Abstract
Nucleosomes compact and regulate access to DNA in the nucleus, and are composed of approximately 147 bases of DNA wrapped around a histone octamer1, 2. Here we report a genome-wide nucleosome positioning analysis of Arabidopsis thaliana utilizing massively parallel sequencing of mononucleosomes. By combining this data with profiles of DNA methylation at single base resolution, we identified ten base periodicities in the DNA methylation status of nucleosome-bound DNA and found that nucleosomal DNA was more highly methylated than flanking DNA. These results suggest that nucleosome positioning strongly influences DNA methylation patterning throughout the genome and that DNA methyltransferases preferentially target nucleosome-bound DNA. We also observed similar trends in human nucleosomal DNA suggesting that the relationships between nucleosomes and DNA methyltransferases are conserved. Finally, as has been observed in animals, nucleosomes were highly enriched on exons, and preferentially positioned at intron-exon and exon-intron boundaries. RNA Pol II was also enriched on exons relative to introns, consistent with the hypothesis that nucleosome positioning regulates Pol II processivity. DNA methylation is enriched on exons, consistent with the targeting of DNA methylation to nucleosomes, and suggesting a role for DNA methylation in exon definition.
To investigate the position of nucleosomes in Arabidopsis thaliana, we sequenced Micrococcal Nuclease (MNase) digested nucleosomal DNA using an Illumina GAII sequencer to achieve a roughly 68-fold coverage of nucleosomes (see Supplementary Methods). The data are displayed in a modified UCSC genome browser (http://apis.pellegrini.mcdb.ucla.edu:8081/Ath-Nucleosomes/, (name = reviewer, password = DkZmv9wJ) along with nucleosome density tracks that allow easy visualization of nucleosome positions throughout the genome (Fig. 1a).
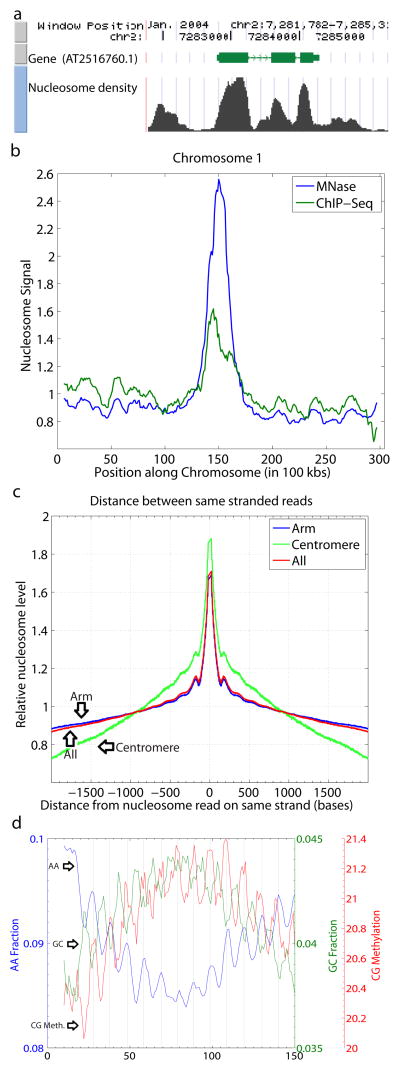
Figure 1. Characterization of A. thaliana nucleosome data.
a, A representative UCSC Browser screenshot of a gene, nucleosome reads found in the region, and the calculated nucleosome densities. b, A chromosomal view of nucleosome read counts and ChIP-seq read counts in 100kb tiles along chromosome 1 shows nucleosome enrichment in pericentromeric regions. c, An autocorrelation of positive stranded reads shows local peaks at positions 174 and 355 bases. d, AA and GC dinucleotide and CG methylation profiles show a 10 base periodicity over nucleosome-bound DNA, with DNA methylation profiles in phase with WW dinucleotide profiles, and out of phase with SS dinucleotide profiles.
To obtain a chromosomal view of nucleosome content, we plotted reads in bins of 100 kilobases tiling the chromosomes. As a control for biases in mapping and sequencing we also sequenced a library of randomly sheared Arabidopsis genomic DNA. We then normalized the nucleosome counts by the number of uniquely mapping genomic DNA counts within each bin along the chromosomes. Nucleosome content was relatively uniform throughout the euchromatic regions of chromosomes, but showed significant enrichment in pericentromeric heterochromatin regions, (Fig. 1b, Supplementary Fig. 1). To confirm these results, we performed ChIP-Seq utilizing an antibody against unmodified histone H3 (Fig. 1b, Supplementary Fig. 1). These observation suggests that nucleosomes are more densely packed in pericentromeric regions that are transcriptionally silent, are rich in transposons and contain heavily methylated DNA, than in the euchromatic arms3.
By examining the relationship between reads that map to opposite strands of DNA, we observed a peak of reads on the reverse strand that occurred approximately 145 –170 bases downstream of the reads on the forward strand (Supplementary Fig. 2a). This broad peak in the general vicinity of the accepted size of a nucleosome suggests that many nucleosomes are well positioned, leading to the repeated sequencing of both the forward and reverse complement of the corresponding nucleosome regions. Similarly, when we plotted the correlation between positive strand reads with other positive strand reads we saw a progressively decreasing correlation from the start of the nucleosome (Fig. 1c, Supplementary Fig. 2b), with smaller peaks spaced at 174 and 355 base pairs from the starting position of the reference reads suggesting some preference for regular spacing of nucleosomes genome wide. Assuming that nucleosomes are wrapped around 147 base pairs of DNA, the average length of the linkers of regularly spaced nucleosomes is about 30 base pairs. Consistent with the higher content of nucleosomes in pericentromeric heterochromatin regions, we found that the peaks at 174 and 355 were more pronounced in these regions than in the euchromatic arms (Fig. 1c), suggesting that pericentromeric nucleosomes have a higher tendency to be in regularly spaced arrays. These results are consistent with earlier evidence for more regular spacing of nucleosomes in Drosophila heterochromatin4.
Similar to findings in animal and fungal high-throughput nucleosome sequencing studies5–8, we found 10 base periodicities in WW (W = A or T) dinucleotides, and SS (S = G or C) dinucleotides that were 5 bases out of phase with the WW dinucleotides (Fig. 1d, Supplementary Figs 3–5). WW dinucleotides are favored at sites where the minor groove faces the histone core, while SS dinucleotides are favored at sites where the minor groove faces away from the histone core9, 10. The out of phase peaks in the frequencies of these dinucleotides leads to optimal bending of DNA, as A/T nucleotides cause a negative base roll and G/C nucleotides cause a positive base roll9, 10.
To study the relationship of DNA methylation with nucleosome positions, we utilized our single nucleotide resolution whole genome bisulfite-sequencing data3. Arabidopsis cytosines are methylated by one of three different DNA methyltransferases depending on their sequence context. CG sites are methylated by MET1, and CHG sites (where H is A, C or T) are methylated by CMT3. Finally, CHH sites are methylated by DRM2, a de novo methyltransferase that is targeted by small RNAs11. Remarkably, all three types of methylation showed a 10 base periodicity on nucleosomal DNA, which was in phase with the WW dinucleotides and out of phase with the SS dinucleotides (Figs 1d, 2a-f, Supplementary Fig. 6). These methylation preferences were not correlated with preferences for CG, CHG or CHH sequences at these locations (Supplementary Fig. 7). Since DNA methyltransferases access the major groove, this methylation would be on DNA which is on the outside of the nucleosome (minor groove facing the histones) and thus more accessible to the DNA methyltransferases. This in turn suggests that DNA might be in part methylated as the DNA is still bound to nucleosomes, leading to the observed 10 base pair periodicity. It has been proposed that chromatin remodeling enzymes are required for DNA methyltransferases to gain access to the DNA, and indeed DRD1, DDM1 and LSH1 are remodelers known to be important factors controlling DNA methylation12. However, our data suggests that nucleosomal DNA may also be a substrate for DNA methyltransferases in vivo, demonstrating a prominent role of the nucleosome in determining methylation patterning throughout the genome.
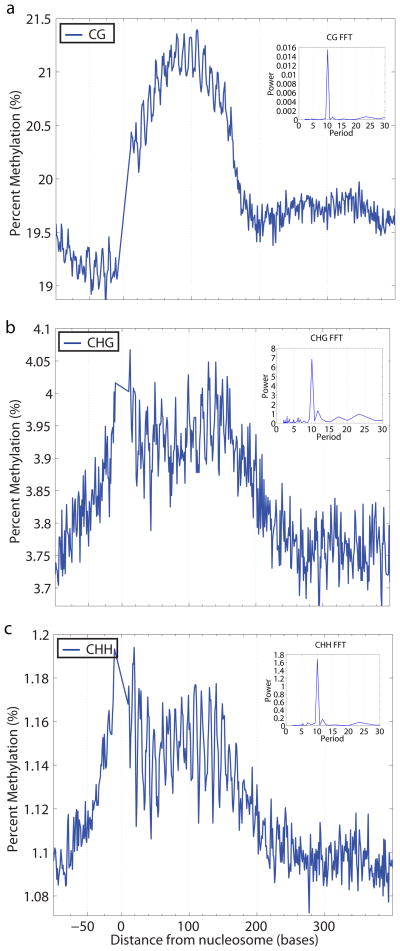
Figure 2. DNA methylation profiles of nucleosome-bound DNA in Arabidopsis.
The weighted average percent DNA methylation was calculated and plotted at each distance from nucleosome start sites (0). a, b, c, CG methylation (a), CHG methylation (b), and CHH methylation (c) each show a 10 base periodicity over nucleosome-bound DNA (1–147 bases). Fast Fourier Transforms (FFTs) can be used to deconstruct inherent frequencies in a complex signal. The FFTs calculated over the region of the nucleosome demonstrate this periodicity in CG (a), CHG (b), and CHH (b) contexts.
We previously reported a 10-nucleotide periodicity in CHH methylation data when performing autocorrelation analysis on the methylation pattern of the whole genome3. Our previous interpretation was that the structure of the DRM2 enzyme might be responsible for this pattern, since the orthologous Dnmt3 enzymes in mammals are known to act as heteromeric complexes in which two methyltransferase active sites have a spacing equivalent to roughly 10 nucleotides of DNA13. However, the current data in which we see 10 base pair periodicities for all types of methylation suggest a more general explanation: that nucleosomes are to some extent dictating access to the DNA and therefore setting the register of methylation for all DNA methyltransferases. Nucleosomal preferences could also partially explain the strong sequence preferences that we previously observed for CHG and CHH methylation3. Highly methylated cytosines tended to be immediately followed by A/T but not C, consistent our finding that DNA methylation is out of phase with CC dinucleotides (Supplementary Fig. 6).
On a larger scale, we also observed that levels of all three types of DNA methylation were higher in nucleosome spanning DNA than flanking DNA, indicating that nucleosome bound DNA is enriched for DNA methylation (Fig. 2, Supplementary Fig. 8). This finding supports the view that nucleosomes are preferentially targeted by DNA methyltransferases. In the case of CMT3, it is predicted that this enzyme is recruited or activated by histone H3 lysine 9 dimethylation, because a knockout of histone H3 lysine 9 methyltransferase mimics a knock out of CMT3, and because the CMT3 chromodomain can bind to methylated histones14. However our data suggest that all types of methylation are enriched on nucleosome bound DNA suggesting that nucleosomes or histone modifications may assist in the recruitment of all of the Arabidopsis DNA methyltransferases.
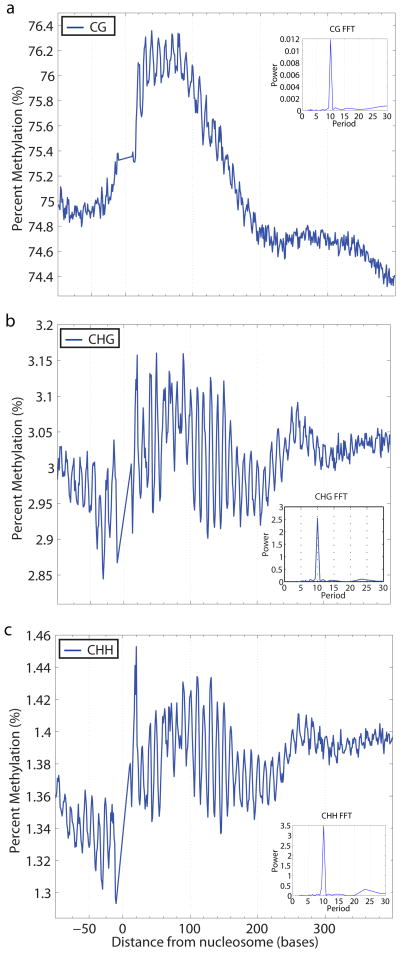
To test whether the patterns of DNA methylation on nucleosomal DNA are also found in human, we utilized previously published MNase nucleosome sequencing data15 together with single nucleotide resolution bisulfite sequencing data on the human embryonic stem cell line HSF1 (see Supplementary Methods). We found that human nucleosome-bound DNA also showed a ten base periodicity in its CG, CHG, and CHH methylation status (Fig. 3). Furthermore, as in Arabidopsis, the overall level of methylation was higher on nucleosome bound DNA than flanking regions. This suggests that nucleosome architecture plays a role in shaping DNA methylation patterning in the human genome, and is consistent with the recent finding of stable anchoring of Dnmt3 DNA methyltransferases to mammalian chromatin16 and more generally with the conservation of DNA methyltransferase function in plants and animals17. We also analyzed DNA methylation patterning on nucleosomes in different regions of the Arabidopsis and human genomes, including genes, promoters, pericentromeric regions, and euchromatic arm regions (Supplementary Fig. 9–17) and found that the 10-nucleotide periodicity was found in all cases, suggesting that the relationship between nucleosome positioning and DNA methylation is general.
Figure 3. DNA methylation profiles of nucleosome-bound DNA in Human.
The weighted average percent DNA methylation was calculated and plotted at each distance from nucleosome start sites (0). a, b, c, CG methylation (a), CHG methylation (b), and CHH methylation (c) each show a 10 base periodicity over nucleosome-bound DNA (1–147 bases). d, e, f, The FFTs calculated over the region of the nucleosome demonstrate this periodicity in CG (d), CHG (e), and CHH (f) contexts.
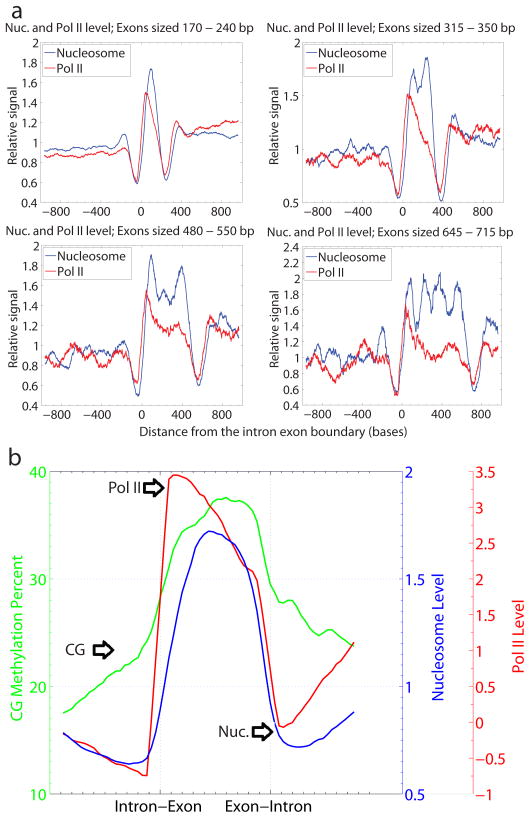
We observed that nucleosomes were much more abundant in exons than in introns (see Fig. 1a for an example), consistent with findings from several recent nucleosome positioning studies in other organisms18–23. The number of nucleosomes per base pair in introns was only 63% of the level found in exons. Furthermore, we found a strong peak of nucleosome start sites at intron-exon junctions and at exon-intron junctions (Fig. 4, Supplementary Fig. 18). The enrichment of nucleosomes in exons was not solely due to higher GC content (Supplementary Fig. 19a-c), or because of consensus splice site sequences (Supplemental Fig. 19d-f), and was confirmed using independent chromatin immunoprecipitation methods (Supplementary Fig. 20). Longer exons contained a higher number of nucleosomes. Examination of exons in the size ranges of 170–240 bp, 315–330 bp, 480–550 bp, and 645–715 bp revealed peaks of one, two, three or four well positioned nucleosomes respectively (Fig. 4a), suggesting that nucleosome are phased within exons.
Figure 4. Nucleosome and PolII levels in Exons.
We performed chromatin immunoprecipitation with an antibody against RNA polymerase II (Pol II), and hybridized the resulting DNA to a whole genome Affymetrix microarray. We normalized these data to randomly sheared genomic DNA to control for probe efficiencies. a, Nucleosomes are phased in exons. Exon size limits were selected such that the exon could house no more than 1, 2, 3, or 4 nucleosomes. Nucleosome midpoints are plotted over these intron-exon boundaries. b, Each exon was divided into 25 equal sized bins and the nucleosome midpoints, Pol II and CG methylation levels are plotted over the exon and flanking introns.
Strong nucleosome enrichment on exons was detected both for genes that are highly expressed, and those not expressed in the shoot tissue used for analysis of nucleosomes (Supplementary Fig. 21), suggesting that DNA sequences position nucleosomes in the absence of active transcription and splicing. Consistent with this hypothesis, utilizing a nucleosome positioning prediction algorithm24 we found similar patterns of nucleosome positioning in introns and exons between the experimental and theoretical datasets (Supplementary Fig. 22). Similarly, using theoretically predicted nucleosome positions, we observed similar patterns of DNA methylation on nucleosome bound DNA (Supplementary Fig. 23), as well as enrichment of predicted nucleosomes in pericentromeric regions (Supplementary Fig. 24).
Because nucleosomes present a barrier to RNA polymerase II (Pol II) transcription, we tested for Pol II occupancy in exons using a chromatin immunoprecipitation microarray approach. We observed significant enrichment of Pol II in exons relative to introns, consistent with the hypothesis that Pol II is paused on exonic DNA (Fig. 4). One possibility is that Pol II stalling on exons could enhance efficient splicing of upstream exons, thus aiding in the fidelity of exon definition. This is consistent with the finding of Pol II enrichment on human exons and suggests that Pol II enrichment on exons might be a common eukaryotic feature20. Furthermore, particular histone modifications have been recently shown to recruit splicing regulators, providing additional possible mechanisms for the regulation of splicing by nucleosome positioning25.
Because of the enhancement of DNA methylation over nucleosomal DNA, and the enrichment of nucleosomes on exons, a prediction is that DNA methylation should be enhanced on exons relative to introns. Indeed, we found depletion of DNA methylation in introns and enrichment in exons in a pattern that was similar to nucleosome occupancy (Fig. 4b). This is consistent with recent findings of enhanced exonic methylation in other plant species, as well as in the Chlamydomonas, honeybee, Ciona and human genomes26, 27. This suggests the possibility that DNA methylation, which frequently exists in the transcribed regions of active genes, could play a conserved role in exon definition or splicing regulation. These findings also reinforce the view that nucleosomal positions play an important role in shaping the methylation landscape of eukaryotic genomes.
Method Summary
Approximately 300ng of MNase digested mononucleosome DNA were used for Illumina library generation following manufacturer instructions. Libraries were sequenced using Illumina Genome Analyzer II following manufacturer instructions. Resulting reads were mapped to the TAIR7 annotation of the Arabidopsis genome and reads that mapped to multiple sites were eliminated. A total of 24.2 million reads were mapped to the forward strand and 23.9 million reads to the reverse strand, generating a 68-fold coverage of nucleosome space.
Pol II ChIP-chip was performed as previously described28. Briefly, crosslinked chromatin was extracted, sonicated, and used in ChIP with an antibody against Pol II CTD (ab817; Abcam, Cambridge, MA). Pol II-bound DNA and input genomic DNA were extracted, amplified, labeled and hybridized to the Afffymetrix tiling microarrays as previously described28. Three biological replicates were performed and the log2 ratios of Pol II-bound RNA over input DNA were calculated using the Tiling Analysis Software (Affymetrix; Santa Clara, CA).
Supplementary Material
Acknowledgments
We thank members of our laboratories for input and guidance, and the Department of Energy, Institute for Genomics and Proteomics and Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research at UCLA for support. R. K. C was supported by NIH training grant GM07104. S.F. was a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. P.C. is funded by Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research at UCLA. M.B was supported by an Alexander-von-Humboldt Fellowship and U.K. by a Heisenberg Fellowship. X.Z. was supported by a Faculty Research Grant (JR-040) from the University of Georgia. Research was supported by NIH grants GM60398 and GM42143, and NSF Plant Genome Research Program #0701745. S. E. J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
S.E.J. and M.P. designed the research. S.F., Y.V.B, H.S., Y.Y., J.H., M.B., P.H., U.K., S.S.M., X.Z. and A.T.C. performed experiments. R.K.C, P.Y.C, F.K., J.K., and S.J.C analyzed data. R.K.C., S. F., S.E.J, and M.P wrote the paper.
Author Information
All sequencing files have been deposited in GEO under accession code GSE21673. The authors declare no competing financial interests.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty–five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun FL, Cuaycong MH, Elgin SC. Long–range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol. 2001;21:2867–79. doi: 10.1128/MCB.21.8.2867-2879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 6.Mavrich TN, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–83. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valouev A, et al. A high–resolution, nucleosome position map of C. elegans reveals a lack of universal sequence–dictated positioning. Genome Res. 2008;18:1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavrich TN, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 10.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 12.Kobor MS, Lorincz MC. H2A.Z and DNA methylation: irreconcilable differences. Trends Biochem Sci. 2009;34:158–61. doi: 10.1016/j.tibs.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindroth AM, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–76. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson R, Enroth S, Rada–Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009 doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon–intron structure. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 21.Kolasinska–Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponts N, et al. Nucleosome landscape and control of transcription in the human malaria parasite. Genome Res. 20:228–38. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan N, et al. The DNA–encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurent L, et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010;20 :320–31. doi: 10.1101/gr.101907.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. Whole–genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.