gray for MPER peptide. (F) X-ray crystal structure of unbound ES2. The left image shows a Cα-ribbon of the unbound ES2 structure. The right graph shows rmsds for different residue ranges of the 2F5 epitope in the unbound ES2 structure (yellow) or in the initial ES2-computational model (gray), with both compared to the target 2F5-bound conformation of gp41. (G) Crystal structure of ES2 in complex with antibody 2F5. The left image shows a Cα-ribbon of the ES:2F5 complex, with the 2F5 Fab colored blue and gray and ES2 yellow. Coloring for the right graph is the same as in F.
This procedure resulted in the design of five epitope scaffolds from parent coordinates 1LGYa, 1KU2a, 2MATa, 1IWLa, and 1D3Bb (refs. 18–22), which we named ES1-ES5, respectively (Fig. 1D and Table 1). On average, eight epitope residues were transplanted and eight additional mutations were made to each of these scaffolds (Fig. S1). In two of the scaffolds (ES2 and ES5), the region encompassing the epitope graft was occluded on the native scaffold oligomer, but in both cases we judged that mutations associated with epitope transplantation would interfere with oligomerization and result in stable monomeric proteins with exposed 2F5 epitopes. Model properties of the resultant 2F5-epitope scaffolds showed main-chain rmsds ranging from 0.7 to 1.3 Å (Table 1 and Fig. S1), indicating reasonable replication of the epitope shape. The nonbound face of the epitope graft, meanwhile, showed up to 70% less solvent-accessible surface area than equivalent residues on the 2F5-bound peptide (Table 1), indicating substantial occlusion of the nonbound face.
Table 1.
Computational design and experimental characterization of 2F5-epitope scaffolds
| ES1 | ES2 | ES3 | ES4 | ES5 | MPER WT | MPER cyc | |
| Computational Design | |||||||
| PDB code of parent | 1LGYa | 1KU2a | 2MATa | 1IWLa | 1D3Bb | NA | NA |
| rmsd gp41660-667Cα (Å) | 0.7 | 0.7 | 0.8 | 0.9 | 1.4 | NA | NA |
| rmsd gp41660-667 MC (Å) | 0.7 | 0.8 | 0.8 | 1.0 | 1.3 | NA | NA |
| DDMP rmsd gp41660-667 | 0.2 | 0.4 | 0.4 | 0.5 | 1.1 | NA | NA |
| Nonbound surface (Å2) | 187 | 176 | 108 | 160 | 79 | 241 | 193* |
| Experimental Characterization | |||||||
| Expression | Bacterial | Mammalian | Bacterial | Mammalian | Bacterial | Synthetic | Synthetic |
| Binding to antibody 2F5 Fab | |||||||
| on-rate (1/Ms) (x105) | 6.52 ± 0.04 | 16.68 ± 0.04 | 1.56 ± 0.01 | 7.43 ± 0.02 | 9.70 ± 0.03 | 7.17 ± 0.04 | 8.48 ± 0.05 |
| off-rate (1/s) (x10-3) | 3.50 ± 0.02 | 1.00 ± 0.00 | 0.547 ± 0.004 | 13.90 ± 0.02 | 2.92 ± 0.01 | 4.62 ± 0.03 | 1.64 ± 0.02 |
| KD (nM) | 5.38 ± 0.04 | 0.600 ± 0.004 | 3.50 ± 0.02 | 18.80 ± 0.03 | 3.01 ± 0.01 | 6.44 ± 0.03 | 1.93 ± 0.02 |
| Thermodynamics (37 °C) | |||||||
| ΔG (kcal/mol) | ND | −12.8 ± 2.3 | ND | −11.0 ± 0.2 | −10.8 ± 0.4 | −10.6 ± 0.1 | −11.8 ± 0.2 |
| ΔH (kcal/mol) | ND | −2.7 ± 0.1 | ND | −10.0 ± 0.2 | −17.6 ± 0.6 | −25.6 ± 0.5 | −18.7 ± 0.3 |
| −TΔS (kcal/mol) | ND | −10.1 ± 2.3 | ND | −0.9 ± 0.2 | 6.8 ± 0.7 | 15.0 ± 0.5 | 6.9 ± 0.3 |
| Antigenic recognition by flexible MPER sera (mean EC50) | |||||||
| Flexible MPER A (serum dilution) | 45.6 ± 13.8 | 19.6 ± 16.4 | 56.0 ± 11.4 | 18.9 ± 3.1 | 90.9 ± 72.5 | 155.3 ± 36.6 | ND |
| Flexible MPER B (serum dilution) | 20.0 ± 31.1 | 2.1 ± 0.6 | 19.0 ± 1.9 | 8.2 ± 8.6 | 85.2 ± 33.8 | 357.4 ± 22.3 | ND |
| MPER flexible loop (serum dilution) | 59.2 ± 30.9 | 38.6 ± 0.9 | 38.4 ± 16.1 | 27.9 ± 0.9 | 723.3 ± 611.4 | 9227.0 ± 1668.8 | ND |
ES1-ES5 are 2F5-epitope scaffolds; MPER WT is a peptide composed of residues 656–670 of gp41 linked to a C9 tag; MPER cyc is a cyclized version of MPER WT. NA, not applicable; ND, not determined.
*Nonbound surface of MPER cyc is based on a structural model.
Biochemical, Biophysical, and Antigenic Characterization of 2F5-Epitope Scaffolds.
Epitope scaffolds were first tested for expression in a mammalian system, which succeeded for scaffolds ES2 and ES4. The remaining scaffolds were expressed bacterially, which, following a refolding step, yielded soluble ES1, ES3, and ES5 scaffolds, although these tended to aggregate. To assess potential utility in elicitation, we tested the scaffolds for binding to antibody 2F5 with surface-plasmon resonance, because high affinity for 2F5 was likely a required property of an immunogen capable of eliciting antibodies that induce the 2F5-recognized shape in gp41. When the epitope scaffolds were directly coupled to surface-plasmon resonance chips, experimental affinities to 2F5 Fab ranged from 0.600 ± 0.004 to 18.80 ± 0.03 nM, which were comparable to 2F5 affinities for free and cyclized peptides of 6.44 ± 0.03 and 1.93 ± 0.02 nM, respectively (Fig. S2A and Table 1).
To evaluate the degree of conformational stabilization of the epitope in the scaffolds, we analyzed the thermodynamics of their interaction with 2F5. Although in general the contributions of configurational entropy are difficult to separate from those of solvation in calorimetry experiments (23, 24), as a first approximation in the cases described here, the latter should be similar for peptides and epitope scaffolds because their interfaces should be nearly identical, assuming contacts made outside the epitope are negligible. Thus, in interaction with antibody 2F5, a more favorable binding entropy for the epitope scaffolds relative to the free peptide is likely to indicate conformational fixation of the epitope. We obtained isothermal titration calorimetry measurements for ES2, ES4, and ES5 as well as for both wild-type and cyclized MPER peptides (Fig. S2B and Table 1); we were unable to obtain accurate measurements for ES1 and ES3, however, likely because of problems with aggregation. For the ES2 scaffold, a -TΔS change at 37 °C of -10.1 ± 2.3 kcal/mol was observed, indicating an overall increase in entropy upon binding and suggesting the graft in ES2 to be rigid. For the ES4 and ES5 scaffolds, -TΔS values of -0.9 ± 0.2 and 6.8 ± 0.7 kcal/mol were observed, respectively; the latter value was close to that observed for the cyclized epitope peptide, suggesting the grafts in these scaffolds were more flexible than that in ES2. In contrast, the wild-type epitope peptide had a -TΔS change of 15.0 ± 0.5 kcal/mol, indicating substantial loss of entropy upon binding, consistent with the expected loss of conformational diversity for an unbound- to bound-peptide transition.
To provide an alternative measure of rigidity of the engrafted epitopes, serum responses generated with the 2F5 epitope as a flexible peptide or when grafted into a flexible β-hairpin loop (in a manner inconsistent with the extended 2F5-bound structure) (25) were used to compare antigenic recognition of the 2F5 epitope as a free peptide or when stabilized as a graft in the epitope scaffolds. As shown in Fig. S2C and Table 1, antigenic recognition of the scaffolded epitope was substantially restricted in ELISAs when compared to wild-type free peptide. The grafts in the ES2 and ES4 context showed the highest restriction, followed by ES1 and ES3, while ES5 showed substantially less restriction. The antigenic discrimination observed is consistent with conformational and accessibility constraints imposed on the engrafted epitope by the scaffolds, constraints that allow only a fraction of the population of antibodies elicited by a flexible epitope immunogen to bind. We thus assessed the correlation between this measure of epitope flexibility and that obtained by isothermal titration calorimetry (Fig. 1E). Antigenic recognition by these sera correlated with entropy of 2F5 recognition (r = 0.79, p = 0.0024), suggesting that entropy of 2F5 recognition (-TΔS) and antigenic recognition by sera generated against the epitope in a flexible context measure related properties of the engrafted epitope.
Crystal Structures of 2F5-Epitope Scaffold ES2: Free and Complexed to Antibody 2F5.
To confirm the accuracy of the computational design, the ES2 scaffold, which showed the highest affinity to 2F5, the most entropically favorable interaction with 2F5, and one of the most restrictive recognition profiles by flexible MPER-elicited sera, was chosen as a representative scaffold for structural characterization. Crystals of the free form of ES2 were obtained and diffracted to 2.8 Å, with two molecules of ES2 present in the asymmetric unit (Table S1). The structure of ES2 is shown in Fig. 1F, with a close-up view superimposed with the parent gp41 peptide in Fig. S2D. Least squares superposition of gp41 residues 660-667 in the two asymmetric-unit copies of crystalline ES2 against the target 2F5-bound epitope showed Cα rmsds of 0.7 Å and difference-distance matrix (26) averages of 0.3 Å (Table S2). More comprehensive superpositions were performed for various subranges of the epitope graft, and, as shown in Fig. 1F, the crystal structure of free ES2 displayed a pattern of mimicry to gp41 very similar to that predicted by the computational model.
The crystal structure of the ES2 scaffold was also determined in complex with the 2F5 antibody at ∼2.5 Å resolution (Fig. 1G and Table S1). Binding by antibody 2F5 induced the epitope graft to adopt even more closely the 2F5-bound conformation of gp41. Superposition of the epitope graft showed a Cα rmsd against residues Leu660–Ala667 of the parent gp41 peptide of only 0.2 Å, an almost fourfold decrease from the same superposition in the free ES2 structure (Fig. 1G, Fig. S2D, and Table S2). Extending the alignment subrange by one residue upstream to Glu659, increased the superposition rmsd for Cα atoms to 0.7 Å, while extending it to Leu669 increased the rmsd to 1.3 Å (Table S2). These results indicate that in the ES2 context, the epitope most closely resembled residues Leu660-Ala667 of gp41, and that over these residues, 2F5 induces the graft in the ES2 scaffold to adopt near perfect resemblance to the target conformation of the 2F5-bound gp41 epitope.
Polyclonal Sera Elicited by Epitope Scaffolds Mimic 2F5 Binding to the gp41 MPER.
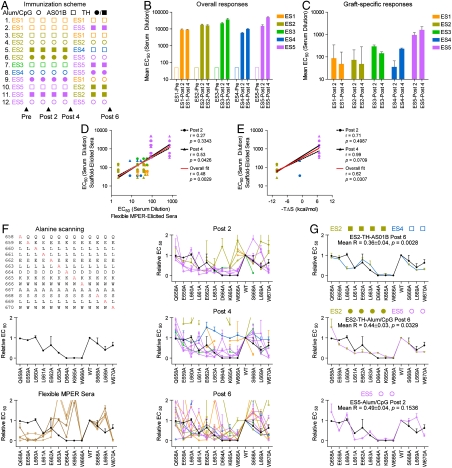
We next investigated epitope-scaffold immunogenicity. Guinea pigs were immunized with the 2F5-epitope scaffolds, either using a single scaffold (homologous immunizations) or in combination (heterologous immunizations) (Fig. 2A). ELISAs were performed on prebleeds as well as after the second, fourth, and sixth immunizations. In all cases, robust immune responses were seen against the entire inoculated scaffold (Fig. 2B and Fig. S3A). To assess the degree to which these robust responses were directed against the engrafted gp41 epitope, homologous post-2 and post-4 immunization-elicited sera were tested for binding to the gp41-peptide epitope by ELISA. As shown in Fig. 2C and Fig. S3B, the responses against the grafts depended on the inoculated scaffold, with ES5-immunized animals showing the highest graft-specific responses, ES3- and ES4-immunized animals displaying intermediate responses, and ES1- and ES2-immunized animals displaying lower graft-specific responses. Graft-specific responses did not correlate with the presence of a T-helper epitope (PADRE) (27), the use of Alum/CpG versus AS01B adjuvant, the degree of graft similarity to the 2F5-bound conformation, nor the degree of occlusion of the nonbound surface. The magnitude of the graft-specific response did, however, correlate with the rigidity of the scaffold graft (p < 0.04), as assessed either by recognition with sera elicited by flexible versions of the MPER (Fig. 2D) or by entropy of 2F5 recognition (Fig. 2E). These results suggest that epitope flexibility enhances immunogenicity, and conversely, that graft rigidity reduces immunogenicity.
Fig. 2.
Immununogencity of 2F5-epitope scaffolds. (A) Immunization scheme. A priori, it was unclear what factors would influence immunogenicity. We therefore utilized a highly redundant sparse matrix with three primary variables: type of epitope scaffold (ES1, orange; ES2, yellow; ES3, green; ES4, blue; ES5, purple); type of adjuvant (Alum/CpG, circles; ASO1B, squares); and the presence (closed symbols) or absence (open symbols) of linked T-help (“TH”; PADRE) (27). Twelve different immunization schemes were evaluated in guinea pigs, four animals per group, with sera sampled prior to immunizations (Pre) and after two (Post 2), four (Post 4), and six (Post 6) immunization cycles. (B) Overall titers. ELISA EC50 values of polyclonal antibody responses against the entire scaffolds are shown, as assessed by binding of Pre, Post 2, and Post 4 serum time points to the whole scaffold. (C) Graft-specific titers. ELISA EC50 values of polyclonal antibody responses against the 2F5-epitope portion of the epitope scaffolds are shown, as evaluated by binding of Pre, Post 2, and Post 4 serum time points to a 2F5-epitope peptide (individual responses are shown in Fig. S3). (D) Graft-specific titers (Post 2, circles and dashed black line; Post 4, triangles and solid black line) elicited by the epitope scaffolds (vertical axis) are compared with recognition of the epitope scaffolds by sera generated by the epitope when immunized in a flexible context (e.g., as free peptide or placed into a flexible loop) (horizontal axis). The overall fit is shown as a red line. (E) Graft-specific titers (Post 2, circles and dashed black line; Post 4, triangles and solid black line) elicited by the epitope scaffolds (vertical axis) are compared with the entropy of 2F5 recognition. Lower observed entropies are expected to result from interactions of 2F5 with more rigid grafts. The overall fit is shown as a red line. (F) Residue-by-residue interrogation of the elicited responses. Single alanine mutants were introduced into a collection of 2F5-epitope peptides spanning residues 658–670 (top left). The effects of these alanine mutants on antibody 2F5 binding were evaluated by ELISA, with changes to the central Asp-Lys-Trp tripeptide ablating binding and other residues displaying more muted responses (middle left, black line). The alanine mutants were also used to interrogate sera elicited by flexible and cyclized peptides (bottom left, dark brown and light brown, respectively), as well as against all Post 2, Post 4, and Post 6 sera (right panels; lines and symbols are colored based on scaffold coloring depicted in A; for Post 6, symbols are colored based on scaffolds used in the final two immunizations). The 2F5 alanine scan profile (black) is shown in all panels for comparison. (G) Optimal responses. Responses to the alanine-mutant epitope peptides were ranked by R-value of the response, as defined by the expression 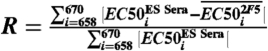 , where i is the residue position at which the MPER was mutated to alanine. Alum/CpG, linked T help, increasing number of immunizations, heterologous immunizations, and use of ES5 all biased toward reduced R-values (Fig. S3C and Table S5). Shown here are results from alanine-scanning for the top three responses along with corresponding R-values and p-values of the immunization schemes (p-values were obtained as described in SI Materials and Methods; because 58 different sera or grouped sera were analyzed, Bonferroni adjustments were calculated to account for multiple comparisons, with individual p-values from each serum comparison to 2F5 multiplied by a factor of 58). The 2F5 alanine scan profile (black) is shown in all panels for comparison.
, where i is the residue position at which the MPER was mutated to alanine. Alum/CpG, linked T help, increasing number of immunizations, heterologous immunizations, and use of ES5 all biased toward reduced R-values (Fig. S3C and Table S5). Shown here are results from alanine-scanning for the top three responses along with corresponding R-values and p-values of the immunization schemes (p-values were obtained as described in SI Materials and Methods; because 58 different sera or grouped sera were analyzed, Bonferroni adjustments were calculated to account for multiple comparisons, with individual p-values from each serum comparison to 2F5 multiplied by a factor of 58). The 2F5 alanine scan profile (black) is shown in all panels for comparison.
To provide insight into graft-specific immune recognition, elicited responses were interrogated with alanine mutants spanning residues 658–670 of gp41 (Fig. 2F). The 2F5 antibody interacted with alanine mutants in a manner inversely proportional to the contact surface area normally observed with each unaltered residue, with alanine mutants to the central Asp-Lys-Trp tripeptide ablating binding. Sera elicited by both free and cyclized peptide showed alanine-interrogated profiles that were significantly different from 2F5, especially for Asp and Lys in the central tripeptide. Sera elicited by epitope scaffolds, meanwhile, showed a variety of responses (Fig. 2F). When elicited responses were sorted by similarity to 2F5 based on a reliability factor (R value) which quantifies ELISA responses (Fig. 2G and SI Materials and Methods), top responses were derived either from heterologous immunizations or from homologous immunizations with ES5, and these were remarkably similar to antibody 2F5.
Monoclonal Antibodies Elicited by Epitope Scaffolds Replicate 2F5 Structure-Specific Recognition of the gp41 MPER.
Having thus shown that with select epitope scaffolds it was possible to elicit polyclonal responses that closely mimicked 2F5 in terms of binding across the gp41 MPER, we next sought to determine the biophysical and structural characteristics of specific monoclonal antibodies present in the mimicked responses. Two groups of five mice were immunized with the 2F5-epitope scaffolds: either with ES5 (five times) or with an ES5 prime (two times) followed by an ES1 boost (three times). Mice that had high titers to the ES2 scaffold and the gp41-peptide epitope were chosen for fusion, and B cell-hybridoma clones were selected on the basis of binding to heterologous scaffolds and free peptide. A total of six monoclonal antibodies were isolated in this manner, three from each mouse group. Surface–plasmon resonance analysis (Fig. S4A) revealed that antibodies from the ES5-ES1 prime-boost group had the highest affinity for peptide and scaffolds, and the two tightest binders from this group, 11f10 and 6a7, which were determined to be isogenic variants (Fig. S4D), were selected for structural characterization. The antigen-binding fragments of each these antibodies was produced and crystallized in complex with a peptide corresponding to gp41 residues 660–667, yielding crystals that diffracted to 2 Å resolution (Fig. 3 and Table S1).
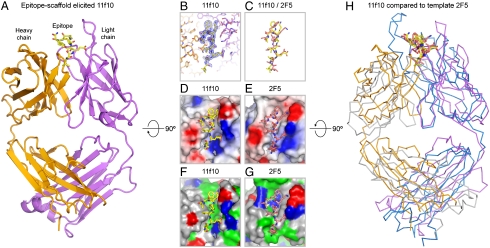
Fig. 3.
Structure of an epitope-scaffold-elicited antibody. The epitope scaffolds developed here are mimics of the target template, chosen in this case to be the 2F5-recognized structure of gp41. Immunization teaches the immune system to make antibody “molds” capable of recognizing or inducing the desired conformation in the target template (even if the template is a flexible peptide, which would otherwise rarely assume the desired conformation). (A) Structure of ES5-ES1-elicited antibody, 11f10 (heavy chain, orange; light chain, purple), in complex with a peptide corresponding to residues 660–667 of gp41 (yellow). (B) Close-up of bound gp41 peptide in the 11f10 complex. Experimental electron density (2Fo-Fc, at 1σ contour) is shown in blue around the bound gp41 peptide (yellow). (C) Comparison of gp41-peptide conformations when bound by antibody 11f10 (yellow) or by template antibody 2F5 (salmon). (D) Electrostatic potential of ES5-ES1-elicited antibody 11f10 displayed at its molecular surface, with electronegative regions in red, electropositive regions in blue, and apolar regions in white. (E) Electrostatic potential of antibody 2F5 displayed at its molecular surface and colored as in D. (F) Antigen-combining surface of antibody 11f10, colored according to residue type (hydrophobic, green; polar, gray; positive, blue; negative, red). (G) Antigen-combining surface of template antibody 2F5, colored as in F. (H) Overlap of elicited and template antibodies (with light and heavy chains colored orange and purple for 11f10, respectively, and colored gray and blue for 2F5, repectively), aligned by superposition of all atoms of the bound gp41-peptides (residues 660–667). Bound gp41 peptides in D–H are colored as in C.
The electron density for the peptide was clearly defined (Fig. 3B), with the exception of Leu660 at the peptide N terminus. Superposition of the 11f10- and 6a7-bound gp41 peptide against 2F5-bound gp41 revealed that the epitope-scaffold-elicited antibodies induced a peptide conformation remarkably similar to that induced by 2F5 (Fig. 3C and Fig. S4B), with rmsds for backbone atoms of 1.1 and 1.0 Å for 11f10 and 6a7, respectively. Side-chain orientations of gp41 in the 11f10 and 6a7 bound structures were also similar, with the same chi-1 and chi-2 side-chain rotamers observed in roughly half of the peptide residues. When electrostatic potentials were mapped onto antibody surfaces, a high degree of similarity was also observed between antibodies 11f10 and 6a7 and antibody 2F5 (Fig. 3 D and E). Although the similarities in the electrostatic surfaces were striking, they did not necessarily correspond to similar residue contacts between the antibodies and gp41 (Fig. 3 F and G and Tables S3 and S4). For instance, Trp 96 of the light chain of 11f10 (and 6a7) packed perpendicular to Trp 666 of gp41, with its Nε atom situated for hydrogen bonding to the π-electrons of the indole ring of Trp 666 of gp41, while in the case of 2F5, interactions with gp41 Trp 666 are mediated by non-Trp residues. Another notable difference between the antibodies is that the tip of the 2F5 CDR H3 loop—likely involved in membrane interactions (15)—had no corresponding partner in 11f10 or 6a7. Nonetheless, similarities were observed between the angles of approach and the spatial orientations of the 11f10 and 6a7 antibodies relative to gp41 as compared to those of 2F5 (Fig. 3H and Fig. S4 B and C). Superposition of the gp41 peptide in the epitope-scaffold-elicited antibodies against the gp41 peptide in the template-2F5 antibody overlays approximately 80% the variable regions of these antibodies, but with a mode of binding that effectively switches the heavy and light chain positions of the elicited and 2F5 antibodies.
Discussion
Structural specificity is a defining characteristic of mature antibody recognition. While antibodies may use other specialized mechanisms of recognition, such as posttranslational mimicry (antibody 412d, ref. 28) or co-membrane binding (antibody 2F5 investigated here, refs. 13, 15, 16), they all bind with high affinity to a specific protein-epitope structure. Here we utilize epitope scaffolds to teach structure-specific recognition of a target epitope. Antibodies elicited with the epitope scaffolds bound to the 2F5 epitope with high affinity, induced a conformation similar to that induced by 2F5, and showed similar angles of epitope approach (Fig. 3 C and H and Fig. S4). Nonetheless, we do not expect these antibodies to recreate fully the antibody properties of 2F5, because the membrane-binding component of 2F5 recognition was not addressed in the design procedure. This component is responsible for a substantial portion of the free energy of 2F5 recognition, without which neutralization is difficult to attain (15, 16, 29, 30).
A number of other groups have also studied the expression of the nominal 2F5-epitope sequence (ELDKWAS) in scaffold systems: a variable loop of the HIV-1 gp120 envelope glycoprotein; a surface loop of human rhinovirus; and a surface loop of bovine papilloma virus (25, 31–33). All of these studies resulted in the elicitation of antibodies targeted to the ELDKWAS epitope, and two of these studies reported the induction of weak neutralizing antibodies. However, none of the studies demonstrated similarity between the elicited polyclonal response and 2F5 binding to gp41, nor the elicitation of monoclonal antibodies able to induce the 2F5-bound conformation in gp41, both of which we demonstrate with the 2F5-epitope scaffolds to provide proof of concept for the “reelicitation” of structure-specific antibodies.
The modular nature of the epitope scaffolds—with the same epitope placed into different acceptor–scaffold backgrounds—provides a means to examine factors that influence elicitation. We observed a range of flexibilities for the engrafted epitope, with flexibility inferred from two different measures: antigenic recognition by sera generated with the epitope in a highly flexible context and thermodynamic measurements of the entropy of 2F5 recognition. While equating binding entropies with conformational entropies is generally not permissible (23, 24), the similarity of the recognition by 2F5 appears to permit such analysis in this particular case. The results with respect to immunogenicity were striking: Notably, the flexibility of the engrafted epitope, as judged by thermodynamics and antigenic recognition, correlated with immunogenicity, with a flexible but otherwise equivalent epitope generating significantly higher immune responses than a rigid one. This finding provides an explanation for prior observations that elicited responses against viral antigens are often focused on flexible loops—as has been classically observed with influenza virus (34) and more recently with adenovirus chimeras (35). One possible mechanism for this finding may relate to the ability of flexible epitopes to utilize more fully mechanisms of induced fit, thereby engaging nascent immunoglobulins on a larger percentage of B cells and initiating their maturation (as has been proposed for the inverse problem of antibody recognition) (36). Perhaps relevant to this, the finding that heterologous immunizations more often elicit graft-specific responses that closely resemble those of the target antibody suggests that successive populations of B cells can be appropriately expanded and affinity matured by successive epitope-scaffold boosts, as we have done with ES5 and ES1 to elicit antibodies highly similar to the 2F5 antibody. Lastly, the epitope-scaffold-elicited antibodies themselves provide insights into the diversity of molecular recognition enabled by the adaptive immune response. Antibodies 11f10 and 2F5 show related recognition chemistries—with similarities in electrostatics (Fig. 3 D and E) and comparable combining site-amino acid chemistries (Fig. 3 F and G)—even though their orientations of heavy and light chains are dramatically different (Fig. 3H) as are their progenitor immunoglobulin genes (Fig. S4D). Thus, in addition to the potential utility of epitope scaffolds in vaccine modalities, the antibodies they elicit provide a means to delineate the diversity of molecular recognition by the adaptive immune response as it fulfills requirements of structure-specific elicitation.
Materials and Methods
A summary of experimental techniques is given here, with full methods and associated references presented in SI Materials and Methods.
Creation of 2F5-Epitope Scaffolds.
The central concept was to employ, as acceptor scaffolds, proteins with preexisting structural similarity to the antibody-bound conformation of the target epitope. The gp41-2F5 structure revealed significant peptide-antibody contacts over the range of gp41 residues from E659 to L669 (refs. 12, 13), so we designed epitope scaffolds to mimic this segment of gp41. Briefly, the program PISCES (37) was used to cull the Protein Data Bank to crystal structures of resolution better than 3.0 Å and of protein chains with more than 50 residues. MAMMOTH (38) was used to search for sequence-independent structural matches based on Cα coordinates, and ROSETTA (39) was used for clash-checking and protein design.
Structural matches to different peptide segments spanning gp41 residues 659–669 were ranked by the ratio of rmsd to the number of superimposed residues (nsup). The best 5% of matches (700 matches with rmsd/nsup < 0.132) were then evaluated for steric clash between the scaffold backbone (side chains removed) and the antibody (all atoms), with the relative orientation of scaffold and antibody determined by structural superposition of the gp41-epitope segment onto the scaffold. The 2F5 scaffolds with the least clashes and which did not require cofactors were retained for further analysis. Many of these scaffolds were oligomeric in their native state; if the oligomeric interface was judged to be stable after epitope transplantation, the clash analysis was repeated with the native oligomers. In the final design stage, epitope residues were transplanted to scaffold positions according to the MAMMOTH structural alignment, and scaffold positions adjacent to epitope or antibody were redesigned to accommodate epitope side chains and to avoid interactions with antibody. Lastly, epitope scaffolds ES1-ES5 were ranked according to rmsd for the core epitope (residues 660–667), because end effects involving residues 659, 668, and 669 often led to changes in overall rmsd when compared to gp41 bound by 2F5 in different crystal forms (see for example, Fig. S1 and Table S2).
Expression, Purification, and Characterization of 2F5-Epitope Scaffolds.
Epitope scaffolds were expressed in a 293F Freestyle transient transfection system (Invitrogen) or bacterially, from inclusion bodies, and purified by chelating and 2F5-affinity chromatography. Biophysical characterization of the 2F5-epitope scaffolds was carried out with surface–plasmon resonance and isothermal titration calorimetry to determine affinities and to characterize the thermodynamics of their interaction with antibody 2F5, respectively. X-ray crystallography was used to verify atomic-level mimicry of the epitope, and ELISA measurements were used to evaluate the interaction of the scaffolds with sera, with EC50s calculated from the entire dilution curve.
Immunogenicity of 2F5-Epitope Scaffolds.
Guinea pigs were immunized in two week intervals with 20 ug of epitope scaffolds as adjuvanted with either Alum/CpG or with AS01B (GlaxoSmithKline). Antibody titers were evaluated by ELISA against either the entire immunized protein or against the engrafted epitope.
Generation and Structural Characterization of Monoclonal Antibodies Elicited with 2F5-Epitope Scaffolds.
Mice were immunized in two week intervals with 20 ug of epitope scaffold in Alum/CpG. Those with the highest titers of graft-specific responses, as assessed by ELISA against peptide or heterologous epitope scaffold, were chosen for production of monoclonal antibody with standard fusion and selection techniques (ProSci). Monoclonal antibodies obtained were characterized by surface–plasmon resonance for affinity to gp41-peptide epitope and heterologous scaffolds, and those with highest affinity were sequenced, crystallized, and analyzed by X-ray crystallography as antigen-binding fragments in complex with the 2F5-epitope peptide (residues 660–667).
Supplementary Material
Acknowledgments.
We thank B.K. Chakrabarti and G.J. Nabel for sera generated from placing the 2F5 epitope into the flexible V3 loop of gp120; Y. Geng and V. Savich (ProSci) for assistance for monoclonal antibody elicitation; H. Katinger for antibody 2F5; J.R. Mascola and K. McKee for neutralization analysis; G.J. Nabel, J.R. Mascola, L. Shapiro, I. Wilson, and members of the Structural Bioinformatics and Biology Sections, Vaccine Research Center, for discussions and comments on the manuscript; J. Stuckey for assistance with figures; and GlaxoSmithKline for AS01B adjuvant. Support for this work was provided by the Intramural Research Program of the National Institutes of Health and by the International AIDS Vaccine Initiative and its Neutralizing Antibody Consortium. Use of SER-CAT at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science.
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (accession codes 3LES and 3LEV for free and 2F5-bound structures of ES2 scaffold, respectively, and accession codes 3LEX and 3LEY for elicited antibodies 11f10 and 6a7, respectively, in complex with a gp41 peptide corresponding to the 2F5 epitope).
See Commentary on page 17859.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004728107/-/DCSupplemental.
References
- 1.Glennie MJ, Johnson PW. Clinical trials of antibody therapy. Immunol Today. 2000;21(8):403–410. doi: 10.1016/s0167-5699(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez-Carneado J, et al. Surface grafting onto template-assembled synthetic protein scaffolds in molecular recognition. Biopolymers. 2000;55(6):451–458. doi: 10.1002/1097-0282(2000)55:6<451::AID-BIP1020>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Huang CC, et al. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13(5):755–768. doi: 10.1016/j.str.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Vita C, Roumestand C, Toma F, Menez A. Scorpion toxins as natural scaffolds for protein engineering. Proc Natl Acad Sci USA. 1995;92(14):6404–6408. doi: 10.1073/pnas.92.14.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muster T, et al. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68(6):4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley JM, et al. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78(23):13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Deng Y, Dey AK, Moore JP, Lu M. Structure of the HIV-1 gp41 membrane-proximal ectodomain region in a putative prefusion conformation. Biochemistry. 2009;48(13):2915–2923. doi: 10.1021/bi802303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biron Z, et al. A monomeric 3(10)-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry. 2002;41(42):12687–12696. doi: 10.1021/bi026261y. [DOI] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 10.Sun ZY, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28(1):52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40(32):9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 12.Julien JP, Bryson S, Nieva JL, Pai EF. Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site. J Mol Biol. 2008;384(2):377–392. doi: 10.1016/j.jmb.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 13.Ofek G, et al. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78(19):10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbato G, et al. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J Mol Biol. 2003;330(5):1101–1115. doi: 10.1016/s0022-2836(03)00611-9. [DOI] [PubMed] [Google Scholar]
- 15.Ofek G, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106(48):20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song L, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci USA. 2009;106(22):9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohno M, et al. The crystal structure of lipase II from Rhizopus niveus at 2.2 A resolution. J Biochem. 1996;120(3):505–510. doi: 10.1093/oxfordjournals.jbchem.a021442. [DOI] [PubMed] [Google Scholar]
- 19.Campbell EA, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. 2002;9(3):527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 20.Lowther WT, et al. Escherichia coli methionine aminopeptidase: Implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis. Biochemistry. 1999;38(24):7678–7688. doi: 10.1021/bi990684r. [DOI] [PubMed] [Google Scholar]
- 21.Takeda K, et al. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 2003;22(13):3199–3209. doi: 10.1093/emboj/cdg324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kambach C, et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96(3):375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 23.Thomson J, Ratnaparkhi GS, Varadarajan R, Sturtevant JM, Richards FM. Thermodynamic and structural consequences of changing a sulfur atom to a methylene group in the M13Nle mutation in ribonuclease-S. Biochemistry. 1994;33(28):8587–8593. doi: 10.1021/bi00194a025. [DOI] [PubMed] [Google Scholar]
- 24.Varadarajan R, Connelly PR, Sturtevant JM, Richards FM. Heat capacity changes for protein-peptide interactions in the ribonuclease S system. Biochemistry. 1992;31(5):1421–1426. doi: 10.1021/bi00120a019. [DOI] [PubMed] [Google Scholar]
- 25.Chakrabarti BK, et al. Expanded breadth of virus neutralization after immunization with a multiclade envelope HIV vaccine candidate. Vaccine. 2005;23(26):3434–3445. doi: 10.1016/j.vaccine.2005.01.099. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa K, Ooi T, Saito N, Isogai Y. Tertiary structure of proteins. 1. Representation and computation of conformations. J Phys Soc Jpn. 1972;32(5):1331–1337. [Google Scholar]
- 27.Alexander J, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1(9):751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 28.Huang CC, et al. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science. 2007;317(5846):1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Julien JP, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84(9):4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwick MB, et al. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J Virol. 2004;78(6):3155–3161. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold GF, et al. Broad neutralization of human immunodeficiency virus type 1 (HIV-1) elicited from human rhinoviruses that display the HIV-1 gp41 ELDKWA epitope. J Virol. 2009;83(10):5087–5100. doi: 10.1128/JVI.00184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X, et al. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine. 1999;17(22):2862–2872. doi: 10.1016/s0264-410x(99)00125-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Huang Y, Fayad R, Spear GT, Qiao L. Induction of mucosal and systemic neutralizing antibodies against human immunodeficiency virus type 1 (HIV-1) by oral immunization with bovine Papillomavirus-HIV-1 gp41 chimeric virus-like particles. J Virol. 2004;78(15):8342–8348. doi: 10.1128/JVI.78.15.8342-8348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289(5796):373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 35.Roberts DM, et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441(7090):239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 36.Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13(5):611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang G, Dunbrack RL., Jr PISCES: A protein sequence culling server. Bioinformatics. 2003;19(12):1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 38.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): An automated method for model comparison. Protein Sci. 2002;11(11):2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 40.Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441(7095):847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 41.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6(5) doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.