Abstract
Microbial niches contain toxic chemicals capable of forcing organisms into periods of intense natural selection to afford survival. Elucidating the mechanisms by which microbes evade environmental threats has direct relevance for understanding and combating the rise of antibiotic resistance. In this study we used a toxic small-molecule, bromoacetate, to model the selective pressures imposed by antibiotics and anthropogenic toxins. We report the results of genetic selection experiments that identify nine genes from Escherichia coli whose overexpression affords survival in the presence of a normally lethal concentration of bromoacetate. Eight of these genes encode putative transporters or transmembrane proteins, while one encodes the essential peptidoglycan biosynthetic enzyme, UDP-N-acetylglucosamine enolpyruvoyl transferase (MurA). Biochemical studies demonstrate that the primary physiological target of bromoacetate is MurA, which becomes irreversibly inactivated via alkylation of a critical active-site cysteine. We also screened a comprehensive library of E. coli single-gene deletion mutants and identified 63 strains displaying increased susceptibility to bromoacetate. One hypersensitive bacterium lacks yliJ, a gene encoding a predicted glutathione transferase. Herein, YliJ is shown to catalyze the glutathione-dependent dehalogenation of bromoacetate with a kcat/Km value of 5.4 × 103 M-1 s-1. YliJ displays exceptional substrate specificity and produces a rate enhancement exceeding 5 orders of magnitude, remarkable characteristics for reactivity with a nonnatural molecule. This study illustrates the wealth of intrinsic survival mechanisms that can be exploited by bacteria when they are challenged with toxins.
Keywords: enzyme evolution, enzyme recruitment, glutathione transferase
Bacterial populations are constantly exposed to a myriad of harmful chemicals from the environment. Antibiotics have been used clinically since the mid-twentieth century to fight bacterial infections. Toxic anthropogenic chemicals are widely utilized for agricultural, medicinal, and industrial purposes. The enormous metabolic and functional diversity possessed by microbes enables them to deploy protective mechanisms to facilitate survival under the selective pressures imposed by these toxic molecules. The recent rise of antibiotic resistance demonstrates that bacteria are capable of rapidly evolving evasive strategies; it has also exposed our lack of knowledge about the evolutionary processes leading to resistance (1).
A resistance phenotype may arise through intrinsic mechanisms or via the acquisition of resistance genes through horizontal transfer (1, 2). In both cases, three biological processes are commonly associated with toxin evasion—the efflux of harmful compounds, the enzymatic inactivation of toxic agents, and the overexpression or modification of target genes. Intrinsic resistance can occur when a cell constitutively expresses a chromosomal gene encoding a latent or weakly active protein that confers a modest level of protection. Such genes represent the prima materia from which high-level resistance can later evolve (3–6). Acquired resistance genes are coding sequences disseminated on mobile genetic elements. These genetic elements are presumed to originate in soil-dwelling, antibiotic-synthesizing microbes as a mechanism of self-protection (2, 7). In the present study, we were interested in discovering the extent to which intrinsic resistance of a naïve bacterial population can play a role in combating the toxicity of a nonnatural small-molecule. Revealing the reservoir of intrinsic resistance genes that are subject to evolutionary recruitment promises to aid our understanding of the processes leading to the emergence of antibiotic resistant pathogens.
We sought to identify the full spectrum of bromoacetate resistance mechanisms available to the model bacterium, Escherichia coli. The reactivity of bromoacetate is likely to mimic that of electrophilic natural products as well as anthropogenic environmental contaminants that microbes may encounter. The clinically significant natural antibiotic fosfomycin, and the fungal natural product terreic acid, are electrophilic molecules that both target an essential nucleophilic cysteine residue in bacteria (8, 9). In addition, commonly utilized pesticides such as methyl bromide, 1,3-dichloropropene, and chloropicrin function by covalently modifying essential cellular nucleophiles (10). We also postulated that the low activation energy required for cleavage of the carbon-bromine bond in bromoacetate, in addition to its small size and ease of cellular entry, would increase the likelihood of discovering enzymes possessing fortuitous dehalogenase activities.
In the first part of our study, we wished to identify coding sequences that could be easily recruited for bromoacetate resistance via a mutation that causes constitutive expression of a single-gene. A genetic selection, using the complete ASKA library of E. coli open reading frames (11, 12), was used to mimic the natural evolutionary process of gene recruitment via derepression (13). This technique increases the intracellular concentration of all library-encoded proteins to reveal latent and weakly active bromoacetate resistance mechanisms. A similar genetic selection strategy has recently been used to identify genomic regions in Pseudomonas aeruginosa capable of conferring resistance to aminoglycosides (14). In the second part of our investigation we sought genes that provide innate protection against bromoacetate, without the need for constitutive overproduction. To this end, we screened the entire Keio collection of approximately 4,000 E. coli single-gene knockouts (15) for mutants hypersensitive to bromoacetate. The Keio collection has previously been used to identify mutants hypersensitive to at least 1 of 22 different antibiotics (16, 17). Similarly, screening of transposon libraries in Acinetobacter baylyi and P. aeruginosa has identified genes whose disruption hypersensitizes these organisms to a variety of antibiotics (18–20). These studies yielded sets of proteins that could be targeted with codrugs to potentiate the activities of extant antibiotics. Our own study provides a genome-wide assessment of the scope of genes available to E. coli to combat the deleterious effects of a nonnatural xenobiotic molecule, which this organism is unlikely to have encountered in the past.
Results and Discussion
The E. coli Genome Harbors Nine Genes that Confer Resistance to Bromoacetate when Overexpressed.
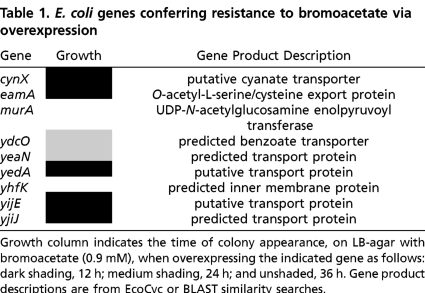
To identify genes that can confer resistance to bromoacetate when overexpressed, the DH5α strain of E. coli was subjected to genetic selection. Bacteria were transformed with a plasmid-borne genomic expression library (11) and cells were challenged for growth on LB-agar plates supplemented with 0.9 mM bromoacetate (the lowest concentration sufficient to inhibit growth for one week at 37 °C), chloramphenicol (to select for transformants), and IPTG (to induce gene expression). Numerous colonies were selected after 12–36 h of growth, and the resistance genes were identified via sequencing of plasmid DNA (Table 1). This experiment uncovered a substantial reservoir of genes that can be readily mobilized to withstand bromoacetate. Eight of the selected genes are predicted to encode transport or transmembrane proteins, while one encodes the essential enzyme UDP-N-acetylglucosamine enolpyruvoyl transferase (MurA). MurA catalyzes the first committed step in cell wall peptidoglycan biosynthesis.
Table 1.
E. coli genes conferring resistance to bromoacetate via overexpression
Growth column indicates the time of colony appearance, on LB-agar with bromoacetate (0.9 mM), when overexpressing the indicated gene as follows: dark shading, 12 h; medium shading, 24 h; and unshaded, 36 h. Gene product descriptions are from EcoCyc or BLAST similarity searches.
The bromoacetate resistance transporters revealed by our genetic selection possess a variety of experimentally defined and putative physiological functions (Table 1). CynX encodes a putative cyanate permease and is located within the cyanate inducible cyn operon, which also contains cynS and cynT. The cynS and cynT genes encode for cyanase and carbonic anhydrase, respectively (21). EamA has been shown to efflux cysteine and O-acetyl-L-serine, an intermediate in the biosynthesis of cysteine (22). Overexpression of eamA is also known to provide resistance to azaserine. Overexpression of yedA and yijE has been shown to confer resistance to toxic purine base analogs, suggesting that their physiological function may be associated with nucleotide transport (23). YdcO is predicted to encode a benzoate transporter, while yeaN, yhfK, and yjiJ encode putative transport or transmembrane proteins with unknown functions (24).
Bromoacetate Alkylates the Active-Site Cysteine of MurA.
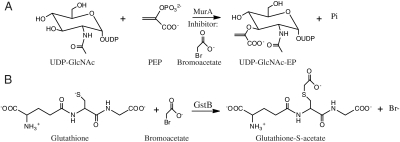
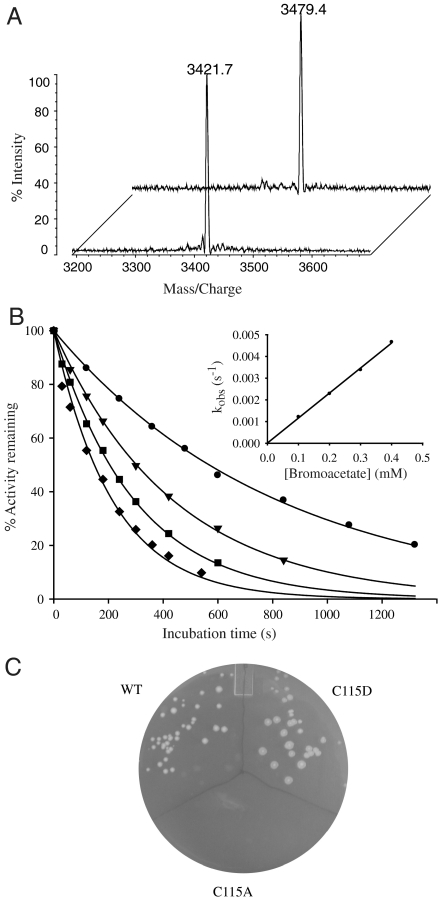
MurA catalyzes the transfer of the enolpyruvoyl moiety of phosphoenolpyruvate (PEP) to the 3′-hydroxyl of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) forming UDP-GlcNAc enolpyruvate (UDP-GlcNAc-EP) (Scheme 1A). This enzyme is the target of the naturally occurring electrophilic antibiotic fosfomycin, which alkylates cysteine 115 in the active site (8). To investigate if bromoacetate also alkylates C115, purified MurA was incubated with bromoacetate and the protein was subjected to tryptic digestion followed by mass spectrometry analysis. The peptide masses obtained from MurA incubated with bromoacetate were compared to the masses obtained from control MurA that was not exposed to bromoacetate. We observed a single difference peak between the two spectra corresponding to the tryptic fragment containing C115 (Fig. 1A). The mass difference of 57.7 Da accounts for alkylation of C115 by bromoacetate, because no other cysteines are contained within this fragment (expected mass difference, 58 Da).
Scheme 1.
(A) MurA catalyzed reaction. (B) GstB catalyzed reaction.
Fig. 1.
MurA inactivation by alkylation with bromoacetate. (A) MALDI mass spectrometry of MurA tryptic digest. The mass peak is from the peptide containing C115 of control (foreground, expected mass 3421.9 Da) and bromoacetate inactivated enzyme (background, mass increase of 57.7 Da). (B) Time-dependent loss of MurA activity in the presence of bromoacetate and 1 mM UDP-GlcNAc. Bromoacetate concentrations were 0.1 mM (•), 0.2 mM (▾), 0.3 mM (▪), and 0.4 mM (♦). Inset is a replot of the first-order rate constants (kobs) vs. bromoacetate concentrations. Data were analyzed as described in SI Material and Methods and yielded a second-order inactivation rate constant (kinact) of 11.4 ± 0.2 M-1 s-1. (C) Growth of E. coli BW25113 when overproducing wild-type, C115D or C115A MurA after 48 h incubation at 37 °C on LB-agar supplemented with bromoacetate (0.9 mM), chloramphenicol and IPTG.
To further characterize the reactivity of MurA with bromoacetate, we investigated the time-dependence of this reaction. MurA was incubated with differing amounts of bromoacetate and the residual MurA activity was measured at various time intervals in a coupled spectrophotometric assay using MurB. The activity of MurA was observed to decrease as a single-exponential function with time (Fig. 1B). The first-order rate constants of inactivation (kobs) at various bromoacetate concentrations were plotted against the bromoacetate concentration to obtain a second-order inactivation rate constant (kinact) of 3.1 ± 0.1 M-1 s-1 in the absence of UDP-GlcNAc (Figs. S1 and S2) and 11.4 ± 0.2 M-1 s-1 (Fig. 1B) in the presence of a saturating amount of UDP-GlcNAc (1 mM) in 100 mM triethanolamine-HCl at pH 8.0. Binding of UDP-GlcNAc to MurA has been shown to induce a conformational change resulting in an increased affinity toward PEP (25). The inactivation rate of MurA with fosfomycin is also dependent upon UDP-GlcNAc (8). Surprisingly, the second-order inactivation rate of MurA by bromoacetate, under saturating UDP-GlcNAc at pH 8.0, is only 9-fold slower than by fosfomycin under saturating UDP-GlcNAc at pH 6.9 (104 M-1 s-1) (26).
Next, we wished to investigate whether inactivation of the essential MurA is the primary mechanism of bromoacetate toxicity in E. coli. It seems reasonable to postulate that overexpression of murA confers resistance by allowing higher levels of transferase activity to be realized in vivo. Alternatively, overexpression of murA might serve to titrate bromoacetate from the cytosol in a single-turnover dehalogenation reaction, thus reducing its ability to react with other cellular targets. To distinguish between these possibilities, we took advantage of the fact that fosfomycin producing Mycobacteria possess a MurA that contains an active-site aspartate in place of cysteine. This natural substitution renders the protein resistant to modification by fosfomycin (27), and we also found that it was resistant to alkylation with bromoacetate (Fig. S3). We observed that overproduction of the C115D variant of E. coli MurA confers bromoacetate resistance to a host bacterium on solid and liquid media, whereas the inactive variant, C115A, does not support growth (Fig. 1C and Fig. S4). This indicates that titration of bromoacetate from the cytosol is not important for the resistance phenotype, but, rather, the availability of active MurA is the factor necessary to support growth.
Screening of the Keio Collection of E. coli Single-Gene Mutants Revealed an Array of Genes Contributing to Intrinsic Bromoacetate Resistance.
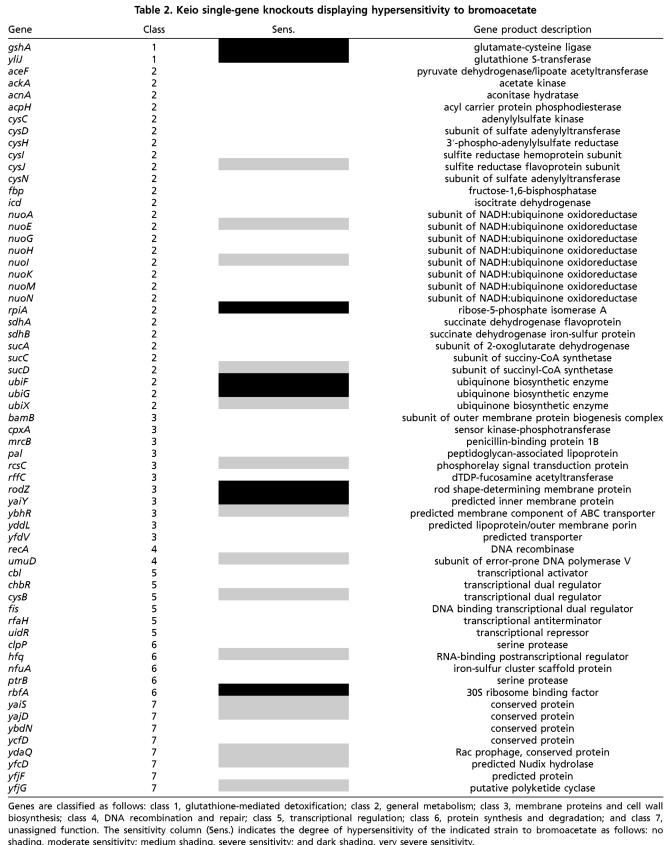
To further define the intrinsic bromoacetate resistome, and to uncover resistance mechanisms that cannot be accessed via overexpression of individual genes, we screened the Keio collection (15) for mutants displaying hypersensitivity to bromoacetate. The Keio collection consists of approximately 4,000 E. coli strains, each possessing a single-gene deletion. We used high-throughput screening to identify mutants that are hypersusceptible to bromoacetate as compared to the parental strain. The Keio collection was screened on LB-agar plates supplemented with a subinhibitory concentration of bromoacetate (0.45 mM). At this bromoacetate concentration, the majority of mutants grow at a rate indistinguishable from the parental strain. Strains possessing deletions in genes contributing to intrinsic bromoacetate resistance are easily identified as displaying a slow growth or no growth phenotype. To account for deletions that cause slow growth in rich media, we compared the growth of each mutant in the presence and absence of bromoacetate. We identified 63 hypersensitive mutants with deletions in genes encompassing a broad range of functions (Table 2).
Table 2.
Keio single-gene knockouts displaying hypersensitivity to bromoacetate
Genes are classified as follows: class 1, glutathione-mediated detoxification; class 2, general metabolism; class 3, membrane proteins and cell wall biosynthesis; class 4, DNA recombination and repair; class 5, transcriptional regulation; class 6, protein synthesis and degradation; and class 7, unassigned function. The sensitivity column indicates the degree of hypersensitivity of the indicated strain to bromoacetate as follows: no shading, moderate sensitivity; medium shading, severe sensitivity; and dark shading, very severe sensitivity.
The mutants are divided into seven classes according to the function of the protein that the deleted gene encoded (24). Class 1 consists of two genes necessary for the glutathione-mediated detoxification of electrophilic molecules. GshA encodes glutamate-cysteine ligase, one of two genes critical for the biosynthesis of the tripeptide glutathione, and yliJ encodes a predicted glutathione transferase. Glutathione transferases catalyze the conjugation of glutathione to electrophilic molecules, thus preventing their reactivity with cellular components. Class 2, the largest class identified, consists of mutants that have general metabolic genes inactivated. Interestingly, many mutants in this class are involved in acetate metabolism, including TCA cycle genes, suggesting flux of bromoacetate through metabolism as a mechanism of detoxification. This is not without precedent as fluoroacetate enters the acetate metabolic pathway, and its toxicity is due to the lethal synthesis of the aconitase inhibitor, fluorocitrate (28). Also prevalent in this class are genes involved in synthesizing and reducing ubiquinone and genes in the cysteine biosynthetic pathway. Ubiquinone is a cofactor critical for the TCA cycle and free cysteine in the cytosol may be significant in conjugating to bromoacetate and reducing its intracellular concentration. Class 3 genes encode membrane proteins and cell wall biosynthetic enzymes, including several predicted transporters. Class 4 contains genes significant for DNA recombination and repair, while class 5 encodes transcriptional regulators, including CysB, a regulator of cysteine biosynthetic genes. Class 6 encompasses genes involved in protein synthesis/degradation and class 7 genes are of unknown function. Eighteen of the hypersensitive mutants we identified have also been shown to be sensitive to at least 1 of 22 clinically used antibiotics (16, 17). These coding sequences most likely represent a common framework of genes crucial to resisting the adverse effects of toxic molecules (Table S1). Notably, we did not identify mutants containing deletions in any genes that were revealed in selections involving the ASKA collection.
The Previously Uncharacterized Glutathione Transferase, YliJ, Catalyzes the Efficient Dehalogenation of Bromoacetate.
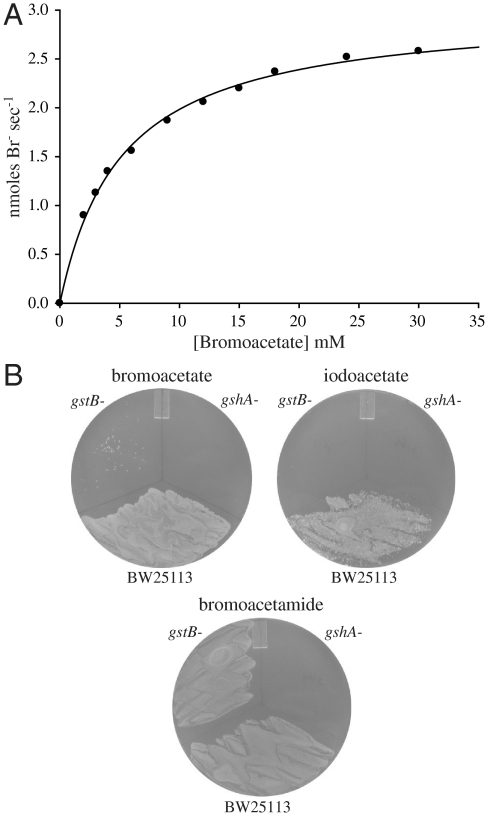
We investigated the ability for the predicted glutathione transferase, YliJ, to catalyze the conjugation of glutathione to bromoacetate. YliJ was purified using a hexa-histidine tag and its glutathione transferase activity toward bromoacetate was examined in a discontinuous assay that detects the appearance of bromide ion (29). YliJ was found to efficiently catalyze the dehalogenation of bromoacetate (Scheme 1B and Fig. 2A). Kinetic analysis of YliJ, herein renamed glutathione S-transferase B (GstB), revealed a kcat value of 27 ± 1 s-1 and a Km value for bromoacetate of 5 ± 0.3 mM, giving a second-order rate constant of 5.4 × 103 M-1 s-1 under saturating glutathione concentrations in 100 mM HEPES at pH 7.0. The high activity of GstB toward a nonnatural molecule is intriguing, so to determine the catalytic proficiency of GstB, we also measured the uncatalyzed rate of dehalogenation (Figs. S5 and S6). The uncatalyzed rate of glutathione conjugation to bromoacetate in 100 mM HEPES at pH 7.0 is 1.4 × 10-2 M-1 s-1, indicating that GstB provides a rate acceleration of greater than 5 orders of magnitude. Notably, overexpression of gstB did not confer resistance to bromoacetate but, rather, decreased the minimum inhibitory concentration, most likely due to the conjugation of glutathione to a critical cellular metabolite. Low gstB expression is sufficient to provide a significant level of bromoacetate resistance because GstB is a highly efficient catalyst. The very severe hypersensitivity of a gstB and a gshA knockout to bromoacetate, compared to the parental strain, is depicted in Fig. 2B.
Fig. 2.
The E. coli genes gstB and gshA provide resistance to bromoacetate. (A) Michaelis–Menten plot of GstB catalyzed conjugation of glutathione to bromoacetate under saturating glutathione. This plot yields a kcat value of 27 s-1 a Km value for bromoacetate of 5 mM and a second-order rate constant of 5.4 × 103 M-1 s-1. (B) Growth of gstB- and gshA- strains on LB-agar supplemented with bromoacetate (0.45 mM), iodoacetate (0.125 mM) or bromoacetamide (0.175 mM) compared to the parental strain (BW25113).
To further characterize GstB, its substrate specificity was examined. We tested the ability for GstB to catalyze the glutathione-dependent dehalogenation of chloroacetate, iodoacetate, bromoacetamide, 2-bromopropionate, and 3-bromopropionate. Remarkably, only iodoacetate was reactive with GstB to any significant degree. In the presence of 2 mM iodoacetate the rate of GstB-mediated dehalogenation was 48.9 μmoles min-1 mg-1 of enzyme. In comparison, the rate of GstB-mediated decomposition of bromoacetate was 10.5 μmoles min-1 mg-1 at the same substrate concentration. The rate of dehalogenation for the other compounds tested (2 mM) was less than 0.07 μmoles min-1 mg-1. The inability for GstB to detoxify bromoacetamide is intriguing given that this molecule is more reactive than bromoacetate and indicates the importance of the carboxylate for GstB substrate recognition. The physiological substrate of GstB is probably a small-molecule containing a carboxylate moiety. The failure of our in vitro assays to detect significant dehalogenation of bromoacetamide by GstB was corroborated in vivo by observing that a gstB knockout is not hypersensitive to this halogenated molecule (Fig. 2B). However, the sensitivity of a gshA knockout to bromoacetamide indicates that an unknown glutathione transferase is capable of detoxifying this compound. The stringency with which GstB recognizes its substrate is interesting because glutathione transferases are commonly characterized as having relaxed substrate specificities (30).
The E. coli genome harbors eight glutathione transferase homologues, of which only four have previously been functionally characterized (31). Glutathione S-transferase (Gst) displays high activity toward the model substrate 1-chloro-2,4-dinitrobenzene (CDNB) and also a low level of glutathione-dependent peroxidase activity toward cumene hydroperoxide, although its physiological function remains unknown (32). YfcF and YfcG display very low glutathione conjugating activity toward CDNB and also show low glutathione-dependent peroxidase activity toward cumene hydroperoxide (33); however, these activities may be vestigial because YfcG was recently shown to be an efficient disulfide bond reductase (34). Stringent starvation protein A (SspA) does not bind glutathione and has no known enzymatic activity. Instead, this protein associates with RNA polymerase and regulates the expression of stress response genes (35). The discovery that GstB is a highly active enzyme, and the discovery that the gstB mutant is the only glutathione transferase mutant hypersusceptible to bromoacetate, suggests that each glutathione transferase has a distinct physiological role.
The results presented here illuminate the scope of resistance genes harbored within the genome of a simple organism. The discovery that nearly 2% of the protein encoding genes in E. coli contribute to, or can be recruited to provide, bromoacetate resistance is striking considering the nonnatural origins of this molecule. Interestingly, our work shows that even highly reactive molecules can have specific effects on microbes by targeting a single essential nucleophile and by being specifically recognized by an inactivating enzyme. Our model system identified three types of resistance mechanisms that are commonly observed in clinically significant antibiotic resistant pathogens—toxin efflux, enzymatic inactivation, and overexpression of a target gene. The breadth of gene functions that were revealed indicates that a robust network exists to maintain intrinsic resistance. The numerous transport proteins identified correlates with findings showing that E. coli transporters display functional redundancy in the efflux of toxic small molecules (36). Transporter polyspecificity provides an important survival strategy, ensuring that a large variety of chemical structures can be efficiently effluxed. The ability for microbes to survive diverse chemical and environmental challenges illustrates the evolutionary advantages of maintaining a large genome size despite the fact that only a small subset of genes are essential during growth on rich media (15, 37). The experimental approach described herein also provides a method to estimate the likelihood that resistance to a particular toxin might arise, and to predict the specific genes that could facilitate evasion. In future studies, it would be interesting to test the predictive powers of our approach by evolving a bromoacetate resistant strain using continuous culture techniques. One could then investigate the extent to which each of the genes identified here are mobilized to combat the toxic challenge.
Materials and Methods
Genetic Selection.
The complete ASKA genomic library was prepared as previously described (12). Electrocompetent DH5α cells were transformed with 100 ng of genomic library. Cells were recovered in SOC media for 1 h at 37 °C and washed twice with M9 minimal media. Washed cells were plated on 30 LB-agar plates supplemented with chloramphenicol (30 μg/mL), bromoacetate (0.9 mM), and IPTG (50 μM). Identical plates excluding IPTG were used as a negative control. Dilution plates on LB with chloramphenicol allowed us to calculate that the transformed library was plated at a density of 1.08 × 105 colony forming units per 100 mm plate. Selected resistance genes were reconfirmed by a second round of selection with purified plasmid DNA, followed by sequencing of isolated plasmids. Although yhfK was selected multiple times in the initial selection, reconfirmation was only successful at a slightly lower bromoacetate concentration (0.8 mM). We found that the ASKA ydcO clone contains 120 bp of genomic sequence preceding the ydcO coding sequence, which would be translated and fused to the N terminus of YdcO.
Screening of the Keio Collection for Mutants Hypersensitive to Bromoacetate.
The Keio collection of kanamycin resistant, single-gene mutants (15) is stored as glycerol stocks in 96-well microtiter plates. Each plate was replicated by transferring a small amount of glycerol stock with a sterile 96-pin replicator into 96-well microtiter plates containing 175 μL of LB media with kanamycin (25 μg/mL) per well. Cultures were grown at 37 °C for 16 h and a sterile 96-pin replicator was used to transfer small amounts of culture from each microtiter plate onto an LB-agar control plate and 2 LB-agar plates supplemented with a subinhibitory concentration of bromoacetate (0.45 mM). The plates were incubated at 37 °C for 16 h and hypersensitive mutants were identified as having a significantly slower growth rate on the LB-agar plates containing bromoacetate as compared to the control LB plate. All mutants identified as hypersensitive were confirmed by first colony purifying each mutant to ensure pure cultures and then approximately 4 × 105 colony forming units were plated on one-third of a 100 mm LB-agar plate and one-third of an LB-agar plate supplemented with 0.45 mM bromoacetate for comparison.
Expression and Purification of GstB.
The gstB gene was PCR amplified using the following primers, forward 5′-TAT GCA TGC TTA CGC TGT GGG GTC GGA A-3′ and reverse 5′-TAT AGA TCT GCT AAC GGG AAT CAT CAC CAC-3′. The PCR product was cloned into the SphI and BglII sites of a modified pQE-70 vector that encodes LacI for repression of gene expression in the absence of IPTG. The gstB clone was transformed into strain BW25113 and expressed as described for murA. GstB was purified as described for MurA except that buffer A consisted of 50 mM HEPES pH 7.7, 40 mM imidazole and 0.5 mM DTT. GstB was dialyzed against 2 L of 20 mM HEPES pH 7.0 containing 1 mM EDTA. Protein concentrations were determined from absorbance readings at 280 nm using a calculated extinction coefficient of 51,450 M-1 cm-1 (ExPASy).
Assay of GstB Activity.
Assay mixtures contained 100 mM HEPES pH 7.0, 1.5 mM L-glutathione and varying amounts of bromoacetate and were initiated by the addition of GstB. Solutions of glutathione and bromoacetate were neutralized with NaOH before use. An identical reaction mixture was generated that did not contain GstB, which served as the blank to correct for both the uncatalyzed hydrolysis of bromoacetate and for small amounts of bromide that were observed in solutions of bromoacetate. Reactions were performed at 25 °C and were immediately quenched at various time points by the addition of N-ethylmaleimide to 20 mM, which blocks thiols. Bromide concentrations were determined by adding 100 μL of a saturated solution of ferric ammonium sulfate in 70% nitric acid and 100 μL of a saturated solution of mercuric thiocyanate in 100% ethanol, incubating for 3 min and measuring the absorbance at 460 nm, essentially as previously described (29). A standard calibration curve was created using NaBr (Fig. S7). The kinetic constants reported are the mean values from two separate experiments with the errors representing the deviation from the mean.
Further details are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
This work was supported, in part, by grants from the James and Ester King Biomedical Research Program (09KN08) and from the National Institute of Diabetes and Digestive and Kidney Diseases (DK081358) to B.G.M.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007559107/-/DCSupplemental.
References
- 1.Walsh CT. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 2.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 3.Davies JE. Origins, acquisition and dissemination of antibiotic resistance determinants. Ciba Found Symp. 1997;207:15–27. [PubMed] [Google Scholar]
- 4.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall BG. Predicting the evolution of antibiotic resistance genes. Nat Rev Microbiol. 2004;2:430–435. doi: 10.1038/nrmicro888. [DOI] [PubMed] [Google Scholar]
- 6.Copley SD. Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat Chem Biol. 2009;5:559–566. doi: 10.1038/nchembio.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 8.Marquardt JL, et al. Kinetics, stoichiometry, and identification of the reactive thiolate in the inactivation of UDP-GlcNAc enolpyruvoyl transferase by the antibiotic fosfomycin. Biochemistry. 1994;33:10646–10651. doi: 10.1021/bi00201a011. [DOI] [PubMed] [Google Scholar]
- 9.Han H, et al. The fungal product terreic acid is a covalent inhibitor of the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) Biochemistry. 2010;49:4276–4282. doi: 10.1021/bi100365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cremlyn RJ. New York: John Wiley & Sons, Inc; 1991. Agrochemicals: Preparation and mode of action. [Google Scholar]
- 11.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library: Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 12.Desai KK, Miller BG. A metabolic bypass of the triosephosphate isomerase reaction. Biochemistry. 2008;47:7983–7985. doi: 10.1021/bi801054v. [DOI] [PubMed] [Google Scholar]
- 13.Hegeman GD, Rosenberg SL. The evolution of bacterial enzyme systems. Annu Rev Microbiol. 1970;24:429–462. doi: 10.1146/annurev.mi.24.100170.002241. [DOI] [PubMed] [Google Scholar]
- 14.Struble JM, Gill RT. Genome-scale identification method applied to find cryptic aminoglycoside resistance genes in Pseudomonas aeruginosa. PLoS One. 2009;4:e6576. doi: 10.1371/journal.pone.0006576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba T, et al. Construction of Escherichia coli K12 in-frame single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamae C, et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol. 2008;190:5981–5988. doi: 10.1128/JB.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu A, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez MJ, Neyfakh AA. Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob Agents Chemother. 2006;50:3562–3567. doi: 10.1128/AAC.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, et al. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci USA. 2009;106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fajardo A, et al. The neglected intrinsic resistome of bacterial pathogens. PLoS One. 2008;3:e1619. doi: 10.1371/journal.pone.0001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung YC, Fuchs JA. Characterization of the cyn operon in Escherichia coli K12. J Biol Chem. 1988;263:14769–14775. [PubMed] [Google Scholar]
- 22.Daßler T, Maier T, Winterhalter C, Böck A. Identification of a major facilitator protein from Escherichia coli involved in efflux of metabolites of the cysteine pathway. Mol Microbiol. 2000;36:1101–1112. doi: 10.1046/j.1365-2958.2000.01924.x. [DOI] [PubMed] [Google Scholar]
- 23.Zakataeva NP, et al. Export of metabolites by the proteins of the DMT and RhtB families and its possible role in intercellular communication. Microbiology. 2006;75:438–448. [PubMed] [Google Scholar]
- 24.Keseler IM, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krekel F, Oecking C, Amrhein N, Macheroux P. Substrate and inhibitor-induced conformational changes in the structurally related enzymes UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) Biochemistry. 1999;38:8864–8878. doi: 10.1021/bi990412o. [DOI] [PubMed] [Google Scholar]
- 26.Schönbrunn E, Eschenburg S, Krekel F, Luger K, Amrhein N. Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA. Biochemistry. 2000;39:2164–2173. doi: 10.1021/bi991091j. [DOI] [PubMed] [Google Scholar]
- 27.Kim DH, et al. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 28.Carrell HL, et al. Fluorocitrate inhibition of aconitase: Relative configuration of inhibitory isomer by X-ray crystallography. Science. 1970;170:1412–1414. doi: 10.1126/science.170.3965.1412. [DOI] [PubMed] [Google Scholar]
- 29.Bergmann JG, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 30.Allocati N, Federici L, Masulli M, Di llio C. Glutathione transferases in bacteria. FEBS J. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- 31.Rife CL, Parsons JF, Xiao G, Gilliland GL, Armstrong RN. Conserved structural elements in glutathione transferase homologues encoded in the genome of Escherichia coli. Proteins. 2003;53:777–782. doi: 10.1002/prot.10452. [DOI] [PubMed] [Google Scholar]
- 32.Nishida M, Kong KH, Inoue H, Takahashi K. Molecular cloning and site-directed mutagenesis of glutathione S-transferase from Escherichia coli. The conserved tyrosyl residue near the N terminus is not essential for catalysis. J Biol Chem. 1994;269:32536–32541. [PubMed] [Google Scholar]
- 33.Kanai T, Takahashi K, Inoue H. Three distinct-type glutathione S-transferases from Escherichia coli important for defense against oxidative stress. J Biochem. 2006;140:703–711. doi: 10.1093/jb/mvj199. [DOI] [PubMed] [Google Scholar]
- 34.Wadington MC, Ladner JE, Stourman NV, Harp JM, Armstrong RN. Analysis of the structure and function of YfcG from Escherichia coli reveals an efficient and unique disulfide bond reductase. Biochemistry. 2009;48:6559–6561. doi: 10.1021/bi9008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams MD, Ouyang TX, Flickinger MC. Starvation induced expression of sspA and sspB: The effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994;11:1029–1043. doi: 10.1111/j.1365-2958.1994.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 36.Tal N, Schuldiner S. A coordinated network of transporters with overlapping specificities provides a robust survival strategy. Proc Natl Acad Sci USA. 2009;106:9051–9056. doi: 10.1073/pnas.0902400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.