Abstract
Phagocytosis of apoptotic cells requires recognition of cell corpses followed by internalization and enclosure within plasma membrane-derived phagosomes. Phagosomes undergo maturation to generate phagolysosomes in which cell corpses are degraded; however, regulation of the maturation process is poorly understood. Here, we identified Rab GTPase 14, which regulates apoptotic cell degradation in Caenorhabditis elegans. rab-14 mutants accumulate many persistent cell corpses owing to defective cell corpse clearance. Loss of rab-14 function affects several steps of phagosome maturation including phagosomal acidification and phagolysosome formation. RAB-14 and UNC-108/RAB2 are recruited to phagosomes at a similar stage and function redundantly to regulate phagosome maturation. Three Rabs, RAB-14, UNC-108/RAB2, and RAB-7, act in sequential steps to control phagolysosome formation. RAB-14 and UNC-108 recruit lysosomes, whereas RAB-7 mediates fusion of lysosomes to phagosomes. Our data reveal the sequential action of Rab GTPases in regulating tethering, docking, and fusion of lysosomes to apoptotic cell-containing phagosomes.
During apoptosis, apoptotic cells are recognized and swiftly engulfed by phagocytes. The internalized cell corpses are enclosed within intracellular vesicles called phagosomes, which undergo a series of maturation steps, leading to the degradation of apoptotic cells in phagolysosomes (1). Recent studies in Caenorhabditis elegans have identified key factors involved in this maturation process, including Rab GTPases, PI3-kinases, and components of the HOPS complex. These factors also regulate vesicular transport and maturation of phagosomes containing opsonized particles or microorganisms, indicating that these processes likely follow a general path that involves similar cellular events and regulatory components (1–3).
RAB5, which governs various early events of endocytic transport, is important for early phagosome maturation (4, 5). In C. elegans, RAB-5 associates with apoptotic cell-containing phagosomes at an early stage and promotes PtdIns(3)P generation through the PI3-kinase VPS-34 (4). Like other Rab GTPases, RAB5 activity is regulated by guanine nucleotide exchange factors (GEFs), which activate Rabs by promoting exchange of GDP for GTP, and GTPase-activating proteins (GAPs), which inactivate Rabs by accelerating GTP hydrolysis (6). GAPEX5, a VPS9 domain-containing RAB5 GEF, is recruited to phagosomes by a microtubule-dependent mechanism and regulates RAB5 activation on apoptotic cell-containing phagosomes in Swiss 3T3 cells (5). In C. elegans, the GTPase-activating protein TBC-2 inactivates RAB-5 to release it from phagosomal membranes, thereby promoting the progression of phagosome maturation through the RAB-5–positive stage (7).
Similar to its role in endocytic trafficking, RAB-7 acts downstream of RAB-5 to control late steps of phagosome maturation (4, 8). The transition of apoptotic cell-containing phagosomes from the RAB-5– to the RAB-7–positive stage was recently shown to be regulated by SAND-1/MON1 and CCZ-1/CCZ1 (9).
In mammalian cells, RAB7 is thought to be involved in the fusion of late endocytic structures or phagosomes with lysosomes (10, 11). However, the function of RAB7 in these processes is poorly understood. RILP (RAB7-interacting lysosomal protein), the only known RAB7 effector, interacts with active RAB7 on membranes of latex bead-containing phagosomes and mediates the tubular growth from phagosomes to late endocytic compartments/lysosomes (11). It is not known whether RILP is involved in maturation of apoptotic cell-containing phagosomes. In C. elegans, HOPS complex components are thought to act downstream of RAB-7 during phagosome maturation, but it is unclear whether any of them act directly as RAB-7 effectors (4, 12). Therefore, the processes through which phagolysosomes are formed remain largely unknown. In addition to RAB-5 and RAB-7, UNC-108/RAB2 is also required for degrading cell corpses (13, 14). Loss of unc-108 function causes similar defects in phagolysosome formation as loss of rab-7 (14), but the role of RAB-7 or UNC-108 in this process remains elusive. Mammals have >60 Rabs and C. elegans has >30 Rabs (15) (www.wormbase.org), but it is not clear how many of these are needed for clearing apoptotic cells in addition to the above three, or how they are coordinated.
Here, we identified Rab GTPase 14 as a component of the apoptotic cell degradation pathway. RAB-14 functions downstream of RAB-5 and in parallel with UNC-108/RAB2 to regulate phagosomal acidification and phagolysosome formation. Three Rabs, RAB-14, UNC-108, and RAB-7, act in sequential steps to regulate phagolysosome formation: RAB-14 and UNC-108 recruit lysosomes, whereas RAB-7 mediates fusion of lysosomes to phagosomes. Our data reveal the sequential involvement of Rab GTPases in regulating phagolysosome formation.
Results
rab-14 Is Required for Cell Corpse Clearance.
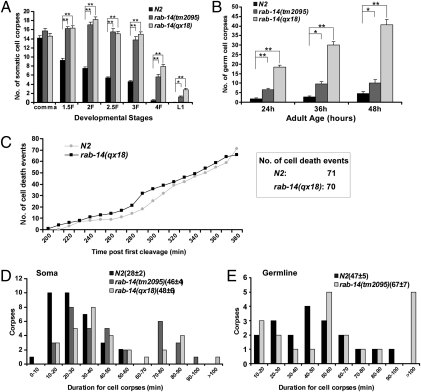
In a forward genetic screen for regulators of cell corpse clearance, we isolated a recessive mutation, qx18, which caused accumulation of apoptotic cells in late embryos (SI Materials and Methods). Compared with wild-type, qx18 mutants contained significantly more cell corpses at most embryonic stages, in L1 larvae and in the adult germline (Fig. 1 A and B). We next examined cell death events occurring between 200 and 380 min after first embryonic cleavage. There were 71 and 70 cell deaths in wild-type and qx18 embryos, respectively, indicating that no excessive cell deaths occurred in qx18 mutants (Fig. 1C). In addition, cell corpse duration was measured by time-lapse recording, and the mean duration of cell corpses in qx18 mutants is 48 min, which is 71% longer than that in wild type (mean 28 min) (Fig. 1D). Germ cell corpses also persisted much longer in qx18 mutants than in wild type. Collectively, these data indicate that cell corpse removal is defective in qx18 mutants.
Fig. 1.
rab-14 mutants are defective in cell corpse clearance. (A and B) Time-course analysis of cell corpse appearance during embryonic (A) and germ-line (B) development in wild-type (N2), rab-14(qx18), and rab-14(tm2095) mutants. The number of cell corpses was scored in the head region of embryos at each developmental stage or in one gonad arm at indicated adult ages. The y axis shows the mean number of cell corpses. At least 15 animals were scored for each strain, and data are shown as mean ± SEM. Data derived from different genetic backgrounds at multiple developmental stages were compared by two-way analysis of variance. Post hoc comparisons were carried out by using Fisher's PLSD (protected least squares difference) test. *P < 0.05; **P < 0.0001. All other points had P > 0.05. (C) The qx18 mutation does not affect cell death occurrence. Embryonic cell deaths occurring 200–380 min after first cleavage were followed in wild type (N2) and rab-14(qx18) mutants. The y axis indicates the number of cell deaths observed at each time point. The box shows the total number of cell deaths observed. (D and E) Four-dimensional microscopy analysis of cell corpse duration in soma (D) and germ line (E) was performed in wild-type (N2), rab-14(qx18), or rab-14(tm2095) mutants. Thirty-three somatic cell corpses or 20 germ cell corpses were monitored in each strain. The numbers in parentheses indicate the mean duration of cell corpses (±SEM). The y axis shows the number of cell corpses within each duration range as shown on the x axis.
We cloned the gene affected in qx18 mutants and found that it corresponds to rab-14, which encodes a Rab small GTPase sharing significant sequence homology with Rab GTPase 14 in fly, mouse, and human (Fig. S1 and SI Materials and Methods). Sequencing of rab-14 in qx18 mutants revealed a C to T transition, which resulted in substitution of the Threonine at codon 67 with Methionine (ACG > ATG; T67M). This mutation affects the phosphate/Mg2+ binding domain PM3, which is conserved in all members of the Ras GTPase superfamily (Fig. S1) (16). In addition, tm2095, a deletion mutation of rab-14, causes accumulation of both somatic and germ cell corpses at various developmental stages (Fig. 1 and Fig. S1A). Expression of full-length rab-14 (genomic DNA or cDNA) under control of the rab-14 promoter (Prab-14rab-14), or mouse RAB14 cDNA driven by C. elegans heat-shock promoters, fully rescued the persistent cell corpse phenotype of rab-14 mutants, indicating that mouse RAB14 can substitute for C. elegans RAB-14 in removing apoptotic cells (Fig. S1A). RAB-14 protein expression was detected in wild-type and qx18 but not tm2095 by using anti-RAB-14 antibodies, indicating that tm2095 is likely a null mutation of rab-14 (Fig. S2A and SI Materials and Methods). However, far fewer germ cell corpses were observed in tm2095 animals than in qx18 mutants (Fig. 1B). To determine whether the elevated accumulation of germ cell corpses in qx18 mutants is indeed caused by loss of rab-14 gene function, we performed RNAi experiments to inhibit rab-14 expression in wild type, rab-14(qx18), and rab-14(tm2095) mutants. rab-14 RNAi treatment resulted in similar numbers of persistent germ cell corpses in both wild-type and tm2095 mutants, suggesting that tm2095 is a null mutation of rab-14 (Fig. S2C). In qx18 mutants, rab-14 RNAi treatment significantly reduced the number of germ cell corpses to a level comparable with tm2095, indicating that the higher number of persistent germ cell corpses in qx18 animals is likely caused by a dominant-negative effect of RAB-14(T67M) (Fig. S2C). In agreement with this, overexpression of RAB-14(T67M), but not wild-type, GDP-locked, or GTP-locked RAB-14 driven by C. elegans heat-shock promoters led to accumulation of cell corpses (Fig. S2B). qx18 and tm2095 embryos at various developmental stages contained similar numbers of somatic cell corpses, which were not obviously enhanced or reduced by rab-14 RNAi treatment (Fig. 1A and Fig. S2D). We used the tm2095 allele in all subsequent experiments as it likely represents a null rab-14 mutation.
RAB-14 Functions in Engulfing Cells to Remove Cell Corpses.
To determine the RAB-14 expression pattern, we generated a GFP RAB-14 fusion protein driven by the rab-14 promoter (Prab-14gfp::rab-14), which fully rescued the persistent cell corpse phenotype of rab-14(tm2095) mutants (Fig. S1A). GFP::RAB-14 was ubiquitously expressed and displayed a punctate staining pattern starting around the 100-cell stage and continuing throughout larval and adult stages (Fig. S3). Strong GFP::RAB-14 expression was found in several engulfing cell types such as pharyngeal muscle cells, hypodermal cells, intestine cells, and gonadal sheath cells (Fig. S3A). Expression of RAB-14 specifically in engulfing cells driven by the ced-1 promoter (Pced-1rab-14) (17), but not dying cells controlled by the egl-1 promoter (Pegl-1rab-14) (18), rescued the cell corpse phenotype of rab-14(tm2095) mutants, indicating that rab-14 functions in engulfing cells to remove cell corpses (Fig. S1A). GFP::RAB-14 was enriched on apoptotic cells, forming a ring-like structure around cell corpses in embryos and gonadal sheath cells, which suggested that RAB-14 may associate with phagosomes (Fig. S3). GFP::RAB-14(Q70L), which locks RAB-14 in a GTP-bound state, also had a punctate staining pattern and clustered around cell corpses (Fig. S3B) (19). Conversely, GFP::RAB-14(S25N), a GDP-bound RAB-14, completely lost the vesicular staining pattern and failed to surround apoptotic cells, indicating that RAB-14 needs to be activated for its targeting to intracellular vesicles and phagosomes (Fig. S3B) (19). Similarly, GFP::RAB-14 carrying the qx18 T67M mutation failed to associate with vesicular structures or target to phagosomal membranes but was diffuse within the cytoplasm (Fig. S3B). Given that the cell corpse phenotype of rab-14(lf) mutants was rescued by expression of wild-type or Q70L RAB-14, but not S25N or T67M RAB-14 (Fig. S1A), these results suggest that RAB-14 must associate with phagosomes to function in cell corpse clearance.
RAB-14 Acts Downstream of RAB-5 Activation to Regulate Phagosome Maturation.
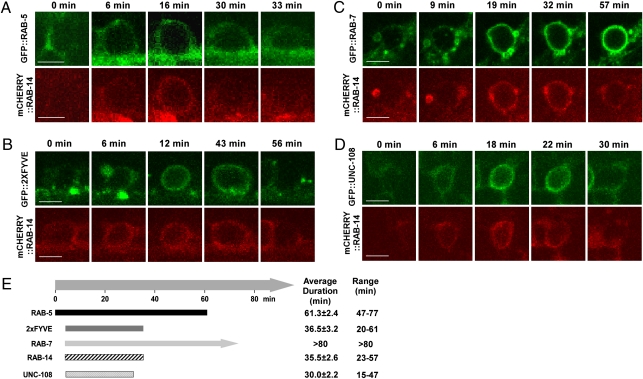
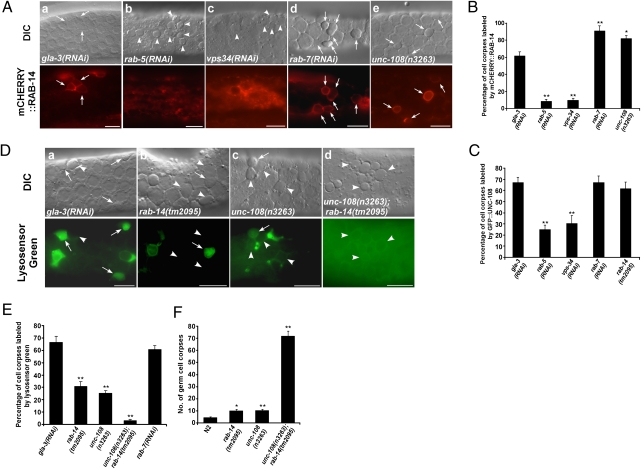
In C. elegans, Rab GTPases act at different steps to regulate the maturation of phagosomes containing apoptotic cells, with RAB-5 and RAB-7 functioning at early and late stages, respectively (4, 8). UNC-108/RAB2 is also involved in this process, but the stage at which it functions remains elusive (13, 14). In sheath cells that internalize apoptotic germ cells, phagosomal recruitment of RAB-5 and the onset of PtdIns(3)P synthesis were followed by recruitment of RAB-14, RAB-7, and UNC-108 at roughly, but not exactly, the same time (Fig. 2). Phagosomal association of RAB-14 and UNC-108 appeared to be transient and shorter than that of RAB-5 and RAB-7 (Fig. 2E). We saw a similar order of Rab recruitment to phagosomes in embryos where cell corpses are cleared much faster and Rab GTPases, especially RAB-5, stayed on phagosomes for a much shorter time (Fig. 1 D and E and Fig. S4). Thus, RAB-14 may be involved in phagosome maturation and may act at the step downstream of RAB-5 activation. Consistent with this, loss of rab-5 or vps-34 function disrupted the phagosomal association of RAB-14, whereas neither RAB-5 recruitment nor PtdIns(3)P generation was affected in rab-14(lf) mutants (Fig. 3 A and B and Fig. S2E). Loss of rab-7 or unc-108 function did not disrupt the phagosomal association of RAB-14 but caused an increase in RAB-14–positive phagosomes. Apoptotic germ cells were surrounded by mCHERRY::RAB-14 in rab-7(RNAi) (91%) and unc-108(n3263) (87%) mutants, respectively, compared with 61% in animals that are wild-type for rab-7 and unc-108 (Fig. 3 A and B). These data suggest that rab-7 and unc-108 are required either for RAB-14 release from phagosomes or for further resolving RAB-14–positive phagosomes. Phagosomal association of UNC-108 was also significantly reduced in rab-5(lf) and vps-34(lf) animals, whereas loss of unc-108 function did not affect phagosomal association of RAB-5 or PtdIns(3)P generation (Fig. 3C and Fig. S2E). Phagosomal association of UNC-108 was not obviously affected in either rab-7(lf) or rab-14(lf) mutants (Fig. 3C). Collectively, these data suggest that RAB-14 and UNC-108 act downstream of RAB-5 activation to regulate phagosome maturation.
Fig. 2.
RAB-14 is recruited to phagosomes at a similar stage to UNC-108 and RAB-7. Confocol fluorescent images of germ corpses in wild-type animals coexpressing Prab-14mcherry::rab-14 and Pced-1gfp::rab-5 (A), Pced-1gfp::2xFYVE (B), Pced-1gfp::rab-7 (C), or Pced-1gfp::unc-108 (D) at different time points are shown. (Scale bars: 5 μm.) (E) Time-course of phagosome recruitment of the five reporters shown in A–D. The time point when RAB-5 was detected on phagosomal membranes was defined as 0 min. Data are shown as mean ± SEM. At least 10 cell corpses were followed. Bars represent mean duration.
Fig. 3.
rab-14 and unc-108 act redundantly to regulate phagosomal acidification. (A) DIC and fluorescent images of the gonad expressing mCHERRY::RAB-14 in gla-3(RNAi) (a), rab-5(RNAi) (b), vps-34(RNAi) (c), rab-7(RNAi) (d), or unc-108(n3263) animals (e). Arrows point to germ cell corpses labeled by mCHERRY::RAB-14; arrowheads indicate unlabeled corpses. (B) Quantification of data shown in A. (C) Quantification of phagosomal labeling by GFP::UNC-108 in indicated strains. (D) DIC and fluorescent images of gonads stained by Lysosensor Green in gla-3(RNAi) (a), rab-14(tm2095) (b), unc-108(n3263) (c), or unc-108(n3263);rab-14(tm2095) (d) animals. Arrows point to germ cell corpses positive for Lysosensor Green; arrowheads indicate unstained corpses. (E) Quantification of data shown in D. (F) Quantification of germ corpses in indicated strains, which were aged 48 h after L4/Adult molt. In B, C, E, and F, data are shown as mean ± SEM. At least 15 animals were scored for each condition. An unpaired t test was carried out by comparing all other datasets with that of gla-3(RNAi) (B, C, and E) or wild type (F). *P < 0.05; **P < 0.0001. All other points had P > 0.05. (Scale bars: 10 μm.)
rab-14 and unc-108 Act Redundantly to Regulate Phagosomal Acidification.
Acidification of the phagosomal lumen is gradually achieved during the maturation process and is crucial for the proper activity of acid hydrolases. Because phagosomal acidification appears to occur downstream of RAB-5 activation (5, 7), we examined whether rab-14 is involved in this process. In animals treated with gla-3 RNAi (which does not affect cell corpse clearance but increases germ cell apoptosis) (20), 66% of germ cell corpses were stained by Lysosensor Green, indicating that the majority of wild-type phagosomes are acidified (Fig. 3 D and E). In rab-14(tm2095) mutants, however, only 31% of phagosomes were positive for Lysosensor Green, indicating that rab-14 is required for phagosomal acidification (Fig. 3 D and E). unc-108(n3263) mutants displayed a similar acidification defect, with only 25% of germ cell corpses stained by Lysosensor Green (Fig. 3 D and E) (14). Strikingly, phagosomal acidification was almost completely blocked in unc-108(n3263);rab-14(tm2095) double mutants with only 3% of apoptotic germ cells labeled by Lysosensor Green, indicating that RAB-14 and UNC-108 act redundantly to control phagosomal acidification (Fig. 3 D and E). In agreement with this, unc-108(n3263);rab-14(tm2095) double mutants accumulated significantly higher numbers of persistent cell corpses in both soma and germ line than either of the single mutants alone, indicating that unc-108 and rab-14 act in parallel to remove cell corpses (Fig. 3F and Fig. S2F).
RAB-14 and UNC-108 Act Sequentially with RAB-7 to Mediate Phagolysosome Formation.
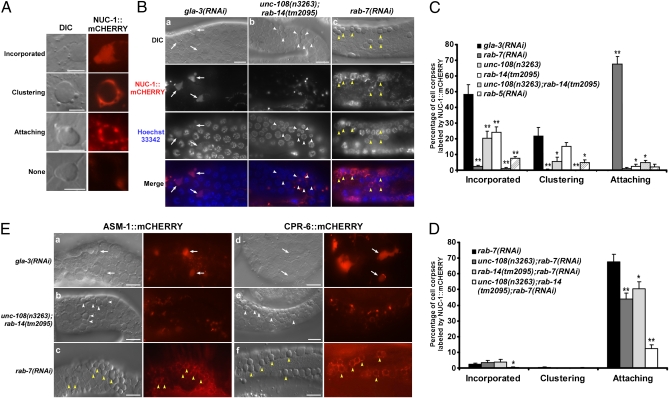
In rab-14(lf) and unc-108(lf) mutants, RAB-7 is found on most phagosomes (Fig. S2E). Similarly, the majority of cell corpses are surrounded by RAB-14 or UNC-108 in rab-7(RNAi) animals (Fig. 3 B and C). Given that RAB-7, RAB-14, and UNC-108 are recruited to phagosomes at a similar stage, these three Rabs may act at closely related steps during phagosome maturation. We focused on phagolysosome formation because both UNC-108 and RAB-7 are implicated in the fusion of phagosomes with lysosomes (8, 14). We followed the recruitment and incorporation of lysosomes into phagosomes by monitoring mCHERRY fusions of lysosomal acid hydrolases. cpr-6 encodes a lysosomal cysteine protease (cathepsin L), which is responsible for protein degradation in lysosomes (21). ASM-1 is homologous to human acid sphingomyelinase, which hydrolyzes sphingomyelin to ceramide and phosphorylcholine within endo-lysosomal compartments (22). NUC-1, the C. elegans homolog of DNase II, is required for resolving TUNEL-positive DNA ends generated during apoptosis, digesting bacterial DNA within the intestinal lumen and completing DNA degradation within phagocytes (23–25). mCHERRY fusions of all three enzymes expressed under control of the engulfing cell-specific ced-1 promoter colocalized well with both Lysotracker Blue and the lysosomal membrane-associated protein CTNS-1::GFP in sheath cells, indicating that the fusion proteins localize to lysosomes (Fig. S5A) (14, 17). mCHERRY fusions of ASM-1, CPR-6, and NUC-1 either clustered around or diffused into the majority of cell corpses in gla-3(RNAi) animals, indicating that they are recruited to and incorporated into phagosomes (Fig. 4). Apoptotic DNA in NUC-1-containing phagosomes gave a weak, diffuse staining pattern with Hoechst 33324 compared with a condensed pattern in phagosomes lacking NUC-1 (Fig. 4B). This indicates that apoptotic DNA is degraded within phagolysosomes. Clustering and incorporation of lysosomal enzymes were significantly reduced in unc-108(lf) or rab-14(lf) single mutants, but completely abolished in animals lacking both, indicating that RAB-14 and UNC-108 play redundant roles in phagolysosome formation (Fig. 4 B–E). Consistent with the notion that phagolysosome formation occurs at late stages of phagosome maturation, this process was blocked by loss of rab-5 activity, which affects early maturation steps (Fig. 4C). Interestingly, in rab-7(RNAi) animals, lysosomal enzymes labeled punctate structures which were attached to, but not incorporated into, phagosomes, suggesting that lysosomes can be recruited to but not fused with phagosomes when rab-7 is lost (Fig. 4). Apoptotic DNA persisted in phagosomes to which NUC-1–positive vesicles were attached but not incorporated (Fig. 4B). By contrast, attachment of NUC-1–positive vesicles was not observed on wild-type phagosomes, which were either surrounded by NUC-1 or contained incorporated NUC-1, suggesting that lysosomes quickly dock and fuse with phagosomes once recruited (Fig. 4C). No attachment of NUC-1–positive vesicles was observed in either unc-108(lf) or rab-14(lf) single mutants, which partially suppressed phagolysosome formation, whereas unc-108(lf);rab-14(lf) double mutants completely blocked lysosome recruitment (Fig. 4C). Collectively, these data suggest that rab-7 is responsible for lysosome/phagosome fusion, whereas UNC-108 and RAB-14 act at an earlier step to recruit lysosomes. To test this, we examined NUC-1::mCHERRY attachment in rab-7(lf) animals that are also defective in either rab-14 or unc-108, or both. Attachment of NUC-1–positive vesicles to phagosomes was partially suppressed in rab-7(RNAi) animals lacking either unc-108 or rab-14, but greatly reduced when both were lost (Fig. 4D). This suggests that RAB-14, UNC-108, and RAB-7 function in sequential steps to regulate phagolysosome formation, with RAB-14 and UNC-108 acting redundantly to recruit lysosomes and RAB-7 mediating fusion of lysosomes to phagosomes. We found that neither RAB-14 nor UNC-108 localized to lysosomes, and their loss of function did not affect delivery of NUC-1, CPR-6, or ASM-1 to lysosomes, suggesting that RAB-14 and UNC-108 act on phagosomal membranes to mediate tethering of lysosomes to phagosomes (Fig. S5 B–D).
Fig. 4.
RAB-14, UNC-108, and RAB-7 act sequentially to mediate phagolysosome formation. (A) Four different staining patterns of NUC-1::mCHERRY on germ cell corpses. Diffuse (Incorporated) or clustered (Clustering) NUC-1 was observed on wild-type phagosomes. NUC-1–positive vesicles attached to cell corpses in rab-7(RNAi) animals (Attaching), whereas no NUC-1 signal (None) was detected around germ cell corpses in unc-108(lf);rab-14(lf) double mutants. (Scale bars: 5 μm.) (B) DIC and fluorescent images of gonads expressing Pced-1nuc-1::mcherry and stained by Hoechst 33342 (which labels DNA) in gla-3(RNAi) (a), unc-108(n3263);rab-14(tm2095) (b), or rab-7(RNAi) (c) animals. Staining of apoptotic DNA became weak and diffuse in phagosomes containing NUC-1 (arrows), but stayed condensed in those lacking NUC-1. White arrowheads show germ cell corpses that failed to recruit NUC-1; yellow arrowheads indicate corpses that contain attached but not incorporated NUC-1. (C and D) Quantification of phagosomes displaying different NUC-1::mCHERRY staining patterns as shown in A in indicated strains. At least 15 animals were scored for each strain. An unpaired t test was carried out by comparing all other data sets with that of gla-3(RNAi) in C or rab-7(RNAi) in D. *P < 0.05; **P < 0.001. All other points had P > 0.05. (E) DIC and fluorescent images of gonads expressing Pasm-1asm-1::mcherry (a–c) or Pced-1cpr-6::mcherry (d–f) in gla-3(RNAi) (a and d), unc-108(n3263);rab-14(tm2095) (b and e), or rab-7(RNAi) (c and f) animals. Arrows indicate germ cell corpses with incorporated fluorescent proteins; white arrowheads indicate unlabeled corpses; yellow arrowheads indicate germ corpses with attached fluorescent protein-containing vesicles. (Scale bars: B and E, 10 μm.)
Discussion
Rab GTPase 14 was identified as a component of latex bead-containing phagosomes by proteomic approaches. It also blocks the maturation of mycobacteria-containing phagosomes in macrophages by promoting their fusion with early but not late endosomes (19, 26, 27). However, its role in the maturation of apoptotic cell-containing phagosomes was unknown. We found that rab-14 mutations in C. elegans affect cell corpse clearance, causing accumulation of persistent cell corpses. RAB-14 acts in engulfing cells to remove apoptotic cells and transiently associates with phagosomes. It functions downstream of RAB-5 activation to regulate phagosomal acidification and phagolysosome formation. Therefore, like RAB-5, RAB-7, and UNC-108/RAB2, RAB-14 promotes apoptotic cell degradation by regulating phagosome maturation. The function of RAB-14 in apoptotic cell removal is likely conserved in mammals because overexpression of mouse RAB14 can efficiently rescue the cell corpse phenotype of C. elegans rab-14 mutants.
The nascent phagosome undergoes a series of fission and fusion events before maturing into acidic, protease-rich phagolysosome (3). We found that three Rabs, RAB-14, UNC-108/RAB2, and RAB-7, function cooperatively and act in sequential steps to regulate phagolysosome formation. RAB-14 and UNC-108 function redundantly in an early tethering step, whereas RAB-7 mediates the docking/fusion of lysosomes with phagosomes. Vesicle tethering, docking, and fusion require proteins that act in a regulated cascade, which is directed and coordinated by Rab GTPases (6). RAB-14 and UNC-108 may recruit the tethering complex specifically to phagosomal membranes to initiate interactions between phagosomes and lysosomes. Subsequent docking and fusion may be achieved by cooperation of tethering factors with components of the docking and fusion machinery such as SNAREs. Given that RAB-7 is found on both phagosomal and lysosomal membranes (Fig. 2C) (8), it may control the docking/fusion step by mediating the pairing of SNAREs from both membranes. The coordination of Rabs in endocytosis or phagocytosis can be accomplished through Rab conversions or Rab cascades in which one activated Rab recruits the GEF for the Rab acting in the next trafficking step (28, 29). In the case of RAB-14, UNC-108, and RAB-7, loss of rab-14 and unc-108 function does not affect RAB-7 activation and its subsequent association with phagosomes (Fig. S2E) (8), so their sequential action in phagolysosome formation is likely achieved through the coordination of effectors instead of controlling the activation of downstream Rabs.
The gradual acidification of maturing phagosomes is thought to be achieved by sequential fusion with progressively acidified endocytic compartments (30). Although RAB-14, UNC-108, and RAB-7 are all required for phagolysosome formation, only RAB-14 and UNC-108 are needed for phagosomal acidification, indicating that the acidification of apoptotic cell-containing phagosomes occur independently of, and likely before, phagolysosome formation (Fig. 3E) (8). As dominant negative RAB7 affects acidification of latex bead-containing phagosomes in macrophages, differential regulatory mechanisms may be used for acidifying phagosomes containing different cargoes (11). How is phagosomal acidification regulated by RAB-14 and UNC-108? In macrophages, human RAB14 is found on both early endosomes and phagosomes containing live mycobacteria, thereby promoting their fusion (19). Because RAB-14 and UNC-108 mainly localize to early endosomes and the trans-Golgi network in sheath cells (Fig. S6), they may control phagosomal acidification by mediating fusion of endosomes with phagosomes, thereby allowing acquisition of vacuolar-type ATPases, the main determinants of phagosomal acidification. Given that both mouse RAB2 and RAB14 can substitute for C. elegans UNC-108 and RAB-14 in removing cell corpses, these two Rabs may act in a similar manner to regulate maturation of apoptotic cell-containing phagosomes in mammals (13).
Materials and Methods
Genetic Analysis.
Strains of C. elegans were cultured at 20 °C. The N2 Bristol strain was used as the wild-type strain. Details of the genetic screen, usage of transgenic and mutant strains, and mapping and cloning of rab-14 are provided as SI Materials and Methods.
Microscopy and Imaging Analysis.
DIC and fluorescent images were captured with a Zeiss Axioimager A1 or a Zeiss LSM 5 Pascal inverted confocal microscope. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. K. S. Ravichandran (University of Virginia, Charlottesville, VA), M. O. Hengartner (University of Zurich, Zurich, Switzerland), Greg Hermann (Lewis and Clark College, Portland, OR) for providing reporters; Drs. Shohei Mitani (Tokyo Women's Medical University, Tokyo, Japan) and Zheng Zhou (Baylor College of Medicine, Houston, TX) and the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN) for strains; Dr. Chonglin Yang for critical reading of the manuscript; and Dr. Isabel Hanson for editing services. This work was supported by National High Technology Project 863.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008946107/-/DCSupplemental.
References
- 1.Kinchen JM, Ravichandran KS. Phagosome maturation: Going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, Yu X. Phagosome maturation during the removal of apoptotic cells: Receptors lead the way. Trends Cell Biol. 2008;18:474–485. doi: 10.1016/j.tcb.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: Aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinchen JM, et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 6.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 7.Li W, et al. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136:2445–2455. doi: 10.1242/dev.035949. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLOS Biol. 2008;6(3):e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: A key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol. 2003;23:6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao H, et al. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in C. elegans. Mol Biol Cell. 2009;20(1):21–32. doi: 10.1091/mbc.E08-04-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Q, et al. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- 14.Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:S3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: Definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 18.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93(4):519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 19.Kyei GB, et al. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. EMBO J. 2006;25:5250–5259. doi: 10.1038/sj.emboj.7601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kritikou EA, et al. C. elegans GLA-3 is a novel component of the MAP kinase MPK-1 signaling pathway required for germ cell survival. Genes Dev. 2006;20:2279–2292. doi: 10.1101/gad.384506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turk B, Turk D, Turk V. Lysosomal cysteine proteases: More than scavengers. Biochim Biophys Acta. 2000;1477:98–111. doi: 10.1016/s0167-4838(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins RW, Canals D, Hannun YA. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal. 2009;21:836–846. doi: 10.1016/j.cellsig.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulston JE. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- 24.Hedgecock EM, Sulston JE, Thomson JN. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- 25.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- 26.Garin J, et al. The phagosome proteome: Insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart LM, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 28.Markgraf DF, Peplowska K, Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581:2125–2130. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- 29.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huynh KK, Grinstein S. Regulation of vacuolar pH and its modulation by some microbial species. Microbiol Mol Biol Rev. 2007;71:452–462. doi: 10.1128/MMBR.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.