Abstract
Members of the Ku superfamily are DNA-end-binding proteins involved in non-homologous end-joining (NHEJ) DNA repair. The published crystal structure of human Ku-DNA complex reveals a heterodimer that forms a ring around dsDNA by means of the Ku core modules. These modules contain a highly conserved seven-stranded β-barrel, which in turn contains an insertion, termed the bridge-region, between its second and third β-strands. The bridge-region adopts an unusual β-strand-rich structure critical for dsDNA-binding and Ku function, but its provenance remains unclear. Here, we demonstrate that the bridge-region of Ku is a novel member of the diverse Zn-ribbon fold group. Sequence analysis reveals that Ku from several Gram-positive bacteria and bacteriophages retain metal-chelating motifs, whereas they have been lost in the versions from most other organisms. Structural comparisons suggest that the Zn-ribbon from Ku bridge-region is the first example of a circularly permuted, segment-swapped Zn-ribbon. This finding helps explain how Ku is likely to bind DNA as an obligate dimer. Further, we hypothesize that retention of the unusual conformation of the turns of the Zn-ribbons, despite loss of the Zn-binding sites, provides clues regarding the mechanism by which the Ku bridge-regions sense the DNA state.
Keywords: NHEJ, zinc finger, zinc ribbon, protein flexibility, segment-swapping
Introduction
Ku proteins are double stranded DNA-binding proteins that are primarily involved in end recognition in a variety of repair, recombination and end protection processes (Brissett and Doherty, 2009; Lieber, 2008; Mahaney et al., 2009; Pitcher et al., 2007; Weterings and Chen, 2008). In eukaryotes Ku is critical in non-homologous end-joining (NHEJ) repair of double-stranded breaks in DNA, V(D)J recombination in vertebrate lymphoid cells and telomere end protection (Downs and Jackson, 2004; Featherstone and Jackson, 1999; Fisher and Zakian, 2005; Kotnis et al., 2008; Lieber et al., 2004; Melnikova et al., 2005; Sandor et al., 2004). Eukaryotic Ku is a heterodimer comprised of two homologous polypeptides, Ku70 and Ku80. Ku70 and Ku80 share an N-terminal von Willebrand factor A (vWA) domain, a central dsDNA-binding core module and a C-terminal helical domain (CT) (Jones et al., 2001; Walker et al., 2001). Ku70 additionally contains a fourth domain, the C-terminal DNA-binding SAP domain (Aravind and Koonin, 2000a), whereas Ku80 possesses an extension involved in DNA-dependent protein kinase catalytic subunit (DNA-PKcs) recruitment (Fig. 1a). With a few exceptions, like the Streptomyces version, prokaryotic and bacteriophage Ku proteins are much smaller and contain only the dsDNA binding core module (Aravind and Koonin, 2001; d’Adda di Fagagna et al., 2003; Doherty et al., 2001; Hudson et al., 2005; Pitcher et al., 2007). The prokaryotic versions of Ku are typically found in conserved mobile operons that additionally encode an eukaryote-type primase, an ATP-dependent DNA ligase and, in certain cases, a predicted nuclease, which together constitute a NHEJ DNA repair system (Aravind and Koonin, 2001; Della et al., 2004).
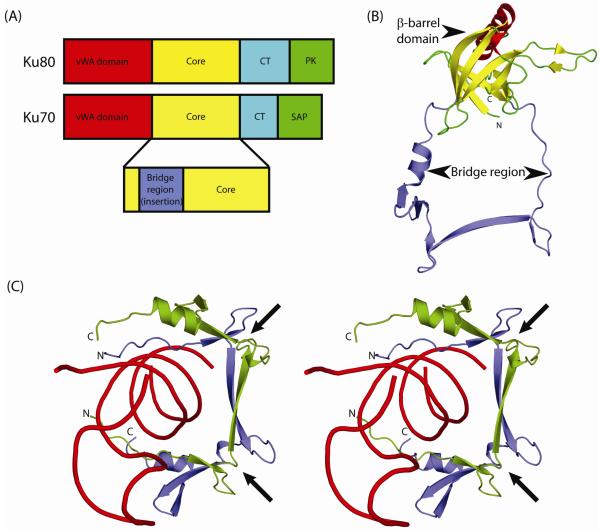
Figure 1.
Domain architecture of Ku.
(A) Eukaryotic Ku is a heterodimer of two similar polypeptides Ku70 and Ku80. Both proteins share a conserved core region (yellow) in addition to an N-terminal von Willebrand factor (vWA; red) domain and a C-terminal helical (CT; cyan) domain. Ku80 contains a C-terminal protein kinase recruitment domain (PK; green) while Ku70 has a SAP domain (green) implicated in DNA binding. Most prokaryotic and bacteriophage versions of Ku contain only the core domain.
(B) Ribbon representation of the human Ku70 core domain (PDB: 1jeq chain A). The Ku core domain is a seven-stranded β-barrel that contains a long insertion, between its second and third β-strands, termed the bridge-region. The β-strands of the β-barrel domain are colored yellow and the bridge region is colored blue.
(C) Stereo ribbon diagram of the dsDNA-encircling bridge-region of human Ku heterodimer (PDB: 1jey). The bridge-regions of Ku70 and Ku80 are colored blue and green respectively, with dsDNA colored red. Arrows indicate the locations of the deteriorated Zn-binding sites.
The crystal structures of the human Ku70/80 heterodimer determined in the presence and absence of dsDNA (PDB 1jeq, 1jey), reveals a toroidal dsDNA-binding region, formed via the dimerization of the core modules Ku70 and Ku80 (Jones et al., 2001; Walker et al., 2001). The encirclement of DNA by the Ku heterodimer covers two turns of dsDNA and the protein-DNA interactions are mostly confined to the sugar-phosphate backbone of dsDNA (Walker et al., 2001). Consistent with this mode of binding, Ku’s recognition of dsDNA is neither sequence nor structure-specific, and it can bind a variety of different dsDNA conformations including blunt ends, 5′- and 3′-overhangs and DNA hairpins (Featherstone and Jackson, 1999; Jones et al., 2001). The dsDNA-binding core module of each subunit is composed of a seven-stranded β-barrel domain that contains a long insertion between its second and third β-strands, referred to as the bridge-region (Fig. 1b). These modules pack against each other making extensive contacts via their respective bridge-regions with each adopting a similar extended conformation that resembles the handle of a basket (Jones et al., 2001) (Fig. 1c). This obligate dimeric state of Ku, together with the presence of only a single Ku representative in several prokaryotic lineages, suggests that the prokaryotic Ku proteins are likely to form homodimers (Aravind and Koonin, 2001).
Currently, sequence and structure classification schemes such as SMART (Letunic et al., 2004), Interpro (Mulder et al., 2002), Pfam (Bateman et al., 2004), SCOP (Andreeva et al., 2004; Murzin et al., 1995) and CATH, (Orengo et al., 1997; Pearl et al., 2005) either consider the bridge-region as an extension of the β-barrel core or classify it as a novel domain with no known affinities. Given that this bridge-region is critical for the toroidal DNA-binding by Ku, and its obligate dimerization, we were interested in determining the origins of this module. Here, we demonstrate using comparative sequence and structural analysis that the Ku bridge-region is a segment-swapped Zn-ribbon that has lost its ability to chelate metal in most organisms, and suggest its inclusion as a new family in the ubiquitous Zn-ribbon fold group (Krishna et al., 2003). This finding also helps interpret the distinct DNA-binding mode of Ku and understand the sequence conservation patterns of certain prokaryotic versions.
Material and Methods
The PSI-BLAST program (Altschul and Koonin, 1998; Altschul et al., 1997) was used to detect homologs of human Ku70 and Ku80 proteins in the NCBI non-redundant (NR; Nov 10, 2009; 10,031,959 sequences; 3,421,245,699 total letters) protein sequence database. Independent, iterative PSI-BLAST searches (inclusion threshold of 0.01) using as query the sequences of the bridge-regions of the human Ku70 and Ku80 proteins (gi: 119580852, range 226-301, gi:10863945, range 257-337) were carried out, until convergence. The retrieved sequences of homologous proteins were clustered using the BLASTCLUST program of the BLAST suite (25% sequence identity), and a representative sequence from each cluster was used to seed new PSI-BLAST searches. Similar searches were performed with the sequences of the complete Ku70/80 β-barrel core domains containing the inserted bridge domain in order to identify remote homologs of the bridge domain that have diverged beyond recognition at the bridge-region but display reasonable sequence similarity at other regions of the core domain. For short proteins such as zinc fingers and disulfide-rich domains, sequence and structure similarity statistics for remote homologs tend to be unreliable due to the short length of their polypeptide chains and biased amino acid composition (Grishin, 2001). Thus, using the complete core domain sequences of Ku70/80 for PSI-BLAST searches helped in reliably identifying very distant homologs of the bridge-region.
Multiple sequence alignments of Ku homologs was constructed using PROMALS3D (Pei et al., 2008) program with default parameters and the resulting alignment was manually refined using the available three-dimensional structure of Ku and the presence of specific conserved zinc-chelating motifs.
The structures of Ku and other Zn-ribbon proteins were retrieved from the PDB (Berman et al., 2000) and structural similarity searches were performed using the DALI (Holm and Sander, 1995) and FATCAT servers (Ye and Godzik, 2004a; Ye and Godzik, 2004b). The structures of the individual bridge regions of Ku (PDB accession code 1jey and 1jeq), as well as segment-swapped versions of Ku that were composed of parts of the Ku70 and Ku80 proteins were used as queries in these structural similarity searches. The structures of the Ku bridge region were superposed on other members of the Zn-ribbon fold group using the Pair Fitting wizard of the molecular visualization program PyMOL (DeLano Scientific) by manually defining equivalent regions.
Results and Discussion
Sequence analysis and identification of a potential Zn-ribbon in the Ku bridge region
Sequences of Ku proteins were retrieved from the NCBI non-redundant (NR) protein sequence database by running iterative PSI-BLAST (Altschul and Koonin, 1998; Altschul et al., 1997) searches until convergence, as described in the Materials and Methods section.
Multiple sequence alignments of Ku proteins reveal that the bridge-region is overall well conserved with very few insertions and deletions seen between domains from different organisms (Fig. 2). Furthermore, this region is present in all known homologs of Ku and we failed to identify even a single version of the Ku β-barrel domain that lacks the bridge region. This suggests that the bridge region is likely to represent a universal interaction surface of these proteins. Interestingly, we observed that Ku proteins of several Gram-positive bacteria, for example Moorella thermoacetica (gi: 83590332; Fig. 2), contained cysteine and histidine residues in pairs (HxxC and CxxC, where x is any residue) in the bridge-region, which showed a perfect match to the Zn-chelating sites of Zn-ribbon domains. When superimposed on the secondary structure these putative Zn-chelating residues are associated with many conserved hydrophobic and polar residues that define the long β-strands of the bridge region (Fig. 2), and recapitulate the location of the cation-chelating residues observed in Zn-ribbon domains. Several other Ku homologs of bacterial origin display partial conservation of the aforementioned Zn-chelating residue pairs, whereas all eukaryotic versions entirely lack the Zn-chelating residues. This suggests that the bridge region originated as a Zn-chelating domain that in course of evolution lost its cation-binding ability.
Figure 2.
Multiple sequence alignment for representative proteins of the Ku superfamily. The first and last residue numbers are indicated before and after each sequence in the alignment. Excluded residues are specified as numbers. The secondary structure diagram based on the crystal structure of the human Ku70/Ku80 (PDB: 1jeq) dimer is indicated above the alignment and the constituents of the Zn-binding site are indicated as green circles. The alignment has been colored as per the 85% consensus, with ‘h’ being hydrophobic, ‘l’ the aliphatic subset thereof, ‘p’ being polar, ‘s’ being small, ‘−‘ being negatively charged, ‘+’ being positively charged, b being bulky, u being tiny and ‘a’ being aromatic residues. The Zn-binding positions are shaded differently in the proteins in which the Zn-chelating residues are retained. The organism abbreviations are -- Aful : Archaeoglobus fulgidus; Aman : Aurantimonas manganoxydans; Atha : Arabidopsis thaliana; BPCorndog : Mycobacterium phage Corndog; Bbre : Brevibacillus brevis; Bhal : Bacillus halodurans; Bjap : Bradyrhizobium japonicum; Bsub : Bacillus subtilis; Drer : Danio rerio; Fsp. : Frankia sp.; Hmod : Heliobacterium modesticaldum; Hsap : Homo sapiens; Lsph : Lysinibacillus sphaericus; Mlot : Mesorhizobium loti; Moth : Moorella thermoacetica; Mtub : Mycobacterium tuberculosis; Ncra : Neurospora crassa; Nsp. : Nocardioides sp.; Nwin : Nitrobacter winogradskyi; Paer : Pseudomonas aeruginosa; Scel : Sorangium cellulosum; Sery : Saccharopolyspora erythraea; Spom : Schizosaccharomyces pombe; Swol : Syntrophomonas wolfei; Tcru : Trypanosoma cruzi
We could detect Ku proteins with intact ligand-binding site only from firmicutes, actinobacteria (the Gram-positive bacteria) and their viruses such as the mycobacteriophage Corndog (Fig. 2). These proteins either contain a conventional Zn-ribbon pattern with a HxxC and CxxC motif or a slightly modified version where one or more of the cysteines have been replaced by acidic residues. These acidic residues could still potentially chelate zinc along with the rest of the intact Zn-contacting ligands. The sole archaeon with a Ku homolog, Archaeoglobus fulgidus, which incidentally also retains Zn-chelating ligands appears to have acquired its Ku via a relatively late lateral gene transfer from Gram-positive bacteria. Within the actinobacteria group, certain organisms (e.g. Frankia; Fig. 2) already display deterioration of the Zn-binding sites. Outside of the firmicutes and actinobacteria the Zn-binding sites have been completely lost. Especially, in proteobacteria, several of which might have multiple Ku homologs, the Zn-ligands have been completely lost (Fig. 2). Several alphaproteobacteria have a Ku operon with two distinct Ku homologs often occurring as tandem genes. Both proteins display deterioration of the Zn-binding sites similar to the eukaryotic Ku70 and Ku80 orthologs. Thus, eukaryotes not only share the presence of a heterodimeric version of the Ku protein with the alphaproteobacteria, but also similarly lack the Zn-ligands. Given these observations and the evidence that the alphaproteobacteria are the precursors of the mitochondria (Andersson et al., 2003), we posit that eukaryotic heterodimeric Ku is a functional equivalent of the corresponding heterodimeric version originally acquired from the mitochondrial precursor.
Structural evidence for the origin of the Ku bridge region from a Zn-ribbon
The overall three-dimensional structure of protein domains remain conserved even after their sequences have diverged substantially during the course of evolution. Hence, it is possible to infer remote evolutionary relationship between protein domains even after their definitive sequence motifs have eroded. The bridge-region in Ku70/Ku80 structure contains two distinctive β-hairpins with peculiar turns. The only cognates of these structures of the Ku70/Ku80 proteins are the knuckles, which are distinctive turns found in Zn-binding domains having a consensus sequence of CPXCG and chelate zinc via the side-chains of cysteine residues (Grishin, 2001; Krishna et al., 2003) (Fig. 3; knuckles colored red). In the Ku70/Ku80 structure, the first knuckle-like region in Ku70 is followed by a long β-strand that forms hydrogen bonds with the corresponding region from the Ku80 bridge-region. This region is then followed by the second zinc knuckle-like region, a short α-helix, two antiparallel β-strands and another short α-helix that is linked to the third β-strand of the β-barrel core by a long loop region (Fig. 1b). Visual examination of the heterodimeric bridge-region of human Ku shows a striking resemblance to members of the Zn-ribbon fold, which however is not apparent from the monomeric bridge structures. Thus, this similarity to the Zn-ribbon fold only emerges in the dimer and can be rationalized only upon assuming a segment-swap between the Ku70 and Ku80 bridge-regions (Fig. 3a). Consistent with this, a structural similarity search of the individual bridge-regions of Ku70 and Ku80 using the DALI (Holm and Sander, 1995) and FATCAT (Ye and Godzik, 2004a; Ye and Godzik, 2004b) servers failed to find reasonable structural similarity with any other protein. However, a search with a segment-swapped domain composed of parts of the Ku70 and Ku80 proteins, could find links to the YfgJ protein from Escherichia coli (PDB: 2jne), transcriptional elongation factor TFIIS (PDB: 1tfi), eukaryotic RNA polymerase II subunit 9 (RPB9; PDB 1qyp) as top hits, in addition to several other Zn-ribbon domains.
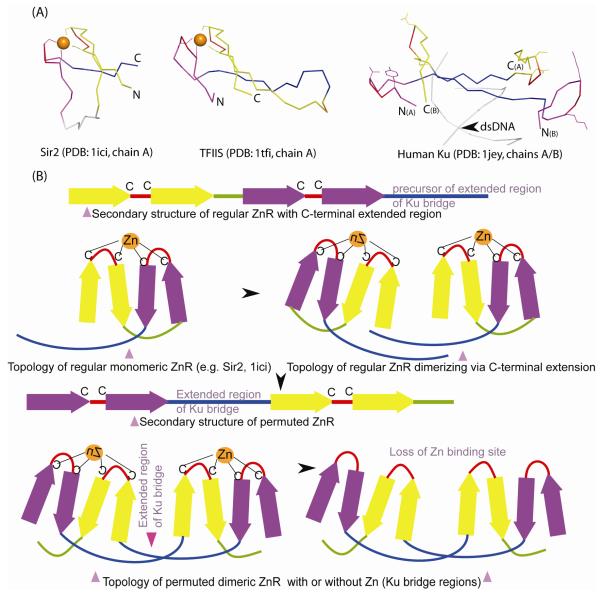
Figure 3.
Structural comparison of Ku bridge-region to Zn-ribbons.
(A) Structural diagrams of Zn-ribbons from Sir2 (left; 1ici: chain B, 119-156), TFSII (middle; 1tfi: chain A, 8-50) and bridge-region of the human Ku heterodimer (right; 1jey: chain A, 287-313; chain B, 277-307) are shown. The structures were superimposed using the program PyMOL by manually defining equivalent residue pairs from regions comprising the zinc knuckles (red, purple) and the β-strands (yellow, blue) and translated for clarity. In some of the alignments, circular permutation needs to be assumed in order to obtain best structural superimposition. Structurally equivalent regions are colored similarly. The zinc chelating residues of Sir2 and TFSII and the equivalent residues from Ku are shown as ball-and-sticks. The zinc ion is indicated as an orange sphere. The primary β-hairpin of the Zn-ribbon is colored purple, the extended region of Ku-bridge in blue, the zinc knuckle in red and the other β-strands in yellow.
(B) A permutation scheme for arriving at the Ku bridge-region structure from a Sir2-like precursor. In its ancestral state, Ku Zn-ribbons, resembling Sir2, likely dimerized via their C-terminal extended regions. A circular permutation resulted in placing the extended region of Ku-bridge between the primary and secondary zinc knuckles (colored purple and yellow) and extensive hydrogen-bonding interactions likely locked this atypical conformation in place. This is then likely to have been followed by the loss of Zn-binding sites, to abolish zinc dependence of NHEJ.
These observations suggest that indeed the bridge region of the Ku domain is derived from a regular Zn-ribbon via assembly of the cation-chelating sites between the knuckles from two independent monomers, instead of between the N- and C-terminal knuckles of the same polypeptide (Fig. 3a, b). The regions encompassing the Zn-chelating knuckles have an unusual backbone and side-chain geometry that are probably not among the most favorable conformations associated with regular turns in globular proteins. This observation, together with the detection of several bacterial versions containing functional Zn-chelating residues typical of Zn-ribbons (Fig. 2), suggest that these structures indeed arose from an ancestral zinc-chelating site, with secondary losses, including in the common ancestor of the eukaryotic Ku70 and Ku80. Strengthening of the hydrogen bonding between the extended regions in the bridge-region appears to have allowed stabilization of the structure even upon loss of metal chelation.
Evolutionary and functional considerations
Despite being a very simple structural scaffold, Zn-ribbons have been utilized in a wide range of biochemical functions, such as nucleic acid binding, protein interaction (including dimerization), and as a cofactor providing a metal in redox reactions (Aravind and Koonin, 1999; Krishna et al., 2003). To better understand how the Zn-ribbon scaffold has been utilized in the Ku bridge-region we compared it to different Zn-ribbons such as those from the transcription factor TFIIS (DNA-binding; 1tfi), primase DNAG (DNA-binding; 1d0q) and Sir2 (dimerization; PDB:1ici) (Min et al., 2001; Pan and Wigley, 2000; Qian et al., 1993). The structures were superimposed using the program PyMOL by defining structurally equivalent residues based on visual inspection. These comparisons reveal that both the segment-swapped units from Ku are structurally similar to Zn-ribbons such as the nucleic-acid-binding TFIIS and the protein-interacting Zn-ribbon of Sir2 (Fig. 3a). The dimerization mode of the Sir2 and segment-swapped units of the Ku bridge domain are very distinct; however, both share a long strand that sweeps across the hairpin dyad (the principal extended segment of the bridge region). In each of the Zn-ribbons used in the comparison the long β-strands trailing off from the knuckles play a critical role in their interactions, just as what is observed in the DNA-protein interactions mediated by the two Zn-ribbon units of the Ku dimer (Fig. 3a) (Min et al., 2001; Qian et al., 1993; Walker et al., 2001). This suggests that the segment swapping notwithstanding the ancestral mode of substrate interaction of the Zn-ribbon fold has been retained in the Ku-bridge region.
Interestingly, the structural alignment generated from our structural superposition, as well as visual examination, suggest a circular permutation with respect to the Zn-ribbon of Sir2 (Fig. 3a, b). Circular permutations are not unprecedented among protein- or nucleic-acid-interacting Zn-ribbons, as well as rubredoxins. For instance, previous analysis has shown that homologous Zn-ribbons, such as those found in the Ile-tRNA synthetase (PDB 1f4l), are related by circular permutation (Krishna et al., 2003). The identification of the Ku bridge-region as a segment-swapped, permuted Zn-ribbon has considerable evolutionary as well as functional implications. In terms of an evolutionary scenario, these observations suggest that in the ancestral state the β-barrel and Zn-ribbon domains probably collaborated as separate polypeptides, which interacted with dsDNA by binding it on opposite sides of the double helix. This was followed by the insertion of the Zn-ribbon into the N-terminus of the β-barrel. The closest relative of the Ku β-barrel domain is the SPOC domain (Andreeva et al., 2004; Ariyoshi and Schwabe, 2003), which, lacks the bridge module and provides a model for the structural prototype of the Ku barrel prior to the Zn-ribbon insertion. However, SPOC is restricted to eukaryotes and is thus unlikely to be the precursor of the Ku β-barrel domain. All extant Ku proteins possess the bridge region suggesting that Zn-ribbon insertion occurred prior to their common ancestor. It is unclear if the permutation of the Zn-ribbon preceded or occurred after the insertion into the β-barrel domain. However, given that all Ku homologs are collinear in their alignment in the region corresponding to the bridge (Fig. 2), it is clear that their common ancestor already possessed the permuted form. A likely development from this permutation was ability of the Zn-ribbons to be reconstituted from the swapped segments of the individual subunits, thereby favoring the emergence of obligate dimerization in Ku proteins.
In functional terms, our findings provide a structural basis for exploring previous proposals about the possible role of the bridge-region in Ku function (Jones et al., 2001; Walker et al., 2001). The identification of the Zn-ribbon in the bridge-region suggests that the reconstitution of a pair of complete Zn-ribbons from two separate Ku polypeptides is central to the mechanism by which Ku dimerizes to form a clamp around DNA ends – the complete DNA binding site is available only when the segment-swapped elements are combined. Further, we predict that the unusual conformations typical of the knuckles of Zn-ribbons is likely to allow the bridge to function as a sensor that allows the Ku dimer to dissociate from both linear and circular dsDNA after NHEJ has been achieved. It is possible that these functions favored the loss of Zn-binding (to eliminate zinc dependence) while retaining the unusual knuckle conformations intact (for sensor function). In light of this, we propose that the Ku domains of Gram-positive bacteria with intact metal chelating residues are indeed likely to retain zinc dependence for their assembly.
Conclusions
Zn-ribbons are amongst the most widespread of small, zinc-stabilized, protein domains (Krishna et al., 2003). Like other small protein domains, which are predominantly supported by metal-chelation or disulfide bonds (cysteine rich domains) and typically lack extensive secondary structure elements, Zn-ribbons are hard to detect, delineate and classify by entirely automatic methods. Further, on number of occasions the metal-binding sites of such proteins have deteriorated during evolution. This divergence, with further structural modifications like circular permutations and insertion into larger domains makes uncovering their evolutionary connection to their classical Zn-binding homologs non-trivial. So far almost all identified cases of deteriorated zinc fingers have been by experts who use a combination of sequence, structural and functional information to identify, delineate and classify such domains (Aravind and Koonin, 1999; Aravind and Koonin, 2000b; Grishin, 2000; Grishin, 2001; Krishna and Grishin, 2004; Krishna et al., 2003; Makarova et al., 2001; Mulder et al., 2002; Murzin et al., 1995; Orengo et al., 1997). Nevertheless, identification of these domains not only helps in elucidating evolutionary puzzles of particular protein families, but also provides key functional insights regarding these proteins.
The bridge-region of the Ku superfamily represents the first example of a segment-swapped version of the Zn-ribbon. Our findings illustrate the remarkable adaptability of this simple protein scaffold and extend the range of functional features associated with the Zn-ribbon domain. Based on the preservation of the Zn-ribbon structure even after loss of the zinc-chelating sites, we propose that it is key for the action of Ku in its conserved role in NHEJ. Our prediction that this atypical Zn-ribbon structure is likely to be the primary sensor for the DNA state during NHEJ lends itself to direct testing and might further illuminate the molecular details of Ku function.
Acknowledgements
LA was supported by the intramural funds of the National Library of Medicine, NIH. SSK is supported by the CSIR (India). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST--a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–7. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358:165–77. doi: 10.1098/rstb.2002.1193. discussion 177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–9. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–70. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000a;25:112–4. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000b;10:R132–4. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–74. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–20. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–41. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissett NC, Doherty AJ. Repairing DNA double-strand breaks by the prokaryotic non-homologous end-joining pathway. Biochem Soc Trans. 2009;37:539–45. doi: 10.1042/BST0370539. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Weller GR, Doherty AJ, Jackson SP. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003;4:47–52. doi: 10.1038/sj.embor.embor709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–5. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- Doherty AJ, Jackson SP, Weller GR. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 2001;500:186–8. doi: 10.1016/s0014-5793(01)02589-3. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–78. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. Ku, a DNA repair protein with multiple cellular functions? Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- Fisher TS, Zakian VA. Ku: A multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005 doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Grishin NV. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J Mol Biol. 2000;299:1165–77. doi: 10.1006/jmbi.2000.3841. [DOI] [PubMed] [Google Scholar]
- Grishin NV. Treble clef finger--a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–14. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–80. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Hudson JJ, Hsu DW, Guo K, Zhukovskaya N, Liu PH, Williams JG, Pears CJ, Lakin ND. DNA-PKcs-Dependent Signaling of DNA Damage in Dictyostelium discoideum. Curr Biol. 2005;15:1880–5. doi: 10.1016/j.cub.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Jones JM, Gellert M, Yang W. A Ku bridge over broken DNA. Structure (Camb) 2001;9:881–4. doi: 10.1016/s0969-2126(01)00658-x. [DOI] [PubMed] [Google Scholar]
- Kotnis A, Du L, Liu C, Popov SW, Pan-Hammarstrom Q. Review. Non-homologous end joining in class switch recombination: the beginning of the end. Philos Trans R Soc Lond B Biol Sci. 2008 doi: 10.1098/rstb.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna SS, Grishin NV. The finger domain of the human deubiquitinating enzyme HAUSP is a zinc ribbon. Cell Cycle. 2004;3:1046–9. [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–50. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–4. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–26. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–50. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0033. RESEARCH 0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova L, Biessmann H, Georgiev P. The Ku protein complex is involved in length regulation of Drosophila telomeres. Genetics. 2005;170:221–35. doi: 10.1534/genetics.104.034538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–79. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P, Copley R, Courcelle E, Durbin R, Falquet L, Fleischmann W, Gouzy J, Griffith-Jones S, Haft D, Hermjakob H, Hulo N, Kahn D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Orchard S, Pagni M, Peyruc D, Ponting CP, Servant F, Sigrist CJ. InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform. 2002;3:225–35. doi: 10.1093/bib/3.3.225. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5:1093–108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Pan H, Wigley DB. Structure of the zinc-binding domain of Bacillus stearothermophilus DNA primase. Structure. 2000;8:231–9. doi: 10.1016/s0969-2126(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, Lee D, Akpor A, Maibaum M, Harrison A, Dallman T, Reeves G, Diboun I, Addou S, Lise S, Johnston C, Sillero A, Thornton J, Orengo C. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–51. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher RS, Brissett NC, Doherty AJ. Nonhomologous end-joining in bacteria: a microbial perspective. Annu Rev Microbiol. 2007;61:259–82. doi: 10.1146/annurev.micro.61.080706.093354. [DOI] [PubMed] [Google Scholar]
- Qian X, Gozani SN, Yoon H, Jeon CJ, Agarwal K, Weiss MA. Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry. 1993;32:9944–59. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- Sandor Z, Calicchio ML, Sargent RG, Roth DB, Wilson JH. Distinct requirements for Ku in N nucleotide addition at V(D)J- and non-V(D)J-generated double-strand breaks. Nucleic Acids Res. 2004;32:1866–73. doi: 10.1093/nar/gkh502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–14. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–24. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Database searching by flexible protein structure alignment. Protein Sci. 2004a;13:1841–50. doi: 10.1110/ps.03602304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004b;32:W582–5. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]