Abstract
MicroRNAs (miRNAs) are a class of small, noncoding RNAs that function as posttranscriptional regulators of gene expression. Many miRNAs are expressed in the developing brain and regulate multiple aspects of neural development, including neurogenesis, dendritogenesis, and synapse formation. Rett syndrome (RTT) is a progressive neurodevelopmental disorder caused by mutations in the gene encoding methyl-CpG–binding protein 2 (MECP2). Although Mecp2 is known to act as a global transcriptional regulator, miRNAs that are directly regulated by Mecp2 in the brain are not known. Using massively parallel sequencing methods, we have identified miRNAs whose expression is altered in cerebella of Mecp2-null mice before and after the onset of severe neurological symptoms. In vivo genome-wide analyses indicate that promoter regions of a significant fraction of dysregulated miRNA transcripts, including a large polycistronic cluster of brain-specific miRNAs, are DNA-methylated and are bound directly by Mecp2. Functional analysis demonstrates that the 3′ UTR of messenger RNA encoding Brain-derived neurotrophic factor (Bdnf) can be targeted by multiple miRNAs aberrantly up-regulated in the absence of Mecp2. Taken together, these results suggest that dysregulation of miRNAs may contribute to RTT pathoetiology and also may provide a valuable resource for further investigations of the role of miRNAs in RTT.
Keywords: Brain-derived neurotrophic factor, massively parallel sequencing, Dlk1-Gtl2 imprinting locus, DNA methylation, cerebella
Rett syndrome (RTT; Mendelian Inheritance in Man 312750) is an autism-spectrum disorder caused primarily by loss-of-function mutations in the X-linked methyl-CpG–binding protein 2 (MECP2) gene (1). After apparently normal development for 6–18 mo, RTT patients acquire severe neurological symptoms including breathing irregularities, ataxia, seizures, autistic traits, and mental retardation (2). They also show regression of head growth and loss of acquired motor skills. Mice with genetic deletion of Mecp2 develop normally until 6 wk after birth, when they exhibit many symptomatic features of RTT patients, and die prematurely by 12 wk of age (3, 4). Functional and electrophysiological studies of mouse models of RTT have confirmed that aberrant Mecp2 levels lead to deficits in neuronal maturation, synaptogenesis, and neural circuit connectivity (5–7). Furthermore, restoration of wild-type Mecp2 expression in Mecp2-null mice at symptomatic stages can functionally rescue a wide spectrum of disease symptoms and prevent premature death (8). These findings therefore indicate that mouse models of RTT may serve as powerful tools for investigating molecular functions of Mecp2 relevant to disease progression.
Both in vitro biochemical analysis and in vivo chromatin immunoprecipitation (ChIP) assays suggest that Mecp2 specifically binds to methylated DNA and may function as a global transcriptional regulator by recruiting chromatin-remodeling complexes or regulating higher-order chromatin structures (9–13). Although a few neuronal genes have been identified as direct targets for Mecp2 (14, 15), our knowledge about Mecp2-regulated genes and pathways in the brain remains limited. Recent studies have identified an unexpectedly large number of genes that are dysregulated in discrete brain regions of mouse models with aberrant Mecp2 levels (16, 17). Notably, Mecp2-dependent changes in gene expression are small in magnitude, generally less than twofold (17). Thus, Mecp2 may be required for fine-tuning gene expression of a cohort of protein-coding genes through direct and/or indirect mechanisms.
MicroRNAs (miRNAs) are a class of endogenous RNAs that are 21–23 nt in length. These small, noncoding RNAs can base pair to cognate sites in 3′ UTR of protein-coding mRNAs to regulate mRNA translation and/or stability negatively (18, 19). A growing body of evidence suggests that miRNAs play critical roles in normal neuronal functions, and dysregulation of miRNAs is implicated in neurological diseases (19–23). Because hundreds of genes can be directly repressed, albeit each to a modest degree, by individual miRNAs (24, 25), abnormal levels of miRNAs may contribute to disease progression of RTT and related neurological disorders by altering protein output from neuronal mRNAs. However, it is technically challenging for conventional microarray-based technologies to quantify miRNA levels accurately in vivo, because of the small size of miRNAs and high sequence homology among family members. Thus, unbiased high-throughput sequencing approaches are needed to investigate Mecp2-dependent regulation of miRNAs.
In this study, we systematically examined expression levels of all annotated mature miRNAs (∼700 miRNAs) in the cerebella of control (Mecp2+/y; hereafter referred to as “WT”) and mutant (Mecp2−/y; “KO”) mice at multiple stages of disease progression using massively parallel sequencing technologies. We also performed in vivo genome-wide mapping of Mecp2 promoter occupancy to identify miRNAs that may be regulated directly by Mecp2. Moreover, computational analysis and luciferase reporter assays demonstrated that multiple de-repressed miRNAs may negatively regulate the 3′ UTR of Brain-derived neurotrophic factor (Bdnf) mRNA, down-regulation of which has been implicated in RTT etiology (26). Collectively, our unbiased high-throughput screening approaches have identified dysregulated miRNAs in a mouse model of RTT, thereby providing a valuable resource for further investigations of the role of miRNAs in RTT.
Results
Massively Parallel Sequencing of miRNAs in WT and KO Mouse Cerebella.
To identify miRNAs dysregulated in a specific brain region of the RTT mouse model, we focused our analysis on the cerebellum, a brain region highly enriched with Mecp2-expressing neurons and critically involved in motor controls. A number of RTT phenotypes (e.g., ataxia, abnormal gait, and hypoactivity) could be attributed to cerebellar dysfunction. We prepared small RNA libraries from four pairs of WT and KO mouse cerebella at pre-/early-symptomatic stages (6 wk) for SOLiD massively parallel sequencing (Applied Biosystems). The SOLiD platform directly sequences individual RNA species in small RNA libraries, thereby providing an unbiased and quantitative measurement of relative concentrations of individual miRNAs in a highly reproducible manner (SI Appendix, Fig. 1) (27). To minimize variations among sequencing assays, we took advantage of the multiplexing capability of the SOLiD system to sequence control and mutant small RNA libraries simultaneously by using primers with distinct barcodes. We sequenced small RNA libraries prepared from pooled WT and KO cerebella with an average of 3 million short reads per sample. These sequence reads then were mapped to all 590 nonredundant mouse miRNA precursor sequences (a as exemplified by mmu-mir-137 in SI Appendix, Fig. 2), and comparable numbers of reads mapped to known genomic loci encoding miRNA precursors were obtained from WT and KO samples (SI Appendix, Table 1). Notably, the passenger strand (also known as “star sequences”) of miRNA precursors, many of which are transient intermediates during miRNA biogenesis, were identified frequently by our sequencing analysis, suggesting that these sequencing assays are of sufficient depth to detect mature miRNAs expressed at low levels. To validate further the accuracy of the SOLiD sequencing system, we used quantitative TaqMan miRNA quantitative PCR (qPCR) assays to examine a panel of mature miRNAs whose expression levels may span a wide range in the mouse brain. A comparison of the threshold cycles determined by qPCR assays and read counts from sequencing revealed good correlation between these two methods (SI Appendix, Fig. 3A). Together, these results indicate that the SOLiD small RNA sequencing assay can quantify the expression levels of mature miRNAs in vivo reliably over a wide dynamic range.
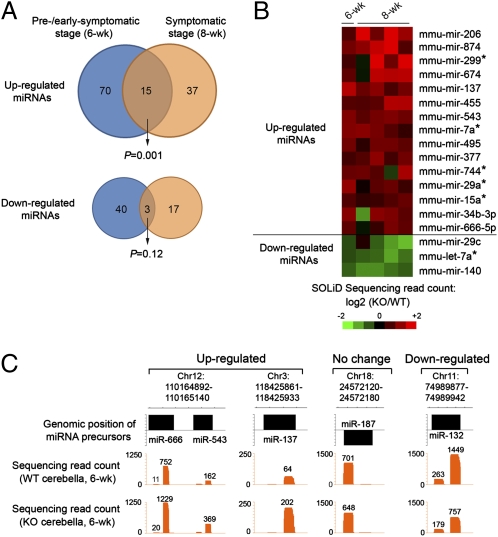
We next identified mature miRNAs differentially expressed in control and mutant cerebella by directly comparing the number of sequencing reads for individual miRNAs. This analysis showed that there were 85 up-regulated and 43 down-regulated mature miRNAs (>1.5-fold and >10 reads) in KO cerebella compared with WT (Fig. 1A and SI Appendix, Table 2). By normalizing to the level of the ubiquitously expressed U6 spliceosomal RNA, TaqMan qPCR assays successfully validated a majority (11 of 13) of randomly selected dysregulated miRNAs identified by SOLiD sequencing (SI Appendix, Fig. 3B). Our sequencing analysis also confirmed several miRNAs previously found to be dysregulated in cultured Mecp2-null postnatal cortical neurons (miR-132) or adult hippocampal neural stem cells (e.g., miR-137, miR-222, and miR-221) (SI Appendix, Table 2) (28, 29), suggesting that Mecp2-dependent regulation of these miRNAs may be conserved in other brain regions. Furthermore, results from both SOLiD sequencing and TaqMan qPCR assays indicated that most dysregulated miRNAs were associated with moderate (less than twofold) changes in expression in absence of Mecp2, in agreement with the finding that Mecp2 is generally involved in fine-tuning the gene expression levels (17).
Fig. 1.
Systematic identification of dysregulated miRNAs in WT and KO cerebella. (A) Venn diagram of the number of up- or down-regulated miRNAs detected in Mecp2-null cerebella. Note that a significant portion of aberrantly up-regulated mature miRNAs is detected consistently in Mecp2-null mice at both the pre-/early-symptomatic stage (6 wk postnatal) and the symptomatic stage (8 wk postnatal). (B) Heat map representation of 18 mature miRNAs that are dysregulated consistently in Mecp2-null mice at both the pre-/early-symptomatic stage (four pairs of 6-wk-postnatal WT and KO littermates sequenced as pooled samples) and the symptomatic stage (four pairs of >8-wk-postnatal WT and KO samples sequenced individually). Color intensity represents the degree of differential expression of miRNAs between WT and KO cerebella. (C) Genomic representation (NCBI build 36) of aggregated sequencing reads mapped to dysregulated miRNAs at the pre-/early-symptomatic (6-wk-postnatal) stage. Sequencing read counts for a given mature miRNA sequence are indicated also.
Because distinct sets of miRNAs could be dysregulated before and after the onset of severe disease symptoms, we performed additional SOLiD small RNA sequencing analysis of two litters of mice (two controls and four mutants) 8 wk after birth to investigate how Mecp2 may contribute to dysregulating miRNAs at the symptomatic stage. This analysis identified 52 up-regulated miRNAs and 20 down-regulated miRNAs (averaged fold change >1.5; three or more of four pairs show the same trend) at the symptomatic stage (SI Appendix, Table 3). Further analysis indicated that 15 mature miRNAs were consistently up-regulated in both the pre-/early symptomatic (6 wk) and symptomatic (8 wk) stages, a number significantly higher than expected by chance (P = 0.001, hypergeometric test) (Fig. 1 A and B). In contrast, only three mature miRNAs were found to be reproducibly down-regulated at both ages (P = 0.12). Moreover, TaqMan miRNA qPCR assays confirmed dysregulated miRNAs detected in two stages (miR-137) or specific to the symptomatic stage (miR-34c and miR-101a) (SI Appendix, Fig. 3C). Interestingly, a cohort of other small, noncoding RNAs (e.g., tRNAs, rRNAs, and snRNAs) also became dysregulated in postnatal KO cerebella (SI Appendix, Table 4), suggesting that Mecp2-dependent regulation of noncoding RNAs is not restricted to miRNAs. Collectively, these results suggest that Mecp2 may be required for regulating expression levels of a cohort of miRNAs at multiple stages of disease progression.
In Vivo Genome-Wide Mapping of Mecp2 Occupancy at miRNA Gene Promoters.
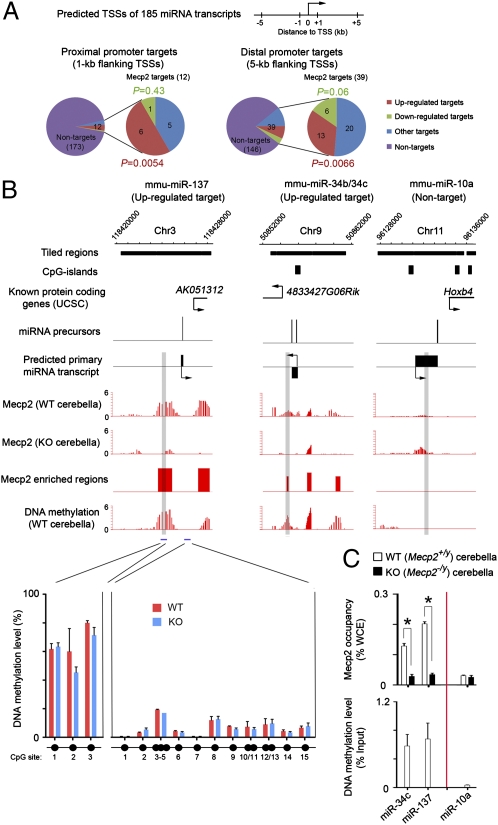
Because KO mice develop the neurological symptoms over a course of several weeks, many dysregulated miRNAs may represent secondary effects caused by long-term deletion of Mecp2. To identify miRNAs directly regulated by Mecp2, we sought to determine Mecp2 occupancy encompassing miRNA gene promoters by ChIP analysis using polyclonal antibodies that were qualified to immunoprecipitate Mecp2 under cross-linking conditions (30) (SI Appendix, Fig. 4). Mature miRNAs are generated from larger primary transcripts (or miRNA transcription units), but transcriptional start sites (TSSs) for most miRNA transcripts are not experimentally defined. To circumvent this problem, we focused on 185 primary miRNA transcripts (specifying at least 336 mature miRNAs) whose TSSs were computationally predicted with high confidence on the basis of histone-modification signatures (H3K4me3 and H3K36me3 for TSSs and transcribed regions, respectively), evolutionary conservation, and available EST data (31). We thus designed customized tiling microarrays to cover the promoter regions of these 185 miRNA transcript units with known/predicted TSSs. Immunoprecipitated DNA and total genomic DNA from postnatal cerebella (6–8 wk) were cohybridized to the customized tiling microarray (NimbleGen) to examine Mecp2 occupancy at genomic regions flanking predicted miRNA gene promoters. Using a sliding window-based statistical algorithm optimized for identifying epigenetic marks with a broad enrichment pattern (DNA methylation and certain histone modifications, e.g., H3K36me3), replicate ChIP-chip experiments showed that Mecp2 was enriched in 1-kb (or 5-kb) genomic regions flanking putative transcription start sites of 12 (or 39) primary miRNA transcripts (1-kb: 6.5%; 5-kb: 21% of 185 predicted miRNA transcripts) (Fig. 2A and SI Appendix, Table 5), as exemplified by miRNA transcripts encoding miR-137 or miR-34b/34c (Fig. 2B). The Mecp2 occupancy within these genomic regions was specific, because Mecp2 ChIP-chip signals were not observed at the same sites in KO cerebella (Fig. 2B). Locus-specific ChIP-qPCR analysis further confirmed these observations (Fig. 2C, Upper). In agreement with the known biochemical function of Mecp2 as a methyl-CpG–binding protein (13), methylated DNA immunoprecipitation followed by the same promoter tiling microarrays (MeDIP-chip) and locus-specific DNA methylation analyses showed that Mecp2-bound regions generally were DNA- methylated (Fig. 2 B and C). In contrast, promoter regions of nontarget miRNAs (e.g., miR-10a) were not DNA-methylated (Fig. 2 B and C). Taken together, these results suggest that Mecp2 binds directly to DNA-methylated promoter regions of a subset of primary miRNA transcripts in mouse postnatal cerebella.
Fig. 2.
MeCP2 binds directly to gene-regulatory regions of a cohort of miRNAs. (A) Number of Mecp2 directly bound miRNA targets that are up-regulated, down-regulated, and unchanged (Other) in 6-wk-postnatal cerebella. MiRNA transcripts without Mecp2-binding sites within 1-kb (proximal) or 5-kb (distal) regions flanking their TSSs are considered nontargets. Note that only up-regulated miRNAs are significantly enriched in both proximal (P = 0.0054, hypergeometric test) and distal (P = 0.0066) Mecp2 miRNA targets. (B) MeCP2 occupancy and DNA methylation (−log10 P value) are shown for two targets (miR-137 and miR-34b/34c) and one nontarget (miR-10a) in WT and KO cerebella. Genomic positions for nearby known protein-coding genes and CpG islands are shown also. Selected regions that are validated by ChIP/MeDIP-qPCR for Mecp2 occupancy and DNA methylation are highlighted in gray. The DNA methylation level within two regions (indicated by blue bars) flanking the miR-137 gene is validated further by bisulphite conversion followed by mass spectrometry (Sequenom). (C) ChIP of endogenous Mecp2 occupancy (Upper) and MeDIP-based DNA methylation analyses (Lower) for two miRNA targets and one nontarget in WT and KO cerebella. Error bars represent SEM. *P < 0.01.
Mecp2 occupancy at DNA-methylated gene promoters may promote transcription silencing or activation by recruiting transcriptional corepressor complexes (e.g., mSin3a/Hdac) or activators, respectively (14, 15, 17). To elucidate the regulatory effect of Mecp2 on miRNA biogenesis at the transcriptional level, we cross-referenced Mecp2 ChIP-chip results to the expression profiles of miRNAs. Six (50.0%) of 12 proximal promoter targets were up-regulated at the pre-/early-symptomatic stage (Fig. 2A, Left and SI Appendix, Table 5), a percentage significantly higher than expected by chance (P = 0.0054, hypergeometric test). In contrast, only one proximal promoter target of Mecp2 was down-regulated (8.3%, P = 0.43). Similar results were obtained for distal promoter targets (Fig. 2A, Right). Notably, although not all mature miRNAs from the same polycistronic transcript showed more than 1.5-fold change in KO cerebella, mature miRNAs in the same transcript that failed to display >1.5-fold difference generally exhibited a similar trend of change in expression. Thus, we concluded that Mecp2 binding at the proximal promoter of miRNA transcripts primarily facilitates transcriptional repression in postnatal mouse cerebella.
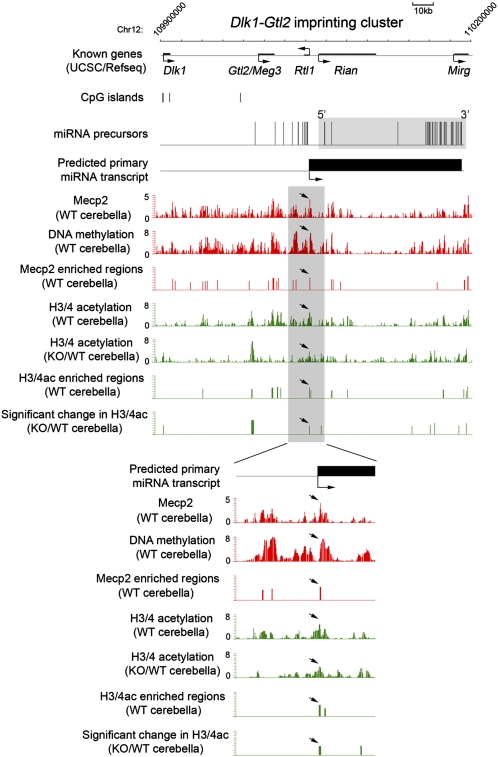
Mecp2 Directly Regulates a Large Cluster of miRNAs Within the Dlk1-Gtl2 Imprinting Domain.
One of the primary miRNA transcripts directly targeted by Mecp2 encodes a large cluster of miRNAs embedded within the Dlk1-Gtl2 imprinting domain on mouse chromosome 12 (32, 33). A recent report has shown that the differential methylation region close to the Gtl2 promoter region is bound by α thalassemia/mental retardation syndrome X-linked (ATRX)/Mecp2/Cohesin proteins, suggesting a role for Mecp2 in transcriptional control of this region (34). Using chromosomal tiling microarrays that cover the entire Dlk1-Gtl2 domain, we performed Mecp2 ChIP-chip analysis in postnatal cerebella (6–8 wk) and identified additional Mecp2-binding sites within this imprinting domain (Fig. 3 and SI Appendix, Fig. 5). MeDIP-chip assays further demonstrated that these Mecp2-enriched regions, including the putative promoter region of the polycistronic miRNA primary transcript (arrows in Fig. 3), were generally DNA-methylated. Therefore, Mecp2 may be involved directly in regulating transcription of these miRNAs. Indeed, sequencing analysis of 6-wk WT cerebellar RNAs indicated that 31 of 41 miRNA precursors within this miRNA cluster had detectable transcription (>10 reads); 22 of 31 transcribed miRNAs showed a consistent trend (sequencing reads mapped to guide and passenger strands exhibited same trend) of increased expression, and two showed a consistent trend of decreased expression in KO cerebella (SI Appendix, Table 6). Moreover, expression of 15 mature miRNAs exhibited a >1.5-fold increase in the absence of Mecp2 (e.g., miR-666 and miR-543 in Fig. 1B), whereas no mature miRNA was down-regulated >1.5-fold (SI Appendix, Table 6). Thus, Mecp2 acts primarily as a negative regulator of this polycistronic miRNA transcript. Interestingly, mature miRNAs generated from this single polycistronic transcript reproducibly exhibited drastically different levels of expression (SI Appendix, Table 6), suggesting the existence of additional miRNA-specific posttranscriptional regulation of precursor processing and/or mature miRNAs stability (35). Such posttranscriptional regulation of miRNA biogenesis also may be the cause of the observed difference in the extent of dysregulation of mature miRNAs derived from the same polycistronic transcript. To test further whether Mecp2 exerts its transcriptionally repressive role at the promoter region of this miRNA transcript unit by recruiting transcriptional corepressors (e.g., histone deacetylases) (13), we performed ChIP-chip analysis of histone acetylation and found a significant increase in histone H3/H4 acetylation levels at the Mecp2-bound promoter region in KO as compared with WT (Fig. 3). Collectively, these results indicate that Mecp2 may regulate directly the expression of this imprinted polycistronic transcript encoding a large number of miRNAs and also reveal complex patterns of miRNA biogenesis at both transcriptional and posttranscriptional levels.
Fig. 3.
Mecp2 binds to specific genomic regions within the Dlk1-Gtl2 imprinting domain. Mecp2 occupancy, DNA methylation, and histone H3/4 acetylation (H3/4ac) in WT and/or KO cerebella (6 wk postnatal) within a 300-kb region of the Dlk1-Gtl2 imprinting domain are shown. Statistically enriched regions (−log10 P > 2) are denoted as vertical bars. Mecp2 enrichment at the putative transcription start site for the polycistronic miRNA transcript is indicated by arrows.
Mecp2-Repressed miRNAs Target the 3′ UTR of Bdnf mRNA.
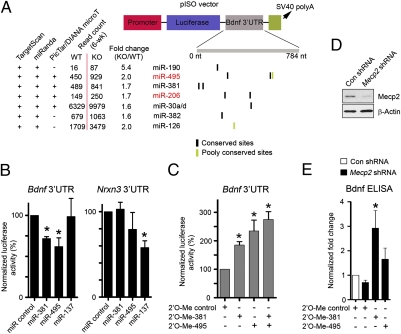
MiRNAs selectively regulate their target mRNAs primarily through base pairing between two to eight nucleotides of miRNAs (also known as seed regions) and the 3′ UTR of mRNAs (36). Computational algorithms analyzing the sequence complementarity between seed regions and evolutionarily conserved domains of 3′ UTRs can provide useful predictions of the potential physiological targets for mammalian miRNAs (36, 37). To assess the functional relevance of Mecp2-regulated miRNAs, we sought to determine the effect of these miRNAs on mRNA targets that potentially were involved in disease progression. Bdnf mRNA and protein levels have been shown previously to be significantly reduced in multiple regions of RTT mouse brain, including cortex and cerebellum, at both early and late symptomatic stage. Importantly, decrease in Bdnf levels has been shown to exacerbate the progression of disease phenotype in RTT mouse models (26, 28). We noted that the Bdnf 3′ UTR was predicted to be targeted by multiple aberrantly up-regulated miRNAs by several independent algorithms (SI Appendix, Fig. 6). For example, TargetScan analysis indicated that Bdnf 3′ UTR contained a total of 20 miRNA-binding sites (11 highly conserved and 9 poorly conserved) for 16 miRNAs (representing 13 distinct seed sequences or miRNA families) aberrantly up-regulated in Mecp2-null cerebella at the pre-/early-symptomatic stage (SI Appendix, Fig. 6). However, only four miRNAs down-regulated in KO cerebella were predicted to target the Bdnf 3′ UTR (two conserved sites and two nonconserved sites). Hence, dysregulated miRNAs may primarily function to regulate negatively Bdnf mRNA translation/stability in Mecp2-null cerebella (16, 26).
To validate experimentally the computationally predicted interaction between dysregulated miRNAs in KO cerebella and Bdnf mRNA, we performed luciferase assays using a Bdnf 3′ UTR-containing reporter (pISO-Bdnf) (Fig. 4A). We first examined the overall effect of miRNAs dysregulated in absence of Mecp2 on the Bdnf 3′ UTR by transfecting the pISO-Bdnf reporter into WT and KO postnatal cerebellar neurons; a roughly 30% decrease in luciferase reporter activities from the Bdnf 3′ UTR reporter was observed in mutant neurons as compared with controls (SI Appendix, Fig. 7A). The human BDNF 3′ UTR has been shown to be repressed by increased levels of miR-30a (38), a miRNA that was expressed in mouse brains and was further elevated in KO cerebella at 6 wk. To determine whether the endogenous miR-30a is involved in suppressing Bdnf expression, we cotransfected the pISO-Bdnf and 2′-O-methyl (2′-O-Me)-modified antisense oligoribonucleotide-based inhibitor into cerebellar neurons. The inhibitor for miR-30a and a closely related family member, miR-30d (2′-O-Me-30a/d), significantly enhanced the pISO-Bdnf luciferase activities in WT neurons (SI Appendix, Fig. 7A), indicating that endogenous miR-30a/d may participate in regulating of Bdnf levels. Interestingly, in KO neurons, even with a higher dose (up to 200 nM), 2′-O-Me-30a/d could restore the pISO-Bdnf luciferase activity only partially, suggesting involvement of additional miRNAs in repressing Bdnf levels (Fig. 4A). We then tested two more miRNAs with multiple evolutionarily conserved putative binding sites within the Bdnf 3′ UTR, miR-381(two sites) and miR-495 (two sites), both of which were aberrantly up-regulated in KO cerebella at 6 wk after birth and were derived from the Mecp2 directly regulated polycistronic transcript within the Dlk1-Gtl2 imprinting domain. Luciferase reporter assays indicated that both miRNAs could down-regulate the pISO-Bdnf luciferase activities significantly (Fig. 4B). Conversely, inhibition of miR-381 and miR-495 resulted in significant up-regulation of Bdnf 3′UTR luciferase reporter activities (Fig. 4C). These effects on the Bdnf 3′ UTR reporter were specific to miR-381 and miR-495, because miR-137 or a control miRNA (a Caenorhabditis elegans-specific miRNA, cel-miR-67), did not regulate the pISO-Bdnf reporter (no binding site for miR-137 or cel-miR-67 is present in pISO-Bdnf) (Fig. 4B). Furthermore, Bdnf ELISAs demonstrated that inhibiting miR-381 and miR-495 in both control and Mecp2-deficient neurons could increase the endogenous level of Bdnf (Fig. 4 D and E and SI Appendix, Fig. 7B). Collectively, these results indicate that multiple aberrantly up-regulated miRNAs in KO cerebella, including miR-30a/d, miR-381, and miR-495, may contribute to down-regulation of Bdnf in RTT brains.
Fig. 4.
Essential roles of Mecp2-regulated miRNAs in regulation of Bdnf expression. (A) Schematic diagram of the pISO luciferase reporter containing the first 800 bp of the Bdnf 3′ UTR (pISO-Bdnf). Also shown are aberrantly up-regulated miRNAs (>1.5-fold) in KO cerebella at the early-symptomatic stage (6 wk postnatal) that are predicted to target the Bdnf 3′ UTR by at least two independent target-prediction algorithms (TargetScan, miRanda, or PicTar/DIANA microT). (B) Luciferase assays in 293T cells transfected with miRNA mimics duplexes (100 nM). Error bars represent SEM. *P < 0.05; n ≥ 4. Note that other miRNAs [cel-miR-67 (specifically expressed in C. elegans) and miR-137] were used as negative controls for pISO-Bdnf. (C) Luciferase assays in WT cerebellar neurons transfected with control (100 nM) or miR-381/495 (100 nM) 2’-O-Methyl oligonucleotide inhibitors. Error bars represent SEM. *P < 0.05; n ≥ 5. (D) Immunoblotting of Mecp2 in postnatal cortical neurons infected with lentiviruses expressing a control shRNA or a shRNA specific for Mecp2. (E) Bdnf ELISAs in postnatal cortical neurons. Error bars represent SEM. *P < 0.05; n = 4.
Discussion
In this study, our genome-wide analyses have established that Mecp2 can contribute to transcriptional regulation of a cohort of miRNAs in a mouse RTT model. Roughly 17% of all known mature miRNAs are found to be considerably dysregulated (>1.5-fold) in KO cerebella before the onset of severe neurological symptoms (6 wk after birth). Combined ChIP-chip and expression analyses support a model in which Mecp2 binding at promoter regions of miRNA transcription units acts primarily as a transcriptional repressor (SI Appendix, Fig. 8A), possibly by recruiting histone deacetylase, because levels of acetylated histone H3 and H4 in Mecp2-null brains increased significantly at many Mecp2-bound regions (Fig. 3) (13). Interestingly, many up-regulated mature miRNAs (18%, 15 of 85 up-regulated at 6 wk after birth; 71%, five of seven up-regulated at both 6 and 8 wk after birth) are from a large polycistronic cluster of miRNAs (encoding a total of 41 miRNA genes) within the Dlk1-Gtl2 imprinting domain. Although currently it is unclear which chromosome (maternal or paternal) is targeted preferentially by Mecp2 within this imprinting domain in the postnatal cerebella, our chromosome-wide analyses of Mecp2 occupancy and DNA methylation levels suggest that Mecp2 may bind directly to multiple DNA-methylated regions within the Dlk1-Gtl2 imprinting domain, including the putative promoter regions of the polycistronic miRNA transcription unit. Because transcription of miRNAs within this cluster has been shown to be regulated by neuronal activity (39), it will be of interest to determine whether Mecp2 connects neuronal activity to transcriptional control of these miRNAs (40, 41). Furthermore, several miRNAs expressed from this cluster are implicated in regulation of dendritogenesis and synapse formation/maturation (20, 39). In support of the notion that dysregulation of synapse-localized miRNAs may contribute to neurological disorders (42), including RTT and other autism-spectrum disorders, multiple Mecp2-regulated miRNAs have been reported to be enriched at synapses (direct targets: miR-341, miR-137, and miR-381; nontargets: miR-29a, and miR-7) (43). Mecp2-regulated miRNAs in neurons therefore may serve as a critical mechanistic link between nucleus-localized Mecp2 and cytoplasmic/synaptic proteins.
Because miRNAs function as negative regulators of mRNAs, Mecp2 may promote expression of neuronal protein-coding genes indirectly by repressing specific miRNAs. We have tested this possibility by focusing on the potential interactions between Mecp2-regulated miRNAs and the mRNA encoding Bdnf, because Bdnf mRNA/protein levels are reduced significantly in KO cerebella (26). Functional analysis suggests that multiple miRNAs aberrantly up-regulated in Mecp2-null cerebella may target the 3′ UTR of Bdnf directly; therefore, it is tempting to propose that additional brain-specific transcripts down-regulated in mutant cerebella and/or hypothalamus (16, 17) also are negatively regulated by derepressed miRNAs. Interestingly, Bdnf and/or neuronal activity, in turn, can induce the expression of miRNAs, including miR-212 and miR-132 (44), both of which are down-regulated in KO cerebella and can repress Mecp2 by targeting the 3′ UTR of Mecp2 mRNA (28, 45), thereby forming a regulatory feedback loop between Mecp2 and these Bdnf-induced miRNAs (SI Appendix, Fig. 8B). Taken together, our study has set the stage for future studies to elucidate fully the physiological mRNA targets of Mecp2-regulated miRNAs and their relative contributions to the course of RTT disease.
Materials and Methods
SOLiD Sequencing and Data Analysis.
Cerebellar RNA samples were prepared from WT and KO littermates at the pre- or early-symptomatic stage (6 wk postnatal) or symptomatic stage (8 wk postnatal) and were sequenced according to the SOLiD small RNA-Seq Kit protocol (Applied Biosystems). Detailed descriptions of the SOLiD sequencing methods and data analysis are in SI Appendix.
ChIP and Mecp2 ChIP-Chip.
To immunoprecipitate chromatin bound by Mecp2 using validated polyclonal antibodies (30), cerebella isolated from four pairs of 6- to 8-wk-old Mecp2+/y and Mecp2-/y littermates were homogenized and cross-linked with 1% formaldehyde for 10 min at room temperature followed by exposure to 0.125 M glycine to stop the cross-linking reaction. ChIP analysis was performed as previously described (46). ChIP-qPCR was performed in an iCycler (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Primer sequences used in real-time PCR are listed in SI Appendix.
For microarray analyses, we amplified input and immunoprecipitated DNA using whole-genome amplification kits (WGA; Sigma). Sample labeling, hybridization, data extraction, and peak detection were performed according to standard procedures by NimbleGen Systems. Two separate hybridizations using biological independent samples (pooled from four pairs of WT and KO cerebella) were performed and yielded similar results. We combined and averaged the signals from replicate arrays. For identification of probes associated with a significant level of Mecp2 signals, a built-in sliding window-based statistical test (in NimbleScan2.5) was used. A detailed description of ChIP-chip data analysis is in SI Appendix.
MeDIP and MeDIP-chip.
MeDIP assays were performed as previously described (46). For microarray analyses, we amplified input and immunoprecipitated DNA by the monoclonal 5-methylcytosine antibody (Eurogentec) using whole-genome amplification kits (WGA; Sigma). Labeling, hybridization, array processing, and data extraction were done following procedures described for ChIP-chip.
Luciferase Reporter Assay and BDNF ELISA.
For luciferase report assays, we cloned Nrxn3 and Bdnf 3′ UTR sequences into the pISO fly luciferase construct (47). miRNA-specific mimics or 2′-O-methyl inhibitors (Dharmacon) and the fly luciferase plasmid were cotransfected into mouse (cortical or cerebellar) neurons using Lipofectamine 2000 (Invitrogen). A TK renilla luciferase reporter (Promega) also was cotransfected as a control for transfection efficiency. Protein lysates were extracted from neurons electroporated (Lonza) with miRNA-specific mimics or 2′-O-methyl inhibitors, and BDNF ELISAs were performed as previously described (26).
Supplementary Material
Acknowledgments
We thank Drs. Keping Hu and Weidong Wang (National Institute on Aging) for providing Mecp2 polyclonal antibodies, members of the Y.E.S. laboratory for helpful discussion, the Epigenetics core facility at University of California–Los Angeles Intellectual Development and Disabilities Research Center for Sequenom DNA methylation analyses, Mavis Swerdel for preparing SOLiD libraries, and the Rutgers University Waksman Genomics Laboratory for the initial SOLiD sequencing experiment. R.P.H. was supported by the New Jersey Commission on Science and Technology. This work was supported by National Institutes of Health Grants R56MH082068, EUREKARO1 (MH048095), and RO3 (DA022262-01), by University of California, Los Angeles Center for Autism Research and Treatment Pilot Grants, and by the International Rett Syndrome Foundation (to Y.E.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) under accession no. GSE24329.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005595107/-/DCSupplemental.
References
- 1.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Chahrour M, Zoghbi HY. The story of Rett syndrome: From clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 4.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 5.Wood L, Gray NW, Zhou Z, Greenberg ME, Shepherd GM. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J Neurosci. 2009;29:12440–12448. doi: 10.1523/JNEUROSCI.3321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dani VS, et al. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird A. The methyl-CpG-binding protein MeCP2 and neurological disease. Biochem Soc Trans. 2008;36:575–583. doi: 10.1042/BST0360575. [DOI] [PubMed] [Google Scholar]
- 10.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 11.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 12.Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 13.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinowich K, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 15.Chen WG, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 21.Wayman GA, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abelson JF, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 26.Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 27.Goff LA, et al. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE. 2009;4:e7192. doi: 10.1371/journal.pone.0007192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein ME, et al. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 29.Szulwach KE, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu K, Nan X, Bird A, Wang W. Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex. Nat Genet. 2006;38:962–964. doi: 10.1038/ng0906-962. author reply 964–967. [DOI] [PubMed] [Google Scholar]
- 31.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seitz H, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin SP, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 34.Kernohan KD, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Mellios N, Huang HS, Grigorenko A, Rogaev E, Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet. 2008;17:3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao J, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: The roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 43.Siegel G, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remenyi J, et al. Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem J. 2010;428:281–291. doi: 10.1042/BJ20100024. [DOI] [PubMed] [Google Scholar]
- 45.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13(9):1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.