Abstract
Chemokines and chemokine receptors are key evolutionary innovations of vertebrates. They are involved in morphogenetic processes and play an important role in the immune system. Based on an analysis of the chemokine receptor gene family in teleost genomes, and the expression patterns of chemokine receptor genes during embryogenesis and the wounding response in young larvae of Oryzias latipes, we identified the chemokine receptor cxcr3a as a marker of innate immune cells. Cells expressing cxcr3a were characterized in fish transgenic for a cxcr3a:gfp reporter. In embryos and larvae, cxcr3a-expressing cells are motile in healthy and damaged tissues, and phagocytic; the majority of these cells has the morphology of tissue macrophages, whereas a small fraction has a dendritic phenotype. In adults, cxcr3a-positive cells continue to specifically express myeloid-associate markers and genes related to antigen uptake and presentation. By light microscopy and ultrastructural analysis, the majority of cxcr3a-expressing cells has a dendritic phenotype, whereas the remainder resembles macrophage-like cells. After challenge of adult fish with bacteria or CpG oligonucleotides, phagocytosing cxcr3a-positive cells in the blood up-regulated il12p40 genes, compatible with their function as part of the mononuclear phagocytic system. Our results identify a marker of teleost mononuclear phagocytic cells and suggest a surprising degree of morphological and functional similarity between the innate immune systems of lower and higher vertebrates.
Keywords: dendritic cell, chemokine receptor, immune system, evolution, il12p40
Despite the fact that the majority of vertebrate species lives in aquatic environments, the analysis of their immune systems has lagged behind that of their land-living relatives. However, in recent years, interest in the organization of immune systems in fish has grown substantially (1–4).
During development of the vertebrate embryo, chemokines and their corresponding chemokine receptors play important roles in morphogenetic processes, including the formation of central and peripheral nervous systems and gonads (5–8). The exceptional functional versatility of the chemokine/chemokine receptor system is also used in the immune system. There, it is not only involved in the early development of lymphocytes and innate effector cells and in the elaboration of secondary lymphoid organs and tertiary lymphoid structures (1, 9–11), but it also directly regulates the immune response (12–14).
Chemokines and chemokine receptors are a key evolutionary innovation of vertebrates (1, 15); however, the species- and group-specific diversifications of this gene family have not yet been fully addressed. It is possible that the diversification of the chemokine/chemokine receptor pairs scales with the complexity of innate and adaptive effector cell populations and their functional integration in the two arms of vertebrate immune systems. In this context, the analysis of lower vertebrates, such as fish, is of particular interest. Fish are thought to lack organized secondary lymphoid organs (4), yet they exhibit robust immune responses that occur in the absence of a recognizable morphological framework. Because chemokines and chemokine receptors are differentially expressed in resting and activated stromal and hematopoietic cells, their activities can be used to define functionally distinct subsets of effector cells. Lower vertebrates possess many cell types that are characteristic of the adaptive and innate immune systems of mammals. The sites of origin and developmental pathways of lymphocytes (1, 16, 17), as well as those of the myeloid lineages (macrophages, neutrophils) (18–22), have been well characterized. This was achieved by use of cell type-specific antibodies recognizing characteristic and evolutionarily conserved cell surface molecules or, for some species—most notably zebrafish—by use of transgenic fish in which the expression of fluorescent reporter proteins is directed by the regulatory regions of specific genes to allow the in vivo visualization and preparative recovery of individual cell types. These studies suggest that, overall, the immune systems of lower and higher vertebrates are surprisingly similar, as is also evident from genetic studies probing the function of key regulators of the immune system (23–25).
Among the least studied effector cell types of the immune system in fish are dendritic cells. This is surprising because dendritic cells play a pivotal role in the regulation of the immune response as professional antigen presenting cells (26), although they are heterogeneous with respect to function and developmental origin (27). Indeed, the distinctions among tissue macrophages, monocytes, and dendritic cells become increasingly blurred, as their developmental and lineage relationships are being explored by genetic means (28). Hence, the characterization of mononuclear phagocytic cells in evolutionarily older types of vertebrates has the potential to provide essential information about their phenotypic and functional diversity.
Here, we report that expression of a fish homologue of the mammalian Cxcr3 gene (29) occurs in mononuclear phagocytic cells and describe their phenotypic and functional characteristics in embryonic, larval and adult stages of the teleost Oryzias latipes.
Results and Discussion
Chemokine Receptor Gene Repertoire of Teleost Fish.
We established the phylogenetic relationships of teleost chemokine receptors using genome sequences of two evolutionarily distant fish species, tetraodon (Tetraodon nigroviridis) (30) and medaka (O. latipes) (31). The overall topology of our phylogenetic tree incorporating mammalian and teleost sequences is similar to that derived from mammalian sequences only (32), and compatible with a recent study of Cc-type chemokine receptors in teleosts (33). Interestingly, it appears that tetraodon and medaka lack recognizable orthologues of Ccr1, Ccr2, Ccr3, Ccr4, Ccr5, Ccr11, CcrL1, and Cxcr6 genes; a previous report (33) used a somewhat different nomenclature (Fig. 1A, Fig. S1A, and Table S1). The teleost genomes are distinguished by the presence of two or three copies of several chemokine receptor genes for which there is only one copy known in mammals; some extra copies might be the consequence of the genome-wide duplication events that are thought to have occurred early in vertebrate evolution (34) and additionally in teleosts (35), but others might have resulted from local duplication events, perhaps as a result of adaptation to distinct environments and/or lifestyles. Indeed, our results provide evidence for species-specific differences between T. nigroviridis and O. latipes. Furthermore, our data show that certain chemokine receptor genes (mostly those of the Ccr type) have evolved after the split of tetraodon and medaka. Overall, the number of chemokine receptor genes encoded by teleost and mammalian genomes is not very different, although fish possess more chemokine receptors of the Cxc type. It will be interesting to additionally (36) examine the complexity of the corresponding Cxc chemokine gene family to determine whether differential expansion has also occurred among the ligands of the known Cxc chemokine receptors.
Fig. 1.
The chemokine receptor gene family in two teleost species. (A) Phylogenetic tree based on derived protein sequences of chemokine receptor genes of human (prefix Hs), mouse (prefix Mm), medaka (prefix Ol), and tetraodon (prefix Tn) genomes. The sequence for the human formyl-peptide receptor FPR1 was used as an outgroup. (B) Expression of chemokine receptor genes in medaka from 5 dpf until larval stage. White box indicates no expression, filled box indicates expression detected by WISH.
The cxcr3 cluster in medaka probably arose as a result of two local duplication events, which, according to conserved synteny relationships with flanking genes (Fig. S1B), appear to have occurred in the last common ancestor of zebrafish (Danio rerio) and medaka (note that tetraodon has only two cxcr3-like genes; the currently available genome assembly of tetraodon precludes synteny analysis). Phylogenetic analysis indicates that the cxcr3a genes represent a diverged form of cxcr3 genes (Fig. S1C); a previously described cxcr3 gene from the grass carp (Ctenopharyngodon idella) (37) corresponds to cxcr3c (Fig. S1C).
Expression of Chemokine Receptor Genes During Embryogenesis.
To examine which of the chemokine receptor genes are expressed in early embryogenesis, and hence before the appearance of mature effector cells of the adaptive immune system, whole-mount RNA in situ hybridization (WISH) studies were performed. In medaka embryos [5-12 d post fertilization (dpf)], only six of 23 chemokine receptor genes are expressed (Fig. 1B, Fig. S2, and Table S1). Interestingly, whereas only one of three paralogues of cxcr3 is expressed (cxcr3a), both paralogues of cxcr4 and ccr9 are coexpressed. The latter have been shown to be important for migration events in the early embryo, regulating the movement of primordial germ cells, neuromasts (38), and lymphoid precursors (1). Hence, based on the differential expression of Cxcr3 in diverse types of mammalian immune effector cells (29, 39), it appears possible that cxcr3a is expressed in cells of the innate immune system of medaka embryos.
Expression of Chemokine Receptor Genes During Wounding Response.
Chemokine and chemokine receptor signaling pairs function in morphogenesis, migration, and response to different kinds of tissue damage. We examined the expression patterns of all chemokine receptor genes identified in the medaka genome, before and shortly after wounding of young larvae (2–5 d after hatching). Initially, we determined the kinetics of leukocyte accumulation at the wounded site by Sudan black staining, which identifies cells of the myeloid lineage. A strong accumulation of positive cells was observed within the first 1 h after the insult; these aggregates remained for at least 24 h (Fig. S3A). Based on these findings, we determined chemokine receptor gene expression during immediate (1 h after insult) and late responses (16 h after insult). For all chemokine receptors examined, the basal levels of expression are very low in the tailfin region (Fig. S3B). However, after wounding, expression increases for a number of genes, although the kinetics appear to be different. The chemokine receptors cxcr4b and cxcr7 were induced in the injured tissue immediately after insult, and their expression levels remained high when examined 16 h later. Interestingly, the expression of the cxcl12a gene, encoding one of their ligands, is also rapidly induced in this region (Fig. S3C). In contrast to other chemokine receptor genes, expression of cxcr4b and cxcr7 might also occur in stromal cells of the fin (40). Other chemokine receptors appear to be expressed in migratory cells; cxcr1, cxcr3a, and xcr1a are expressed shortly after wounding, whereas expression of cxcr5, ccr9a, and ccr9b occurs later.
Development and Function of cxcr3a-Expressing Cells in Embryos and Larvae.
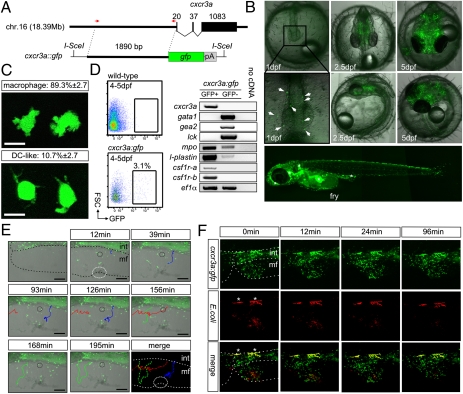
The aforementioned experiments indicated that five chemokine receptor genes (cxcr3a, cxcr4b, cxcr7, ccr9a, and ccr9b) are expressed during embryonic development and the wounding response in larvae. Because the functional roles of cxcr4, cxcr7, and ccr9 chemokine receptor genes have been well characterized in fish, we focused on cxcr3a in our subsequent experiments. To facilitate the temporospatial expression analysis of cxcr3a, we generated transgenic reporter lines (Fig. 2A). Comparison with expression patterns obtained by RNA in situ analysis indicated that the transgenic reporter faithfully reflects the expression of the endogenous cxcr3a gene (compare Fig. 2B vs. Fig. S2; adult stages are detailed later). In transgenic fish, fluorescent cells were detected at 1 dpf and were located in the rostral blood island (2), compatible with the expression in embryonic macrophages; with time, fluorescent cells became more numerous until they were found in many parts of the larvae (Fig. 2B). As observed by in vivo imaging of single cells in transgenic embryos, the majority of GFP-positive cells have the morphology of macrophages, whereas a small fraction of cxcr3a-expressing cells display the morphological features of dendritic cells (Fig. 2C). As determined in larval stages, cxcr3a-positive cells are highly motile (Movie S1). To examine the molecular phenotype of cxcr3a-expressing cells more closely, we isolated them by FACS (Fig. 2D) for subsequent RT-PCR analysis; approximately 30 to 60 GFP-positive cells could be recovered per embryo. These cells expressed endogenous cxcr3a, as expected, and also expressed myeloid-associated markers, such as mpo, csf1r, and l-plastin, but lacked expression of genes characteristic of red blood and T cells (Fig. 2D). By contrast, GFP-negative cells do not express cxcr3a, as expected; additional differences are the lack of expression of csfr1a and csfr1b, only low levels of expression of mpo and l-plastin, and high levels of erythroid and lymphoid markers (Fig. 2D). As cxcr3a is expressed immediately after wounding (Fig. S3B), it was of interest to examine the behavior of cxcr3a-expressing cells during this process using the GFP reporter system. We observed three types of cell behavior in transgenic larvae (Fig. 2E and Movie S2). Some GFP-positive cells move only within a small area (black circles, Fig. 2E and Movie S2), whereas others traverse the region of interest seemingly without regard for the lesion (red and green trajectories, Fig. 2E and Movie S2). Other cells appear to respond to the wound and directly approach the tissue defect (blue trajectory, Fig. 2E and Movie S2). This analysis indicates that cells expressing cxcr3a are functionally heterogeneous with regard to their response to a tissue lesion; interestingly, in zebrafish, evidence for functionally distinct types of macrophages has been obtained (41). The cxcr3a:gfp transgenic line also enabled us to examine whether these cells participated in the immune response to septic insults. To this end, a septic lesion was introduced into the region of the abdominal fin of young larvae (2–5 d after hatching) with red fluorescence-expressing Escherichia coli bacteria. Within approximately 30 min, most bacteria were cleared from the lesion (Fig. 2F and Movie S3). The presence of yellow cells indicates that cxcr3a-expressing cells are capable of phagocytosing foreign material, as confirmed by high-resolution imaging (Movie S4).
Fig. 2.
Characterization of cxcr3a-expressing cells in O. latipes embryos and larvae. (A) Schematic diagram of the transgenic construct indicating the position of the 1.9-kb promoter fragment. (B) Fluorescence microscopy of transgenic embryos at the indicated time points (in dpf). Positive cells identifiable at 1 dpf are indicated. In the panel depicting the fry, merged from four pictures, regions with autofluorescent pigment cells are indicated by an asterisk. Note that the pattern of fluorescence is similar to the distribution of cxcr3a-mRNA as determined by whole-mount in situ hybridization (Fig. S1). (C) Morphology of GFP-positive cells. A total of 664 cells from 13 embryos (51 ± 17 cells per embryo) were observed by live imaging of 5 dpf embryos and categorized as macrophages (two representative examples are shown, Upper) or dendritic-like cells (Lower). (D) Flow cytometric profiles of WT and transgenic embryos (Left) and representative results from RT-PCR reactions. GFP-positive cells express myeloid-associated markers, whose expression is greatly reduced or not detectable in the GFP-negative fraction. By contrast, genes specifically expressed in the erythroid (gata1, embryonic alpha globin 2 [gea2]) and T cell (lck) lineages are exclusively expressed in GFP-negative cells. Note that the flow cytometric analysis is not quantitative, because only mechanical disruption but no enzymatic digestion of tissues was used. Results are representative of two independent cell sortings from pooled embryos and subsequent duplicate cDNA analyses. (E) Still photographs taken at various time points after wounding in the membranous fin (mf) of transgenic larvae. The region of insult is marked by a white dotted circle. Three different migration behaviors are depicted by different colors. Blue and green cells migrate from the intestine (int) region into the insult (Movie S2). (Scale bar, 100 μm.) (F) Time course of bacterial clearance in transgenic larvae. Regions of autofluorescent cells are indicated by an asterisk. Note the appearance of yellow cells, indicative of phagocytosis by GFP-positive cells; also see Movies S3 and S4.
Gene Expression Signature and Morphology of Adult cxcr3a-Expressing Cells.
In adult tissues, GFP-expressing cells have the light-scatter characteristics of myelomonocytic cells (Fig. 3A and Table S2) and express endogenous cxcr3a (Fig. 3B), indicating that the reporter expression reflects endogenous gene expression. Next, we examined the gene expression profiles of cell populations isolated from different tissues, such as blood, kidney (the major hematopoietic site in adult fish), and the spleen (the only known organized secondary lymphoid tissue). We found that cxcr3a-positive cells lack markers specifically expressed in red blood cells and lymphocytes but express myeloid-associated markers, such as csfr1, mpo, and pu.1; as expected for cells involved in antigen uptake and presentation, expression of genes encoding Toll-like receptors, mhc class II, irf family members, and nfkb components was detectable (Fig. 3 C and D and Fig. S4 A and B). flt3, encoding a transmembrane receptor tyrosine kinase and a specific marker for dendritic cells in mammals, was also expressed in cxcr3a-positive cells (Fig. 3 C and D and Fig. S4). GFP-positive cells isolated from different tissues differ with respect to the expression of some genes, such as irf4 and tlr9 (Fig. S4B) and several chemokine receptors (Fig. S4C), pointing to potential developmental and/or functional heterogeneity. Immature thymocytes express rag1, cd4, and cd8a, but not cxcr3a; conversely, cxcr3a-positive cells are negative for these T cell markers (Fig. S5).
Fig. 3.
Gene expression profiles and morphology of cxcr3a-positive cells in adult O. latipes. (A) Light-scatter properties of adult GFP-expressing cells isolated from whole kidney marrow. Note that the majority of cells (green dots) falls into the myelomonocytic gate (red circle). Similar results were obtained for GFP-expressing cells isolated from blood and spleen. Results are representative of three independent experiments from pooled tissues. (B) Cells expressing cxcr3a are confined to the cxcr3a:gfp-positive fraction of blood cells from adult fish. Single myelomonocytic cells were separated according to fluorescence and examined for cxcr3a-expression by RT-PCR. (C) Expression profiles of GFP-positive and GFP-negative cells isolated from whole kidney marrow of adult cxcr3a:gfp medaka fish. Results are representative of two independent cell sorting experiments from pooled tissues and subsequent duplicate cDNA analyses. (D) Expression profiles of GFP-positive and GFP-negative cells isolated from blood of adult cxcr3a:gfp medaka. Results are representative of duplicate cDNA analyses. (E) Distribution of cell types (mean ± SD) among GFP-positive and GFP-negative cells isolated from blood of adult cxcr3a:gfp medaka as revealed by Wright-Giemsa staining. A total of 805 GFP-positive cells (from a total of five fish; i.e., five independent cell sorting experiments) and 227 GFP-negative cells of myelomonocytic morphology (from three fish; i.e., three independent cell sorting experiments) were evaluated from cytospin preparations. (Scale bar, 3 μm.) (F) EM images of GFP-expressing cells obtained from myelomonocytic cells of kidney and spleen (Left) and blood (Right) of adult fish processed according to the two protocols described in SI Materials and Methods. Note the presence of electron-dense cytoplasmic extensions and large numbers of cytoplasmic vacuoles. (Scale bars, 1 μm.)
These results suggest that cxcr3a-expressing cells in adult fish might comprise immune effector cells of the myeloid lineage, including dendritic cells. To substantiate this conclusion, we examined the morphology of GFP-positive cells after fluorescence-activated sorting from blood which is the most easily accessible and richest source in medaka (Table S2). As shown in Fig. 3E, the predominant population (approximately 80%) has a dendritic phenotype with long cytoplasmic extensions and an excentric nucleus; the less frequent cells (15%) have a macrophage-like morphology distinguished by agranular cytoplasm and a high ratio of cytoplasm to nuclei; indeed, a cxcr3 homologue was previously found to be expressed in trout head kidney macrophages (42). By contrast, cells with the morphological characteristics of neutrophils and monocytes predominate in myelomonocytic cells of the GFP-negative fraction. High-resolution ultrastructural analysis of cxcr3a-positive cells by EM confirms that the majority of cells in blood and hematopoietic tissues exhibit a phenotype compatible with dendritic cells (Fig. 3F). Hence, by means of the cxcr3a:gfp reporter, myelomonocytic cells of dendritic phenotype can be isolated and substantially enriched from fish tissues. Using antibodies directed against GFP, the presence of cxcr3a-expressing cells can also be studied in situ. Indeed, cxcr3a-positive cells are present in the major lymphoid organs (thymus and spleen) and in the central nervous system; in the skin of adult fish, they form a dense network of cells (Fig. S6).
Functional Characteristics of Adult cxcr3a-Expressing Cells.
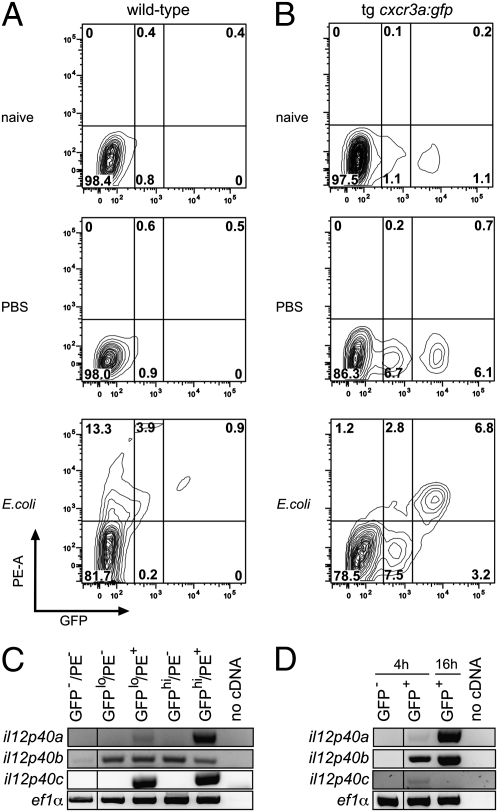
Next, we examined the phagocytic properties of GFP-positive blood cells. To this end, adult fish received intracoelomic injections of red fluorescently labeled inactivated E. coli bacteria. Two hours later, blood cells were collected and cells in the myelomonocytic gate were examined for red and green fluorescence. After injection of bacteria into WT fish, a significant fraction of the blood myelomonocytic cells exhibited red fluorescence (Fig. 4A), indicating that these cells gain access to intracoelomic material. In naive transgenic cxcr3a:gfp fish, two populations of GFP-positive cells (designated GFPlow and GFPhigh) could be distinguished. Their proportions increased upon injection of sterile salt solution, possibly reflecting a wounding response (Fig. 4B, Top and Middle); this increase is augmented by injection of bacteria (Fig. 4B, Bottom). Notably, the GFPlow population phagocytoses to a lesser extent than the GFPhigh cells. To examine whether phagocytosing cells become activated, we examined the expression levels of il12p40 genes, which encode key effectors of activated dendritic cells in mammals (43). Like the situation in the carp (44), the genome of medaka fish contains three paralogues of the mammalian IL12p40 gene (Fig. S7A). Expression of il12p40 genes in GFP-negative and GFP-positive cells isolated from the blood of naive animals is not detectable (Fig. S7B). By contrast, expression of il12p40 genes increases after antigen challenge. Two hours after exposure to bacteria, expression of two genes (il12p40a and il12p40c) becomes clearly detectable in phagocytosing GFP-positive cells, but not in GFP-negative or nonphagocytosing GFP-positive cells. The response of the il12p40b gene appears to be different; first, it is expressed at low levels in both GFP-negative and GFP-positive cells (the latter exhibiting a somewhat higher level), and second, no difference between phagocytosing and nonphagocytosing cells are apparent (Fig. 4C). Next, we examined whether GFP-positive blood cells are also activated by CpG oligonucleotides, a known ligand of the Tlr9 receptor that is highly expressed by these cells (Fig. 3 and Fig. S4). Compared with GFP-negative cells, all three il12p40 paralogues are expressed at higher levels at 4 h after injection, with il12p40b showing the strongest response; interestingly, whereas expression levels of il12p40a and il12p40b genes in GFP-positive cells are even higher at 16 h after challenge, the expression of il12p40c gene is no longer detectable (Fig. 4D). Whether differential responses of il12p40 paralogues are of functional significance remains to be seen.
Fig. 4.
Functional analysis of cxcr3a-positive cells. (A) WT adult fish received intracoelomic injections of red fluorescently labeled bacteria (or vehicle alone), and 2 h later, blood cells were analyzed by flow cytometry (gated for cells with light scatter characteristics of myelomonocytic cells). The proportions of cells are indicated (representative patterns for three animals are shown). Results are representative of at least three independent experiments. (B) cxcr3a:gfp transgenic adult fish received intracoelomic injections of red fluorescently labeled bacteria (or vehicle alone), and 2 h later, blood cells were analyzed by flow cytometry as in A. The proportions of myelomonocytic cells in the individual gates are indicated (representative patterns for at least three animals are shown). Note that the proportions of both GFPlow and GFPhigh cells increase in injected fish and that the majority of phagocytosing cells are GFP-positive. (C) Phagocytosis induces expression of il12p40 genes. Adult cxcr3a:gfp transgenic fish received intracoelomic injections of red fluorescently labeled bacteria, and 2 h later, blood cells were separated by flow cytometry according to their fluorescence characteristics. GFP-negative cells express erythroid (gata1) and lymphocyte (lck) lineage markers, whereas GFP-positive cells do not (cf. Fig. 3C). GFP-positive cells are separated into those expressing low (lo) and high (hi) levels and whether they have phagocytosed red fluorescently labeled bacteria (PE+ or PE−). Results for the three il12p40 paralogues (Fig. S7) are shown. Results are representative of two independent experiments (i.e., cell sorting followed by duplicate cDNA analyses). (D) CpG oligonucleotides induce il12p40 gene expression. Adult cxcr3a:gfp fish were given intracoelomic injections; GFP-negative and GFP-positive blood cells were isolated by flow cytometry 4 h after injection and processed for RT-PCR analysis. For GFP-positive cells, the results for the 16-h time point after injection are also shown. Results are representative of two independent experiments (i.e., cell sorting followed by duplicate cDNA analyses).
Conclusions.
Here, we report on the development, phenotype, and function of the cxcr3a-expressing subset of mononuclear phagocytic cells in the teleost O. latipes. In embryonic, larval, and adult stages, the transgene marks phenotypically and functionally diverse cell types that bear the hallmarks of cells involved in antigen uptake and presentation. Whether cxcr3a expression marks all types or only a subset of mononuclear phagocytic cells in fish remains to be investigated. Our studies provide the starting point for more detailed analysis of these cells in teleosts. For instance, the information about cxcr3a expression can be exploited for the development of a genetic marking system to clarify the lineage relationship among phenotypically distinguishable cxcr3a-positive cells. Moreover, as our transgenic reporter provides a facile means for their detection in and isolation from complex mixtures of cells and tissues, detailed in vivo and in vitro studies of these cells are now possible. Interestingly, a large fraction of cxcr3a-positive cells in adult fish resembles dendritic cells in higher vertebrates. This conclusion is based on their morphological features, a characteristic pattern of gene expression (including the cytokine receptor flt3 and the chemokine receptor cxcr3a), and their activation by bacteria and CpG oligonucleotides. However, whether such cells indeed represent one or more of the much better characterized subsets of mammalian dendritic cells or belong to an evolutionarily more primitive mononuclear phagocytic type of cell is currently unclear. Nonetheless, given the increasingly recognized phenotypic and functional overlap among different subsets of mononuclear phagocytic cells (28, 45), studies in lower vertebrates might inform investigations addressing the lineage relationships among mammalian mononuclear phagocytic cells. Earlier work has hinted at the potential presence of dendritic cells in fish, when cells with their morphological characteristics were described in tissue sections (46). Indirect evidence derived from cloning of genes encoding characteristic surface molecules also suggests the existence of dendritic cells in teleosts (47–50) and in cartilaginous fish (50), the most primitive group of jawed vertebrates. More recently, Lugo-Villarino et al. have identified and functionally characterized myelomonocytic cells resembling dendritic cells in zebrafish (51). Collectively, these studies suggest that cells equivalent to mammalian dendritic cells might be an ancient component of the vertebrate immune system.
Materials and Methods
Phylogenetic Analyses.
Details of phylogenetic analyses can be found in SI Materials and Methods.
Animal Husbandry.
The Cab strain of WT medaka was used for the experiments. Embryos were staged according to Iwamatsu (52).
Transgenic Line.
Details on the construction of cxcr3a:gfp transgenic lines can be found in SI Materials and Methods.
Wounding Assay.
The tail or membranous fins were cut or punctured, respectively, and Sudan black staining was performed at various time points thereafter according to Le Guyader et al. (22).
CpG Injection.
Adult transgenic F3 fish (approximately 1 g body weight) were given intracoelomic injections of 5 μg type B CpG oligonucleotide (ODN 1668) or vehicle (control). Four or 16 h after immunization, GFP-positive cells were isolated from blood.
Phagocytosis Assay.
Fish were given intracoelomic injections of 20 μg of heat-inactivated E. coli (K-12 strain) conjugated with Alexa-594 (Molecular Probes). Two hours later, blood cells were analyzed by flow cytometry.
EM.
GFP-positive cells were fixed in 4% paraformaldehyde and 4% glutaraldehyde solution and processed as detailed in SI Materials and Methods.
In Situ Hybridization Analysis.
WISH in medaka was performed with digoxigenin-labeled RNA riboprobes as described previously (53). Probes are listed in Table S1.
FACS and RT-PCR.
GFP-positive cells from embryos, fry, and adult kidney, spleen, and blood from transgenic cxcr3a:gfp or rag1:gfp (17) were sorted with a FACSAria cell sorting system (BD Biosciences). Total RNA isolation and first-strand cDNA synthesis were performed as previously described (1). RT-PCR analyses were performed using the HotStar HiFidelity polymerase kit (Qiagen). For single-cell RT-PCR assays, GFP-positive cells were directly sorted into reverse transcriptase mix (Superscript II; Invitrogen) supplemented with 0.1% Triton X-100 and primers for ef1α (5′-CTTGAACCAGCTCATCTTGTCGC) and cxcr3a (5′-GCCAGGTAACGGTCAAAGCTGATGC). The resulting cDNAs were used for multiplex PCR, and in a second round amplified separately using primers for ef1α and cxcr3a. The source sequences and relevant primer sequences are listed in Table S1.
Live Microscopy of cxcr3c:gfp Fry.
Details for live microscopic analyses can be found in SI Materials and Methods.
Immunohistochemistry.
Paraffin sections (5–7 μm) were processed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank D. Bönsch, D. Schmitt, O. Kuzmenko, and U. Heinzmann for excellent fish care; P. Kindle for help with microscopy; M. Freudenberg (Max Planck-Institute of Immunobiology, Freiburg, Germany) for provision of CpG oligonucleotides; Y. Takahama (University of Tokoshima, Japan) for rag1:gfp transgenic fish; T. Czerny (University of Applied Sciences, Vienna) for the pSgfp vector; and members of the Boehm laboratory for helpful discussions. Financial support for these studies was provided by the Max-Planck Society and the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000467107/-/DCSupplemental.
References
- 1.Bajoghli B, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–197. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 2.de Jong JLO, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 3.Meeker ND, Trede NS. Immunology and zebrafish: Spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Zapata A, Diez B, Cejalvo T, Gutiérrez-de Frías C, Cortés A. Ontogeny of the immune system of fish. Fish Shellfish Immunol. 2006;20:126–136. doi: 10.1016/j.fsi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: A migration from immunology to neurobiology. Prog Neurobiol. 2008;84:116–131. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raz E, Reichman-Fried M. Attraction rules: Germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;16:355–359. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: a fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- 8.Tiveron MC, Cremer H. CXCL12/CXCR4 signalling in neuronal cell migration. Curr Opin Neurobiol. 2008;18:237–244. doi: 10.1016/j.conb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Campbell DJ, Kim CH, Butcher EC. Chemokines in the systemic organization of immunity. Immunol Rev. 2003;195:58–71. doi: 10.1034/j.1600-065x.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 10.Förster R, Pabst O, Bernhardt G. Homeostatic chemokines in development, plasticity, and functional organization of the intestinal immune system. Semin Immunol. 2008;20:171–180. doi: 10.1016/j.smim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Nitta T, Murata S, Ueno T, Tanaka K, Takahama Y. Thymic microenvironments for T-cell repertoire formation. Adv Immunol. 2008;99:59–94. doi: 10.1016/S0065-2776(08)00603-2. [DOI] [PubMed] [Google Scholar]
- 12.Bajénoff M, Germain RN. Seeing is believing: A focus on the contribution of microscopic imaging to our understanding of immune system function. Eur J Immunol. 2007;37(Suppl 1):S18–S33. doi: 10.1002/eji.200737663. [DOI] [PubMed] [Google Scholar]
- 13.Ruddle NH, Akirav EM. Secondary lymphoid organs: Responding to genetic and environmental cues in ontogeny and the immune response. J Immunol. 2009;183:2205–2212. doi: 10.4049/jimmunol.0804324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villablanca EJ, Mora JR. A two-step model for Langerhans cell migration to skin-draining LN. Eur J Immunol. 2008;38:2975–2980. doi: 10.1002/eji.200838919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordström KJV, Fredriksson R, Schiöth HB. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol. 2008;8:9. doi: 10.1186/1471-2148-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kissa K, et al. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Iwanami N, Hoa VQ, Furutani-Seiki M, Takahama Y. Noninvasive intravital imaging of thymocyte dynamics in medaka. J Immunol. 2007;179:1605–1615. doi: 10.4049/jimmunol.179.3.1605. [DOI] [PubMed] [Google Scholar]
- 18.Hall C, Flores MV, Crosier K, Crosier P. Live cell imaging of zebrafish leukocytes. Methods Mol Biol. 2009;546:255–271. doi: 10.1007/978-1-60327-977-2_16. [DOI] [PubMed] [Google Scholar]
- 19.Hanington PC, et al. Development of macrophages of cyprinid fish. Dev Comp Immunol. 2009;33:411–429. doi: 10.1016/j.dci.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 21.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 22.Le Guyader D, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 23.Petrie-Hanson L, Hohn C, Hanson L. Characterization of rag1 mutant zebrafish leukocytes. BMC Immunol. 2009;10:8. doi: 10.1186/1471-2172-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schorpp M, et al. Conserved functions of Ikaros in vertebrate lymphocyte development: Genetic evidence for distinct larval and adult phases of T cell development and two lineages of B cells in zebrafish. J Immunol. 2006;177:2463–2476. doi: 10.4049/jimmunol.177.4.2463. [DOI] [PubMed] [Google Scholar]
- 25.Trede NS, et al. Zebrafish mutants with disrupted early T-cell and thymus development identified in early pressure screen. Dev Dyn. 2008;237:2575–2584. doi: 10.1002/dvdy.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinman RM. Dendritic cells: Understanding immunogenicity. Eur J Immunol. 2007;37(Suppl 1):S53–S60. doi: 10.1002/eji.200737400. [DOI] [PubMed] [Google Scholar]
- 27.Segura E, Villadangos JA. Antigen presentation by dendritic cells in vivo. Curr Opin Immunol. 2009;21:105–110. doi: 10.1016/j.coi.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Callahan MK, Huang D, Ransohoff RM. Chemokine receptor CXCR3: an unexpected enigma. Curr Top Dev Biol. 2005;68:149–181. doi: 10.1016/S0070-2153(05)68006-4. [DOI] [PubMed] [Google Scholar]
- 30.Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 31.Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- 32.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Chang MX, Wu SG, Nie P. Characterization of C-C chemokine receptor subfamily in teleost fish. Mol Immunol. 2009;46:498–504. doi: 10.1016/j.molimm.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 35.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 36.Peatman E, Liu Z. Evolution of CC chemokines in teleost fish: a case study in gene duplication and implications for immune diversity. Immunogenetics. 2007;59:613–623. doi: 10.1007/s00251-007-0228-4. [DOI] [PubMed] [Google Scholar]
- 37.Chang MX, Sun BJ, Nie P. The first non-mammalian CXCR3 in a teleost fish: gene and expression in blood cells and central nervous system in the grass carp (Ctenopharyngodon idella) Mol Immunol. 2007;44:1123–1134. doi: 10.1016/j.molimm.2006.07.280. [DOI] [PubMed] [Google Scholar]
- 38.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2009;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Vanbervliet B, et al. The inducible CXCR3 ligands control plasmacytoid dendritic cell responsiveness to the constitutive chemokine stromal cell-derived factor 1 (SDF-1)/CXCL12. J Exp Med. 2003;198:823–830. doi: 10.1084/jem.20020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dufourcq P, Vriz S. The chemokine SDF-1 regulates blastema formation during zebrafish fin regeneration. Dev Genes Evol. 2006;216:635–639. doi: 10.1007/s00427-006-0066-7. [DOI] [PubMed] [Google Scholar]
- 41.Mathias JR, et al. Characterization of zebrafish larval inflammatory macrophages. Dev Comp Immunol. 2009;33:1212–1217. doi: 10.1016/j.dci.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol. 2008;181:3310–3322. doi: 10.4049/jimmunol.181.5.3310. [DOI] [PubMed] [Google Scholar]
- 43.Cooper AM, Khader SA. IL-12p40: An inherently agonistic cytokine. Trends Immunol. 2007;28:33–38. doi: 10.1016/j.it.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Huising MO, et al. The presence of multiple and differentially regulated interleukin-12p40 genes in bony fishes signifies an expansion of the vertebrate heterodimeric cytokine family. Mol Immunol. 2006;43:1519–1533. doi: 10.1016/j.molimm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–426. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 46.Lovy J, Wright GM, Speare DJ. Comparative cellular morphology suggesting the existence of resident dendritic cells within immune organs of salmonids. Anat Rec (Hoboken) 2008;291:456–462. doi: 10.1002/ar.20674. [DOI] [PubMed] [Google Scholar]
- 47.Doñate C, et al. CD83 expression in sea bream macrophages is a marker for the LPS-induced inflammatory response. Fish Shellfish Immunol. 2007;23:877–885. doi: 10.1016/j.fsi.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 48.Lin A-F, et al. The DC-SIGN of zebrafish: Insights into the existence of a CD209 homologue in a lower vertebrate and its involvement in adaptive immunity. J Immunol. 2009;183:7398–7410. doi: 10.4049/jimmunol.0803955. [DOI] [PubMed] [Google Scholar]
- 49.Lovy J, Savidant GP, Speare DJ, Wright GM. Langerin/CD207 positive dendritic-like cells in the haemopoietic tissues of salmonids. Fish Shellfish Immunol. 2009;27:365–368. doi: 10.1016/j.fsi.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Ohta Y, et al. Homologs of CD83 from elasmobranch and teleost fish. J Immunol. 2004;173:4553–4560. doi: 10.4049/jimmunol.173.7.4553. [DOI] [PubMed] [Google Scholar]
- 51.Lugo-Villarino G, et al. Identification of dendritic antigen-presenting cells in the zebrafish. Proc Natl Acad Sci USA. 2010;107:15850–15855. doi: 10.1073/pnas.1000494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Aghaallaei N, Bajoghli B, Czerny T. Distinct roles of Fgf8, Foxi1, Dlx3b and Pax8/2 during otic vesicle induction and maintenance in medaka. Dev Biol. 2007;307:408–420. doi: 10.1016/j.ydbio.2007.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.