Abstract
Intestinal health requires the coexistence of eukaryotic self with the gut microbiota and dysregulated host-microbial interactions can result in intestinal inflammation. Here, we show that colitis improved in T-bet−/−Rag2−/− mice that consumed a fermented milk product containing Bifidobacterium animalis subsp. lactis DN-173 010 strain. A decrease in cecal pH and alterations in short chain fatty acid profiles occurred with consumption, and there were concomitant increases in the abundance of select lactate-consuming and butyrate-producing bacteria. These metabolic shifts created a nonpermissive environment for the Enterobacteriaceae recently identified as colitogenic in a T-bet−/−Rag2−/− ulcerative colitis mouse model. In addition, 16S rRNA-based analysis of the T-bet−/−Rag2−/−fecal microbiota suggest that the structure of the endogenous gut microbiota played a key role in shaping the host response to the bacterial strains studied herein. We have identified features of the gut microbiota, at the membership and functional level, associated with response to this B. lactis-containing fermented milk product, and therefore this model provides a framework for evaluating and optimizing probiotic-based functional foods.
Keywords: Enterobacteriaceae, intestinal inflammation, microbiota, probiotics, colitis
Intestinal health requires the coexistence of eukaryotic self with the gut microbiota (1). This balance is maintained by the intestinal epithelium, mucosal immune system, and gut microbes. In inflammatory bowel disease (IBD), interactions between a host's immune system and gut microbiota are dysregulated. Studies of the fecal microbiota of IBD patients reveal distinct differences in community membership between healthy and affected individuals (2, 3). An association between gut microbiome profiles and diseases such as IBD (2–4), obesity (5), type 1 diabetes (6), metabolic syndrome (7), and irritable bowel syndrome (8, 9) is emerging from studies of murine disease models and human study populations. The relationship between microbial communities and disease states raises questions as to how microbial communities can be restructured to prevent and treat disease.
Specific probiotics (live microbes that can provide a health benefit to the host) (10), prebiotics (selectively fermentable substances that confer benefits to the host) (11), and antibiotics all represent modalities to alter the composition and function of the gut microbiota (12, 13). The role of antibiotics in the management of IBD remains controversial, with disparate results across different trial populations (14). Furthermore, recent metagenomic studies suggest that antibiotics have sweeping effects on the intestinal microbiota (15, 16). The distal gut microbiome also appears to be a reservoir for antibiotic resistant microbes (17). Thus the use of antibiotics to alter microbial communities is not as predictable or as specific as desired, and could promote the selection and/or outgrowth of antibiotic-resistant strains. Some probiotics may represent an alternative for modulating the gut microbiome. The beneficial effects of specific probiotics have been ascribed to their ability to alter the intestinal microbiota, support colonization resistance against pathogens, and influence host immune responses (18). Defining how consumption of beneficial microbes ameliorates intestinal inflammation in murine models with defined genetics and pathophysiology may facilitate an understanding of who will benefit from probiotic interventions and how probiotics can be optimized for maximal benefit.
The microbiota colonizing the gastrointestinal tract has coevolved with the host to be mutualistic. However, many gut microbes have the capacity to cause or promote disease. Both host and microbial factors influence bacterial opportunism and deficiencies in beneficial microbes may also contribute to chronic intestinal inflammation. Decreased counts of Faecalibacterium prausnitzii have been observed in fecal samples from IBD patients (19, 20), and low counts of these bacteria appear to predict postsurgical recurrence (21). A decreased relative abundance of Bifidobacterium in the gut microbiota of IBD patients has also been observed (22).
Using both culture-dependent and independent methods, we have recently characterized the fecal microbial communities in a murine model of IBD driven by deficiency of T-bet, a T-box family transcription factor, in the innate immune system; and identified culturable colitogenic bacteria that work in concert with gut microbial communities to drive intestinal inflammation (23). In this study, we found that intestinal inflammation improved in the majority of mice that consumed a fermented milk product containing Bifidobacterium animalis subsp. lactis DN-173 010 (B. lactis), Streptococcus thermophilus, two strains of Lactobacillus delbrueckii subsp. bulgaricus, and Lactococcus lactis subsp. cremoris. Marked shifts in cecal pH (decreased pH) and short chain fatty acid (SCFA) profiles (increased acetic acid, propionic acid, and butyric acid and decreased lactic acid) occurred with consumption, and there were concomitant increases in the abundance of select lactate-consuming and butyrate-producing bacteria. These metabolic shifts created a nonpermissive environment for the Enterobacteriaceae recently identified as colitogenic in a T-bet−/−Rag2−/− ulcerative colitis mouse model (23). In addition, 16S rRNA-based analysis of the T-bet−/−Rag2−/− fecal microbiota suggest that the structure of the endogenous gut microbiota plays a key role in shaping host response to probiotics. We have identified features of the gut microbiota associated with response to a B. lactis-containing fermented milk product, and therefore this model provides a framework for evaluating and optimizing probiotic-based functional foods.
Results
B. lactis-Containing Fermented Milk Product Improves T-bet−/−Rag2−/−Colitis and Requires Live B. lactis.
T-bet−/−Rag2−/− mice consumed a B. lactis-containing fermented milk product (BFMP) or nonfermented milk product (MP) starting at 4 wk of age for 4 wk; a sham feeding and handling control was performed using sterile water (sham). Mice were provided with additional BFMP, MP, or water (100 mg/per mouse provided in its cage). Colitis scores were decreased in the BFMP group (1.67 ± 1.53) compared with MP (4.11 ± 1.53; P < 0.0001) and sham (5.43 ± 0.98; P = 0.0001) (Mann–Whitney test) (Fig. 1A). 12-wk-old T-bet−/−Rag2−/− mice consumed the product for 4 wk, as in Fig. 1A, to determine whether similar effects would be observed in more severe disease. Colitis scores were lower in BFMP group (1.77 ± 1.96) as compared with MP (4.36 ± 2.66; P = 0.015) and sham (8.38 ± 1.04; P < 0.001) (Mann–Whitney test) (Fig. 1B). The BFMP reduced colitis scores in the majority of mice both at early stages and at late stages of disease when intestinal inflammation was more severe.
Fig. 1.
BFMP improves T-bet−/−Rag2−/−colitis and requires live B. lactis. (A) T-bet−/−Rag2−/− mice consumed a BFMP (n = 18), MP (n = 19), or sham (n = 7) daily from 4 to 8 wk of age. (B) T-bet−/−Rag2−/− mice consumed the BFMP (n = 13), MP (n = 11), or sham (n = 13) daily from 12 to 16 wk of age. (C) T-bet−/−Rag2−/− mice consumed BFMP (n = 17), irradiated and sterile BFMP (n = 19), or sham (n = 6). Colitis scores (y axis). Each circle shows data from one mouse. Horizontal bars represent mean. P values, Mann–Whitney test.
To determine whether these observations were dependent upon live bacteria in the BFMP, we generated BFMP without live bacteria by irradiation (4.4 × 104 Gy). The product was only orally instilled in these experiments—ensuring that it was free of live bacteria and unlikely to support the growth of environmental bacteria as might occur if placed in the cage. Colitis scores in the live BFMP were lower than those of the irradiated product (3.0 ± 1.75 vs. 4.44 ± 1.13, P = 0.03) or sham (6.0 ± 1.22, P = 0.018) (Mann–Whitney test) (Fig. 1C).
Consumption of B. lactis-Containing Fermented Milk Product Creates a Nonpermissive Environment for Colitogenic T-bet−/−Rag2−/− Enterobacteriaceae.
Select Enterobacteriaceae instigate inflammation in T-bet−/−Rag2−/− mice (23). BFMP consumption decreased fecal levels of Enterobacteriaceae sevenfold (P = 0.0132, Mann–Whitney test) as compared with MP by RT-quantitative PCR (Fig. 2A). We recently identified T-bet−/−Rag2−/− derived strains of Klebsiella pneumoniae and Proteus mirabilis as specific Enterobacteriaceae that were capable of inducing colonic inflammation in concert with the endogenous gut microbiota (23). Quantitative culture-based fecal counts of both K. pneumoniae and P. mirabilis markedly decreased in response to BFMP consumption (P = 0.0031 K. pneumoniae, P = 0.02 P. mirabilis, Mann–Whitney test) (Fig. 2B). For seven mice, K. pneumoniae was below our limit of detection as was P. mirabilis for four mice (Fig. 2B).
Fig. 2.
Consumption of B. lactis-containing fermented milk product creates a nonpermissive environment for colitogenic T-bet−/−Rag2−/− Enterobacteriaceae. (A) qPCR using primers targeting Enterobacteriaceae. Pre- and postconsumption levels for MP (●) or BFMP (○) (y axis), expressed as log10(equivalent cells/g feces). Each circle indicates data from one mouse; horizontal bars show the mean. P values, Mann–Whitney test. (B) Quantitative fecal culture pre- and post- BFMP and MP (as in A). Colony counts for K. pneumoniae (○) and P. mirabilis (□); detection limit is shown. Each symbol shows data from one mouse; bars show the mean; P values, where significant, are shown. (C) Cecal pH for T-bet−/−Rag2−/− that consumed BFMP, MP, or sham and for sham Rag2−/− mice. Mean values from three independent experiments are shown (n = 5 mice/sample). Error bars represent SD. (D) qPCR and RT-qPCR of fecal samples pre- and postconsumption of MP or BFMP. Data expressed as log10(equivalent cells/g feces).
Because acidic pH can inhibit the growth of the Enterobacteriaceae, we measured the cecal pH. Indeed, the cecal pH of T-bet−/−Rag2−/− mice was markedly lower 8 h after BFMP consumption (P < 0.0001, one-way ANOVA) (Fig. 2C). Survival of lactic acid producing bacteria from the BFMP could account for the decreased pH. Fecal levels of the four lactic acid producing species present in the BFMP: S. thermophilus, L. bulgaricus, L. lactis, and B. lactis were measured by qPCR and RT-qPCR. The BFMP species were detected in most of the postconsumption BFMP group and all changes observed in response to BFMP were significant (B. lactis, P = 0.006, S. thermophilus, P = 0.0139, Wilcoxon signed rank test; L. bulgaricus, P = 0.0001, L. lactis, P = 0.0002, Mann–Whitney test) (Fig. 2D).
Altered SCFA Profiles and Increased Levels of Specific Lactate-Consuming and Butyrate-Producing Bacteria Define the Nonpermissive Environment for the Colitogenic T-bet−/−Rag2−/− Enterobacteriaceae.
The detection of lactic acid producing bacteria suggested that lactic acid may be responsible for the low cecal pH observed, so we measured SCFA levels. Acetic, propionic, and butyric acid levels increased (P < 0.0001, one-way ANOVA), whereas lactic acid decreased (Fig. 3A). This observation was counterintuitive, as the BFMP strains are lactic acid producers. However, the decreased lactic acid and increased butyric acid could come from increased endogenous lactic acid-consuming and butyrate-producing bacteria. qPCR and RT-qPCR of fecal material of T-bet−/−Rag2−/− mice pre- and postconsumption revealed increases in the lactate-consuming Desulfovibrio spp (P = 0.0089) and of the lactate-consuming, butyrate-producing Anaerostipes caccae subgroup (P = 0.02) and Eubacterium hallii (P = 0.004) (Mann–Whitney test) in response to the BFMP as compared with the MP (Fig. 3B). These shifts were specific, as there were not significant shifts in other butyrate producers, such as the Blautia coccoides subgroup, Clostridium leptum subgroup, or the Roseburia subgroup (Fig. 3B).
Fig. 3.
Altered SCFA profiles and increased levels of specific lactate-consuming, butyrate-producing bacteria define a nonpermissive environment for colitogenic T-bet−/−Rag2−/− Enterobacteriaceae. (A) Volatile and nonvolatile SCFA levels of cecal contents for BFMP, MP, and sham T-bet−/−Rag2−/− mice and sham Rag2−/− mice. Bars show mean values from three independent experiments (n = 5 mice/ sample). Error bars indicate standard deviation. (B) qPCR and RT-qPCR using primers targeting the indicated bacteria genera or species. Pre- and postconsumption levels for MP (●) or BFMP (○) shown along the y axis (log10(equivalent cells/g feces)). Each circle indicates data from a single mouse. Horizontal bars show the mean. P values were calculated by Mann-Whitney test or Wilcoxon signed-rank test. (C) Growth kinetics of T-bet−/−Rag2−/−-derived strains of K. pneumoniae and P. mirabilis over a 3-h time course in media with pH and SCFA concentration adjusted to model cecal contents of T-bet−/−Rag2−/− mice that consumed BFMP or MP.
Bacteriostatic and bactericidal activities have been ascribed to volatile SCFAs (24, 25), and thus the cecal conditions of T-bet−/−Rag2−/− mice that consumed the BFMP might be inhospitable to K. pneumoniae and P. mirabilis, as suggested by Fig. 2 A and B. We prepared media that modeled the cecal conditions of the mice that consumed the BFMP or MP with respect to pH and volatile and nonvolatile SCFA concentrations (VFA and non-VFA, respectively). Media with the mean cecal pH of these two groups but without the SCFAs were also prepared. K. pneumoniae growth was inhibited by media with the pH of the BFMP group (pH 4.50) (Fig. 3C), and these slowed growth kinetics were observed up to pH 5.8. In contrast, P. mirabilis growth appeared selectively inhibited by media that contained both the lower pH and VFA and non-VFA of the BFMP group (Fig. 3C). Growth inhibition for P. mirabilis was significant at the 3 h time point (P = 0.03, one-way ANOVA), and decreased growth at 3 h was also significant for K. pneumoniae when both BFMP group pH media were compared with the higher-pH MP group media (P = 0.005, Mann–Whitney test). Thus, both in vivo and in vitro experiments suggest that consumption of this BFMP creates a nonpermissive environment (low pH and altered SCFA profiles) for colitogenic T-bet−/−Rag2−/− Enterobacteriaceae.
Structure of the Gut Microbiota Influences Response to B. lactis-Containing Fermented Milk Product.
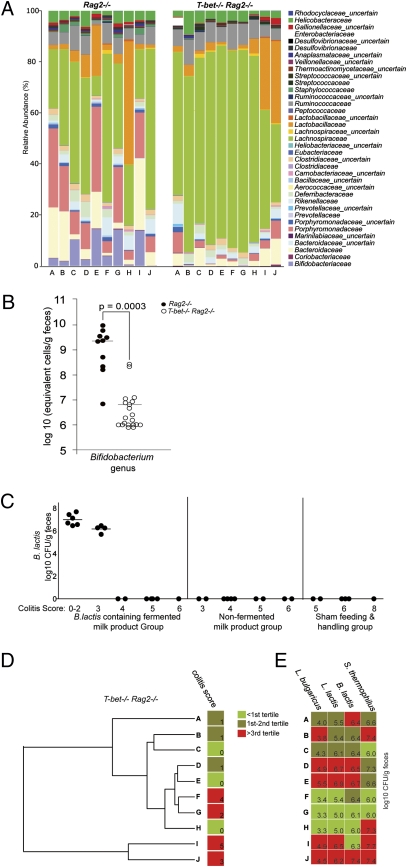
To identify additional features of the microbiota that could explain why T-bet−/−Rag2−/− mice respond to the BFMP, we performed a 16S rRNA gene-based survey of the fecal microbiota of T-bet−/−Rag2−/− and Rag2−/− mice before BFMP consumption (n = 10/group, age 4 wk). Multiplex pyrosequencing of amplicons generated from the V5 and V6 region of the 16S rRNA gene was performed (n = 20 samples; 7579 ± 2379 reads/sample). Compared with Rag2−/− controls, T-bet−/−Rag2−/− samples had a significantly lower proportional representation of operational taxonomic units (OTUs) belonging to the families Bifidobacteriaceae (P = 0.022), Porphyromonadacaeae (P = 0.011), Prevotellaceae (P = 0.011), and Staphylococcaceae (P = 0.0037) (Mann–Whitney test with Bonferroni correction) (Fig. 4A). In contrast, the proportional representation of the Lachnospiraceae (P = 0.05, Mann–Whitney test with Bonferroni correction) was higher in T-bet−/−Rag2−/− samples vs Rag2−/− (Fig. 4A). The reduced relative abundance of the Bifidobacteriaceae was particularly striking, as it suggests a deficiency in T-bet−/−Rag2−/− mice. The BFMP may replete this deficiency, leading to decreased intestinal inflammation.
Fig. 4.
Structure of gut microbiota influences response to B. lactis-containing fermented milk product. (A) Distribution of family-level phylotypes in T-bet−/−Rag2−/− and Rag2−/− fecal microbiota at 4 wk of age (preconsumption). Percent relative abundance is plotted. (B) Bifidobacterium levels (qPCR) from 4-wk-old T-bet−/−Rag2−/− (n = 20) (○) and Rag2−/− (n = 10) (●) fecal samples. (C) B. lactis counts (log10CFU/g feces) (y axis) of sufficient quantity from mice in Fig. 1. Mice grouped by product administered and colitis score (x axis). Each circle represents a single mouse; horizontal bars show the mean. (D) Family-level hierarchical clustering analysis of T-bet−/−Rag2−/− fecal microbial communities before product consumption. Each sample is labeled corresponding to its relative abundance data in A, and colitis scores postconsumption are heat map color coded based on tertile distribution. (E) Postconsumption recovery levels of BFMP strains are heat map color coded based on tertile distribution for the corresponding sample in A and D.
To validate and quantify the reduced proportional representation of bifidobacteria, we measured fecal Bifidobacterium levels from T-bet−/−Rag2−/− mice (n = 20) at 4 wk of age (the onset of colitis detectable by histology) and healthy age-matched Rag2−/− mice (n = 11) by qPCR. There was a 316-fold mean reduction in Bifidobacterium genus levels in T-bet−/−Rag2−/− relative to Rag2−/− mice (P = 0.0003, Mann–Whitney test) (Fig. 4B). Thus T-bet−/−Rag2−/− mice were indeed relatively deficient in bifidobacteria.
We asked whether there was an association between repleting these decreased bifidobacterial levels, as measured by recovery of live B. lactis from the feces of mice that consumed the BFMP, and colitis score. B. lactis was recovered from mice with colitis scores of 0–3 but was below detection (102.5 CFU/g feces) in mice with higher scores and was not detected in controls (Fig. 4C). This observation raised the question as to whether subsets of mice could be identified, before product consumption, that would or would not respond to the BFMP. A hierarchical cluster analysis using the family level OTU assignments (Fig. 4D) revealed that T-bet−/−Rag2−/− with colitis score ≥2 (F, G, I, and J) clustered together in two subsets. Both samples I and J had markedly elevated proportional representation of the Lactobacillaceae (27.9% and 31.9%, respectively, approximately two standard deviations above the mean) relative to the other T-bet−/−Rag2−/− mice. The F and G subset were also notable for low levels of the strains following 4 wk of BFMP (Fig. 4E). In aggregate, these data suggest that the structure of the gut microbiota may influence and predict response to the BFMP.
Discussion
Here, we have used a murine model of colitis to gain insight into how a defined probiotic-containing food reduces intestinal inflammation. In these studies, we focused on the microbiota and its response to host consumption of BFMP containing B. lactis, L. lactis, L. bulgaricus, and S. thermophilus. We found that T-bet−/−Rag2−/− mice have a relative deficiency of Bifidobacterium, that supplementation with a BFMP reduced intestinal inflammation, and that recovery of live B. lactis from mice correlated with optimal response. Consumption of this BFMP resulted in an increase of certain lactate-consuming and butyrate-producing bacteria, decrease in cecal pH, and increases in select cecal SCFA. These conditions proved inhospitable to T-bet−/−Rag2−/−-derived members of the Enterobacteriaceae that we have recently identified as colitogenic. This study provides a gut microbe-based framework for evaluating responses to probiotic interventions in IBD.
How probiotics and commensal bacteria communicate with each other and host immune cells to ensure intestinal homeostasis still remains unclear. SCFAs, in particular butyric acid, is a molecule of interest as BFMP consumption led to increased levels of butyrate and butyrate-producing commensal anaerobes. This may result from metabolic cross-feeding between the BFMP strains and resident butyrate-producing Firmicutes (26). SCFAs affect more than microbes. Both GPR41 and GPR43 bind SCFAs and are expressed in the colonic mucosa (27), and butyrate is an important energy source for colonic epithelial cells (28). Studies of GPR43−/− mice demonstrate a role for this G-protein–coupled receptor in regulating inflammatory responses in a murine model of colitis, suggesting that host sensing of SCFA is important for host–microbial homeostasis (29). Furthermore, proinflammatory cytokines, such as TNF-α, can impair butyrate oxidation by the colonic mucosa, and increasing luminal levels may be beneficial (30).
In addition to butyric acid, propionic and acetic acid were also elevated in the cecal contents of BFMP-treated mice. These SCFAs inhibit Escherichia coli (E. coli) and Salmonella spp. growth in vitro (31) and may impair growth by their effects on pathways, resulting in methionine depletion and homocysteine accumulation (32). The cecal pH and SCFA profiles of mice that consumed this BFMP had a clear growth-inhibitory effect on the colitogenic Enterobacteriaceae of T-bet−/−Rag2−/− mice in vivo and in vitro. These observations may help to inform selection of IBD patients likely to benefit from specific strain-based treatments. Adherent-invasive E. coli colonize ileal lesions and contribute to chronic inflammation in a subset of Crohn's disease patients (33, 34). Consumption of this BFMP may create unfavorable conditions for adherent-invasive E. coli, potentially reducing colonization and inflammation. Gram-negative aerobes and, in particular, the Enterobacteriaceae, are increasingly being recognized as microbial “inflammatory allies” contributing to the dysbiosis of IBD (35), and thus B. lactis DN-173 010-based functional foods may represent an approach to keep gastrointestinal Enterobacteriaceae in check to promote health.
Probiotic bifidobacteria and lactobacilli can influence both microbial and host physiology. Certain bifidobacteria may influence Enterobacteriaceae by decreasing their virulence gene expression, such as shiga toxin 2 expression in enterohemorrhagic E. coli 0157:H7 (36) and the expression patterns of the Salmonella pathogenicity islands SPI1 and SPI2 (37). Probiotic strains may also exert direct effects on the host mucosa. Oxidative stress driven by reactive oxygen species production is a key feature of inflammation in infectious enteritis and IBD, and probiotic lactobacilli may ameliorate this oxidative stress, e.g., by expression of superoxide dismutase (38, 39). Probiotic lactic acid-producing bacteria may also shift cytokine balance in intestine inflammation, decreasing host IL-6 levels and increasing IL-10 (40). As such, the dramatic effects of this BFMP on intestinal inflammation in the T-bet−/−Rag2−/− colitis model likely are not solely restricted to the microbiota.
Although the BFMP with live bacteria was most effective at lowering the colitis score, the irradiated product showed a trend toward lowering intestinal inflammation. The presence of bacterial products (e.g., DNA and cell wall constituents), which engage the host immune system through pattern recognition receptors, may furnish an explanation. Bacterial DNA from certain strains exerts anti-inflammatory effects (41). Exopolysaccharides from B. lactis have been shown to modify the composition of the microbiota in fecal batch culture (42). Functional foods may confer beneficial effects through a panoply of structural and metabolic microbial features, and thus further research is needed to determine the contribution of the specific microbial components in this BFMP to the reduction of intestinal inflammation observed.
Understanding both the microbial community and host response differences underpinning the spectrum of responses to probiotic-based treatments in inflammation will be essential for realizing the full potential of these treatments. Our results demonstrate that this BFMP is effective in T-bet−/−Rag2−/− mice whose disease is in part driven by colitogenic members of the Enterobacteriaceae. In addition, the data from our 16S rRNA sequencing and qPCR-based experiments suggest that the structure of a host's gut microbiota influences response to probiotics. Thus, the development of gut microbe-based biomarkers for both identification of those individuals likely to benefit from probiotics and for monitoring response to probiotics seems an attainable goal. The BFMP used in these studies is a complex mixture of live bacterial strains, bacterial products, and fermentation products that exert effects on gut microbes as well as host cells. Characterization of both gut microbiomes and host responses to functional foods will benefit from emerging metagenomic and metatranscriptomic techniques, as further light is shed on how chronic inflammation resolves in response to consumption of these products.
Materials and Methods
Animal Husbandry.
T-bet−/−Rag2−/− mice, their husbandry, and chow have been described (23, 43). Animal studies and experiments were approved and carried out according to Harvard University's Standing Committee on Animals as well as National Institutes of Health guidelines.
Study Product.
The test product was a fermented milk (Activia; Danone); details are given in SI Materials and Methods for product strain details. The control product was a milk-based nonfermented dairy product without bacteria and with 1.6% lactose per serving. Control product details are provided in SI Materials and Methods. Products were provided by Danone Research. A 100-mg quantity was orally instilled daily at the same time daily and, if indicated, 100 mg per mouse in its cage was provided for additional consumption.
Production Sterilization.
The BFMP was sterilized with 4.4 × 104 Gy in a gamma irradiator. To confirm product sterility after irradiation, the product was cultured under aerobic and anaerobic conditions.
Histology.
Colons were prepared for histology, and histopathology was evaluated in a blinded fashion (with respect to genotype and experimental protocol) by J.N.G. as previously described (43).
Fecal Collection, Storage, and RNA and DNA Extraction.
Fecal pellets were collected into tubes and weighed. RNAlater (Ambion) was added to the pellets, samples were homogenized and then stored at −80 °C until RNA and DNA were extracted as described previously (44).
Quantification of Bacteria Equivalents by qPCR and RT-qPCR.
Determination of bacterial concentration was based on the quantification of either DNA or RNA molecules using primers targeting 16S rRNA sequences (Tables S1–S3). Additional details are provided in SI Methods and Materials. The qPCR and RT-qPCR reactions were performed according to previously published work (44, 45). Primers and their annealing temperature are found in Tables S1–S3.
Primer Design.
Primers are described in Tables S1–S3, and additional details are given in SI Methods and Materials.
Fecal Collection and Culture of Gram-Negative Aerobes.
From each mouse, four to six pellets were collected. Pellets were resuspended in sterile PBS, and 10-fold serial dilutions were generated, plated on MacConkey's medium, and incubated in ambient air at 37 °C overnight. Biochemical assays with the API-20E panel (bioMerieux) confirmed that colony morphology correlated with identification as K. pneumoniae and P. mirabilis.
Cecal pH and SCFA Measurement.
Cecal contents were collected after the animals were killed. An aliquot was removed for pH measurement and the remaining sample was flash frozen in N2(l). SCFA analyses were performed using a Shimadzu GC-14A gas chromatograph (Shimadzu Scientific Instruments) and a SPB-1000 capillary column (Supelco). VFA and non-VFA standard acid mixes were used to identify and quantify the acids present in each sample. Conjugate base concentrations were weight corrected and converted to conjugate acid concentrations using the cecal pH and Henderson–Hasselbalch equation. Additional details on VFA and non-VFA extractions are provided in SI Materials and Methods.
Growth Kinetics in Modeled Media.
Brain–heart infusion medium was prepared to generate the following media. Cecal pH of BFMP mice (BHI pH 4.50), MP mice (BHI pH 6.07), cecal pH of BFMP mice with VFA and non-VFA (BHI pH 4.50, 99.2 mM acetic acid, 31.05 mM propionic acid, 43.87 mM butyric acid, and 2.08 mM lactic acid), and cecal pH of MP mice with VFA and non-VFA (BHI pH 6.07, 2.35 mM acetic acid, 1.34 mM propionic acid, 0.37 mM butyric acid, and 12.9 mM lactic acid). VFA and non-VFA were added to the media, and the pH was subsequently adjusted. We defined an upper limit of pH 5.8 for which these growth kinetic effects were observed.
Metagenomic Sequencing and Analysis.
The V5 and V6 regions of the 16S rRNA gene were targeted for amplification and multiplex pyrosequencing with error-correcting barcodes. Sequencing was performed using a Roche FLX Genome Sequencer at DNAvision. Data were preprocessed to remove sequences with low-quality scores (7,579 ± 2,379 high-quality reads per sample, mean read length: 278). Taxonomy was assigned using RDP classifier v.2.01 and similar sequences were binned into operational taxonomic units (OTUs) using cd-hit with minimum pairwise identity of 97%. Sample clustering was performed using the complete linkage hierarchical clustering (hclust from R http://sekhon.berkeley.edu/stats/html/hclust.html).
B. lactis Quantitative Culture.
Quantitative fecal culture was performed using Rogosa media supplemented with 1% irradiated sterile fermented milk product. Plates were incubated for 5 d (anaerobic conditions). Gram stains were performed and morphotypes consistent with Bifidobacterium as well as control were screened by PCR (46). B. lactis counts were then calculated from the quantitative fecal culture and PCR results and weight corrected based on fecal dry weight.
Statistical Analysis.
Error bars represent standard deviation. Statistical tests used follow the stated P value. Prism (GraphPad) was used for statistical calculations. The Bonferroni correction was applied as noted for multiple comparison correction.
Supplementary Material
Acknowledgments
We thank D. Zhang for histology expertise and M. Montesalvo and staff of the University of Massachusetts Lowell Radiation Laboratory for use of their Gamma Cave Irradiator Facility. This work was supported by National Institutes of Health Grants CA112663 (to L.H.G.) and K08AIO78942 (to W.S.G.) and by a grant from Danone Research.
Footnotes
Conflict of interest statement: L.H.G. is a member of the Board of Directors of and holds equity in the Bristol Myers Squibb Corporation. P.V., C.B., A.K., and J.E.T.v.H.V. are employees of and hold equity in Groupe Danone.
Data deposition: The 454 pyrosequencing reads reported in this paper have been deposited in the NCBI Short Read Archive (accession no. SRX025834.5).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011737107/-/DCSupplemental.
References
- 1.Hill DA, Artis D. Maintaining diplomatic relations between mammals and beneficial microbial communities. Sci Signal. 2009;2:pe77. doi: 10.1126/scisignal.298pe77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, et al. MetaHIT Consortium. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye J, et al. Bacteria and bacterial rRNA genes associated with the development of colitis in IL-10(-/-) mice. Inflamm Bowel Dis. 2008;14:1041–1050. doi: 10.1002/ibd.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Codling C, O'Mahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55:392–397. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 9.Parkes GC, Brostoff J, Whelan K, Sanderson JD. Gastrointestinal microbiota in irritable bowel syndrome: Their role in its pathogenesis and treatment. Am J Gastroenterol. 2008;103:1557–1567. doi: 10.1111/j.1572-0241.2008.01869.x. [DOI] [PubMed] [Google Scholar]
- 10.Food and Agriculture Organization of the United Nations, World Health Organization . Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Geneva: World Health Organization; 2001. [Google Scholar]
- 11.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 12.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des. 2009;15:1528–1536. doi: 10.2174/138161209788168146. [DOI] [PubMed] [Google Scholar]
- 14.Rubin DT, Kornblunth A. Role of antibiotics in the management of inflammatory bowel disease: A review. Rev Gastroenterol Disord. 2005;5(Suppl 3):S10–S15. [PubMed] [Google Scholar]
- 15.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME. Probiotics and immunity. J Gastroenterol. 2009;44:26–46. doi: 10.1007/s00535-008-2296-0. [DOI] [PubMed] [Google Scholar]
- 19.Willing B, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 20.Sokol H, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 21.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 23.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitte colitis. Cell Host Microbe. 2010 doi: 10.1016/j.chom.2010.08.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoades ER, Short SG. Susceptibility of Serratia, Pseudomonas, and Enterobacter to acetic acid. Antimicrob Agents Chemother (Bethesda) 1970;10:498–502. [PubMed] [Google Scholar]
- 25.Sun CQ, et al. The effect of pH on the inhibition of bacterial growth by physiological concentrations of butyric acid: Implications for neonates fed on suckled milk. Chem Biol Interact. 1998;113:117–131. doi: 10.1016/s0009-2797(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 26.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown AJ, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 28.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nancey S, et al. Tumor necrosis factor alpha reduces butyrate oxidation in vitro in human colonic mucosa: A link from inflammatory process to mucosal damage? Inflamm Bowel Dis. 2005;11:559–566. doi: 10.1097/01.mib.0000161918.04760.f3. [DOI] [PubMed] [Google Scholar]
- 31.Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. 1991;70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 32.Roe AJ, O'Byrne C, McLaggan D, Booth IR. Inhibition of Escherichia coli growth by acetic acid: A problem with methionine biosynthesis and homocysteine toxicity. Microbiology. 2002;148:2215–2222. doi: 10.1099/00221287-148-7-2215. [DOI] [PubMed] [Google Scholar]
- 33.Barnich N, Denizot J, Darfeuille-Michaud A. Pathol Biol. 2010. E. coli-mediated gut inflammation in genetically predisposed Crohn's disease patients. in press. [DOI] [PubMed] [Google Scholar]
- 34.Carvalho FA, et al. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carey CM, Kostrzynska M, Ojha S, Thompson S. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods. 2008;73:125–132. doi: 10.1016/j.mimet.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Bayoumi MA, Griffiths MW. Probiotics down-regulate genes in Salmonella enterica serovar typhimurium pathogenicity islands 1 and 2. J Food Prot. 2010;73:452–460. doi: 10.4315/0362-028x-73.3.452. [DOI] [PubMed] [Google Scholar]
- 38.Truusalu K, et al. Eradication of Salmonella Typhimurium infection in a murine model of typhoid fever with the combination of probiotic Lactobacillus fermentum ME-3 and ofloxacin. BMC Microbiol. 2008;8:132. doi: 10.1186/1471-2180-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watterlot L, et al. Intragastric administration of a superoxide dismutase-producing recombinant Lactobacillus casei BL23 strain attenuates DSS colitis in mice. Int J Food Microbiol. 2010 doi: 10.1016/j.ijfoodmicro.2010.03.037. in press. [DOI] [PubMed] [Google Scholar]
- 40.de Moreno de Leblanc A, Perdigón G. The application of probiotic fermented milks in cancer and intestinal inflammation. Proc Nutr Soc. 2010;69:421–428. doi: 10.1017/S002966511000159X. [DOI] [PubMed] [Google Scholar]
- 41.Rachmilewitz D, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520–528. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 42.Salazar N, et al. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int J Food Microbiol. 2009;135:260–267. doi: 10.1016/j.ijfoodmicro.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 43.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuda K, et al. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol. 2009;75:1961–1969. doi: 10.1128/AEM.01843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70:7220–7228. doi: 10.1128/AEM.70.12.7220-7228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mäkivuokko HA, Saarinen MT, Ouwehand AC, Rautonen NE. Effects of lactose on colon microbial community structure and function in a four-stage semi-continuous culture system. Biosci Biotechnol Biochem. 2006;70:2056–2063. doi: 10.1271/bbb.60022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.