Summary
The history of the discovery of the microphthalmia locus and its gene, now called Mitf, is a testament to the triumph of serendipity. Although the first microphthalmia mutation was discovered among the descendants of a mouse that was irradiated for the purpose of mutagenesis, the mutation most likely was not radiation-induced but occurred spontaneously in one of the parents of a later breeding. Although Mitf might eventually have been identified by other molecular genetic techniques, it was first cloned from a chance transgene insertion at the microphthalmia locus. And although Mitf was found to encode a member of a well-known transcription factor family, its analysis might still be in its infancy had Mitf not turned out to be of crucial importance for the physiology and pathology of many distinct organs, including eye, ear, immune system, bone, and skin, and in particular for melanoma. In fact, near seven decades of Mitf research have led to many insights about development, function, degeneration, and malignancies of a number of specific cell types, and it is hoped that these insights will one day lead to therapies benefitting those afflicted with diseases originating in these cell types.
Introduction
The living world presents us with a grandiose display of shades and colors. In domestic species, these shades and colors were selected by us, mostly to delight us; in nature, they were selected by evolution, to attract, impress, confuse, or simply to blend in. Color traits, as any traits, are ultimately influenced by genes, some of which acting directly on coloration and others indirectly by influencing how environmental factors are used for coloration. Much of our knowledge on how these genes work comes from the analysis of spontaneous or induced mutations. Many of these mutations affect only pigmentation, but some lead to combinations of phenotypes in multiple tissues, even tissues of distinct embryological origin. It is these pleiotropic mutations that have especially intrigued geneticists interested in pigmentation.
One of the prototypical pleiotropic pigmentation loci is the microphthalmia locus which has now become important for many divergent fields, as attested by the reviews by Steingrimsson, Liu and Fisher, and Hoek and Goding that accompany this article. In fact, microphthalmia plays prominent roles for instance in the biology and pathology of pigmentation of the skin and its appendages, in eye development and degeneration, and in hearing research. The first mutation at this locus was discovered by Paula Hertwig in Berlin among the descendants of an irradiated mouse and was first published in 1942 (Hertwig, 1942)(see also Steingrimsson, this issue). Later, many other mutant alleles of the locus have been found in mice and other vertebrates (reviewed in Steingrimsson et al., 2004). The corresponding gene, Mitf, was cloned in 1992 and first published in 1993 (Hodgkinson et al., 1993). The gene is expressed in most cell types and has major roles in neural crest-derived and neuroepithelium-derived melanin-bearing pigment cells. Melanocytes, for instance, are regulated by Mitf at the level of specification, proliferation, survival, migration, differentiation, replenishment during feather and hair cycles, and malignant transformation. Retinal pigment epithelium cells are regulated by Mitf at the level of developmental specification and proliferation and consequently in their function in retinal physiology in the adult. Mitf also regulates mast cell development and function which may include protection against microbial infections, and it regulates osteoclasts in their role in bone remodeling. It does all this by encoding a basic-helix-loop-helix-leucine zipper transcription factor, MITF, that forms dimers capable of binding specific DNA sequences in the regulatory regions of a large number of target genes that in turn control the different cellular processes (see Cheli et al., 2010, for a recent review). MITF does not only form homodimers, however, but can also heterodimerize with TCFEB, TCFE3 and TCFEC, three related proteins with which it forms a small subfamily. The common ancestor of the corresponding genes is found in invertebrates including Tripedalia (jellyfish) (Kozmik et al., 2008) and Drosophila (Hallsson et al., 2004), where its function may be in eye development, and in C. elegans (Rehli et al., 1999), where its function is still unknown.
In the past, the acronym Mitf has also been applied to unrelated genes including one called mouse intestinal trefoil factor (mITF, now called Tffe)(Tomita et al., 1995) and one encoding a secreted and allergenic ribonuclease of aspergillus fumigatus (MITf, GenBank # X58278.1, also known under several other names including AspF1). In addition, the abbreviation is used widely outside of biology (my favorite is as acronym for “Most Important Things First”). In this short review, I will describe the history of the discovery of the microphthalmia locus and, to the best of my recollection, the identification of the corresponding Mitf gene.
The discovery of the microphthalmia locus
As mentioned, the first mutation at the microphthalmia locus was described by Paula Hertwig (Fig. 1). Paula was born in 1889 in Berlin, Germany, as the second child of the eminent anatomist and biologist Oscar Hertwig and his wife, Marie. Paula received her Ph.D. in 1916 from the Anatomical-Biological Institute of the Medical Faculty in Berlin but published her first study already in 1911, on changes in mitotic figures of Ascaris eggs after irradiation. A pioneer in many regards, she was the first woman to habilitate in Berlin, the first woman member of the Medical Faculty of the University of Halle, and this university’s first woman Dean. During her long career, she focused on the study of the biological effects of radiation, an interest that was prompted by the use of X-rays for the temporary sterilisation of women as practiced in the beginning of the 20th century. Using mostly mice for her studies, she isolated mutants that became useful models for the investigation of human diseases of the skin, eye, ear, and skeleton. In fact, her studies led her to become one of the early voices who warned of potential damages that could arise from the application of X-rays particularly to reproductive organs. She was also politically engaged, especially before World War II when she served as a member of the last Prussian Parliament. She reduced her political activity to what she considered minimally required during the reign of national socialism in Germany, but helped to shape public organizations again after the war in the German Democratic Republic until she retreated from politics with the rise of the Socialist Unity Party (SED). She became emeritus in 1957, moved to the Black Forest area in 1972, and died in 1983 (see Gerstengarbe, 2003, for a short biography).
Fig. 1.
Paula Hertwig at the microscope (reproduced with permission from BIOspektrum)
Genetical research was well supported in Germany when in 1942, Hertwig published, in her native German, the first account of white, small-eyed (microphthalmic) mice along with five other novel mutants (Hertwig, 1942). The microphthalmic mice arose among the offspring of a cross of an inbred albino mouse with a male, labeled ♂ 944, that descended from an irradiated male. Male 944, however, was not himself considered mutated as his offspring obtained with five of his own daughters were all normal (“…so dass der Bock 944 als unbelastet gerechnet wurde”, Hertwig, 1942). Nevertheless, a son obtained from a mating of male 944 with the above mentioned inbred albino mouse, when crossed with two of his sisters, produced offspring from one of the two that violated the expected 3:1 split of pigmented versus albino coats: there were 33 pigmented and 46 white ones. Hertwig described how she first thought that all white ones were simply albinos and how she only later realized that the eyes of some of them were very small and their lids unopened. In fact, she writes, she only recognized in subsequent breedings that she had discovered a new recessive gene with pleiotropic effects. She concluded that the mutation likely arose in one of the gametes of male 944, or alternatively was derived from the albino female with which this male was crossed.
The mutant homozygotes had unpigmented eyes of variably small size, bent whiskers, lacked eruption of upper and lower incisures, and died soon after weaning. Heterozygotes showed only reduced eye pigmentation at birth but no evidence for coat color defects. It might be for these reasons that Hertwig called the new gene “Microphthalmus” (symbol “m”), and hence named it according to the eye phenotype, rather than the coat phenotype which is much more common among the subsequently isolated microphthalmia alleles. Neverthless, Hertwig writes that with the discovery of m, “for the first time, a second recessive factor for absence of pigmentation [besides albino] has been found in mice, or in rodents in general” (Hertwig, 1942). Perhaps by using this seemingly redundant formulation (any second discovery can be done only once and hence is always done “for the first time”), Hertwig may have wanted to especially emphasize the importance of the pigmentary part of the phenotype of Microphthalmus. In 1948, Hans Grüneberg, who had obtained three microphthalmia mice in 1946 “through the good offices of Prof. R.A. Fisher (Cambridge)” and had analyzed them independently, suggested to use the symbol mi because m had previously been used for the mutation misty (Grüneberg, 1948).
Microphthalmia turned out to be a highly interesting locus not only because of its pleiotropic effects on seemingly unrelated tissues such as the retinal pigment epithelium, melanocytes, and long bones, but also because some mutant alleles show complex inter- and even intra-allelic interactions. Interallelic complementation, for instance, has been observed with Microphthalmia-white (Miwh) and many other alleles, including mi (Konyukhov and Osipov, 1968; Steingrimsson et al., 2003). Miwh homozygotes are white and have small eyes similar to mice homozygous for mi, but Miwh/mi compound heterozygotes have eyes of normal size, though they remain unpigmented (for a more detailed description, see also Steingrimsson, this issue). These observations have led to the hypothesis that the locus is complex, meaning that the complete phenotype as seen in Hertwig’s original microphthalmia mutation might have resulted from defects in at least two very closely linked genes (Hollander, 1968). An opposite phenomenon, interallelic enhancement or aggravation, is seen when mice carrying the microphthalmia-spotted (misp) allele are crossed with mice carrying other alleles such as mi (Steingrimsson et al., 2003). Contrary to what its name suggests, the misp allele does not confer spotting by itself, not even when homozygous, and mi/+ heterozygotes either have no coat phenotype, as originally observed by Hertwig (Hertwig, 1942), or may show only minor white spots on head or belly, as observed by Grüneberg (Grüneberg, 1948). Compound heterozygotes (mi/misp), however, have an entirely white coat, though normal-sized, pigmented eyes (Steingrimsson et al., 2003). In fact, the name “spotted” in misp is derived from its discovery in a colony segregating Miwh, when a novel spotting phenotype spontaneously appeared that was unlike that seen in Miwh heterozygotes (Wolfe and Coleman, 1964).
The phenomenon of interallelic enhancement has recently prompted a search for additional mutations that might influence the phenotype in compound heterozygotes with misp. This led to the intriguing discovery of an intraallelic ( or intragenic) mutation in misp that largely corrects the phenotypic interallelic enhancement associated with the original misp allele (Steingrimsson et al., personal communication). Moreover, microphthalmia mutations also interact with mutant alleles of other genes regulating pigmentation, such as Kit (Beechey and Harrison, 1994; Hou et al., 2000; Diwakar et al., 2008; Wen et al., 2010). Evidently, deeper insights into this complexity could only be provided by the molecular analysis of the corresponding gene or genes.
Cloning of the microphthalmia gene
The gene residing at the microphthalmia locus might have been identified by a number of different strategies. Nowadays, with the availability of the mouse genome sequence and the mapping of the locus to a short interval, one might have chosen a transgenic rescue strategy with bacterial artificial chromosomes (BACs) containing candidate genes from the interval. Even if more then one gene had been involved, one might have expected at least a partial rescue with single BACs unless the gene or genes in question were not functionally represented on such BACs (but now we know that BAC rescue works even with an Mitf gene that is not fully intact, Bauer et al., 2009). When the microphthalmia gene was cloned, however, BAC rescue was not readily available, and other strategies had to be evaluated. It was known, for instance that mi-mutant mast cells can be grown much as wild-type mast cells when kept in the presence of growth factors such as IL-3. Upon removal of IL-3, however, mi-mutant mast cells stop incorporating BrdU, in contrast to wild-type mast cells (Ebi et al., 1990). Hence, one might have tried to rescue IL-3-deprived mutant mast cells with cDNA expression libraries prepared from wild-type mast cells, with the caveat that overexpression of Mitf, for instance, might have accelerated cell death, rather than rescued the mutant cells, as later found by the group of Kitamura (Tsujimura et al., 1997). In fact, the strategy that was finally successful was based on cloning genomic flanking regions of transgene arrays that by mere chance had inserted into the microphthalmia locus and reproduced the microphthalmia phenotype (Hodgkinson et al., 1993; Hughes et al., 1993).
The project that led to the identification of the Mitf gene in our laboratory was initiated by Harold Gainer, National Institue of Neurological Disorders and Stroke (NINDS), and James Battey, then at NINDS and now Director of the National Institute of Deafness and Other Communication Disorders (NIDCD). They were interested in the question of promoter regulation of the arginine vasopressin (Avp) and oxytocin (Oxt) genes. In the mouse, these two genes are juxtaposed on chromosome 2, with their 3’ ends pointing towards each other and separated by only approximately 3.5 kb of sequence. Hence, Yoshinobu Hara, a visiting fellow in Gainer’s laboratory, and Battey constructed transgenes which linked the putative upstream regulatory regions plus exon 1 and a small part of exon 2 of Avp, and separately the respective regions of Oxt, with sequences for the bacterial β-galactosidase and the human AVP 3’ end. Using these constructs for standard pronuclear injection, I generated, in the Spring of 1988, ten living Oxt and eight living Avp transgenic founders. Unfortunately, with respect to the original question of the neuropeptides’ gene regulation, the experiment was a complete failure: none of the transgenic offspring showed Xgal labeling because the β-galactosidase portion of the transgene contained a frameshift mutation that was not recognized before the mice were made. Moreover, in situ hybridization showed that none of the offspring expressed transgenic β-galactosidase RNA in brain nuclei where the respective endogenous genes were known to be expressed (though some showed expression in other brain areas). Because it was theoretically possible that transgene expression in the expected brain areas might have been weak in mice heterozygous at the transgene locus and only visible in homozygotes, Yoshinobu intercrossed the offspring of each line. But even this did not yield the hoped-for results. Instead, it led to the surprise observation of white mice appearing among the offspring of the Avp line originating from founder #1034 which Yoshinobu called vga-9 (#1034 was the ninth mouse analyzed from the first injection session using the vasopressin-beta-galactosidase transgene). My initial reaction was that these white mice were the result of an inadvertant cross with an albino mouse. In retrospect, I reacted just as Hertwig did when she was confronted with overly numerous white offspring. Like Hertwig, I also needed a second look to see that the white mice had, in fact, small red eyes. Yoshinobu, however, had done his homework and pointed out that the white mice looked similar to mi mice. Little did I know at the moment that this observation was the beginning of a new field of research and a switch of my career from 20 years of virology to now nearly 20 years of developmental biology. Also, there is a fine irony here. Had the experiment not been such a failure, the idea of systematically generating transgene-homozygotes might never have come up, and given the high costs of keeping laboratory mice, the vga-9 line might have been terminated long before the appearance of any white and small-eyed mouse.
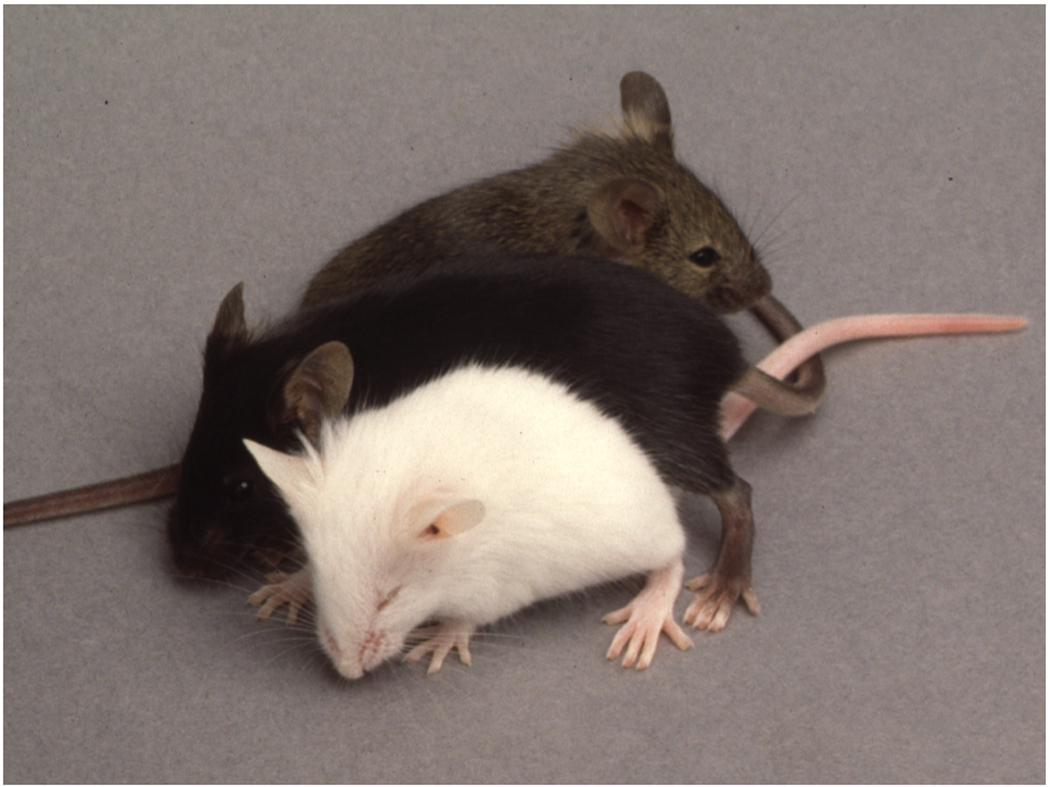
Admittedly, my interest in these mice was not very high in the beginning. They just represented a nuisance, and we spent precious time breeding them without analyzing them. Nevertheless, at a 1990 Mouse Molecular Genetics Meeting at Cold Spring Harbor Laboratory, I must have mentioned them to Lesley Forrester, then working with Alan Bernstein in Toronto on questions of mast cell biology, and so we sent her the mice to test for mast cell deficiencies and for complementation with her mi/+ mice. The results, obtained in 1991, were clear: vga-9 homozygotes had a severe mast cell defect, and some of the F1 mice obtained from crosses between vga-9 and mi heterozygotes looked like mi/mi homozygotes (Fig. 2), all of them carrying the vga-9 transgene. This convinced us that vga-9 was indeed a mutation at the microphthalmia locus. At some point I also mentioned the white mice to Karen Steel, then at the MRC Institute of Hearing Research in Nottingham, who asked me whether I had tested their hearing. I must have looked nonplussed but Karen patiently explained that our white mice might have a hearing problem similar to that she had found in mice homozygous for viable dominant spotting (KitWv)(Steel et al., 1987). This prompted a collaboration with Masayoshi Tachibana working at NIDCD who found that the inner ear of vga-9 mice showed a lack of melanocytes in the stria vascularis of the cochlea and a severe degeneration of outer hair cells and abnormalities in inner hair cells (Tachibana et al., 1992). Lastly, I told Nancy Jenkins from the Mammalian Genetics Laboratoy at the ABL-Basic Research Program of the National Cancer Institute of the mice when she visited the main campus in November 1991. Nancy asked whether we had already collected additional microphthalmia alleles. Of course, the virologists we still were, we had not done so, but I sensed that this might soon change.
Fig. 2.
The vga-9 transgene insertion is allelic with microphthalmia-mi. Vga-9/+ hetero-zygotes were crossed with mi/+ heterozygotes, yielding white, microphthalmic mice that resembled mi/mi homozygotes and carried the vga-9 transgene (bottom mouse). The cross also generated pigmented mice that carried at least one wild-type microphthalmia allele (upper two mice). Photograph courtesy of Lesley Forrester.
At around the same time we tried, unsuccessfully, to clone genomic sequences flanking the transgene insertion by inverse PCR. Using a different strategy, however, we had previously been able to clone one side of a transgene insertion in another line, now known as Meox1Tg(Mx1-TAX)2627Arnh (Mankoo et al., 2003; Skuntz et al., 2009). Colin Hodgkinson, a visiting postdoctoral fellow in the laboratory, decided to design a similar strategy as that employed for the Meox1 insertion to clone the flanking sequences of the vga-9 mutant. Colin recognized that there were great risks inherent in this project. Others had observed that transgene insertions following pronuclear injection of DNA may generate large genomic deletions and re-arrangements, and even the successful cloning of genomic sequences flanking the insertion of a transgene array was no guarantee to identify the relevant gene. But Colin persisted and with skill and a bit of luck, he cloned in 1992 not just one but both flanking regions right away. In fact, luck continued to be on his side and in a few months, everything was done. Probes corresponding to the two cloned flanks were both present on a wild type genomic clone of about 6.7 kb, meaning that a deletion potentially associated with the insertion of the 300 kb transgene array could not have been larger than this clone. With the molecular genetic powerhouse of Nancy Jenkins and Neal Copeland just 40 miles away, it was natural for my small group to enter into a collaboration that turned out to be most fruitful. Using Southern analysis of DNA from interspecific hybrids, Karen Moore in the Jenkins/Copeland laboratory mapped probes derived from the cloned flanks to chromosome 6 where mi was known to reside. Moreover, Southern analysis of DNA from other species besides mouse, so called “zoo blots”, suggested that one of the probes might contain exonic sequences, and in fact, on northern blots prepared with mRNAs from melan-c cells, this probe labeled a single band of about 5.5 kb. This allowed Colin to rapidly obtain a first clone, MC-1, from a melan-c cDNA library, and using this clone to obtain a second overlapping clone, MC-2. Sequencing showed that together, these two clones contained what then was considered the entire open reading frame of a novel member of the E-box binding basic-helix-loop-helix-leucine zipper family of transcription factors (Hodgkinson et al., 1993). With the cloning of the human homolog, we later termed this factor MITF (Tachibana et al., 1994).
As stated above, finding a gene interrupted by a transgene, even if this gene is expressed in the cell types affected by an insertional mutation, does not prove its causal involvement in the phenotype. If, however, an additional allele at the locus showed a mutation in the same gene, we would become confident that we hit upon the right gene. The analysis of other microphthalmia mutants was mostly done by Eirikur Steingrimsson in the Jenkins/Copeland laboratory. In fact, DNA was available from mice carrying two distinct additional alleles, the original mi allele and microphthalmia-white spot (miws), and Eirikur found both of them mutated in Mitf. The allele miws showed a discrete intragenic deletion in the amino-terminal coding region of the gene. The allele mi showed no alteration by Southern analysis, but by RT-PCR, two coding region mutations were identified. One was a silent A–G transition at position 1137, which is in exon 9, and the other a 3 nucleotide deletion resulting in the loss of an arginine codon in a run of four conserved arginines at the carboxyl end of the basic domain. Based on work in related proteins, MITF was expected to form dimers with its helix-loop-helix-leucine zipper domain and contact DNA with its alpha-helical basic domain. The lack of a single amino acid in the basic domain would place the basic alpha helix out of register with the adjacent helix 1 and so render the mutated protein likely incapable of binding DNA (Hodgkinson et al., 1993). Much subsequent work confirmed that this was indeed the case (Hemesath et al., 1994; Steingrímsson et al., 1994). Also, to date, all other known microphthalmia alleles in mice, by my current count 36, have mutations in Mitf. Furthermore, alterations in the homologs in other species including zebrafish (Lister et al., 1999), quail (Mochii et al., 1998), rat (Opdecamp et al., 1998; Weilbaecher et al., 1998), hamster (Hodgkinson et al., 1998), dog (Karlsson et al., 2007), and man (Tassabehji et al., 1994) are also associated with pigmentary phenotypes, and, in mammals, may lead to deafness (in humans called Waardenburg syndrome IIa). Moreover, the gene is expressed in the cells affected (Opdecamp et al., 1997; Nakayama et al., 1998), including developing and adult melanocytes, developing retinal pigment epithelium cells, mast cells, and osteoclasts, although it is also found in many other cell types that do not show overt phenotypes in Mitf mutants. And lastly, Mitf is integrated into the very signaling pathways known to regulate the affected cell types (Opdecamp et al., 1997; Hou et al., 2000; Nguyen and Arnheiter, 2000; reviewed in Arnheiter et al., 2006). The identification of the different Mitf mutations also provided a rational basis for the phenotypic variations between different alleles (Steingrímsson et al., 1994). Furthermore, the cloning of a single gene responsible for the phenotype laid to rest the hypothesis that the full mi phenotype might depend on mutations in more than one gene.
Interestingly, without our knowledge, another group had also identified a transgenic insertional mutation that created a microphthalmia-like phenotype that was not complemented by the mi mutation. The analysis of this insertion was more complex than ours as only one transgene/genomic junction fragment mapped to chromosome 6 while the other mapped to chromosome 1. The chromosome 6 junction was reported to contain a sequence with similarity to human TCFE3 and in fact corresponded to exon 8 of Mitf. A partial cDNA obtained from heart, when used as a probe for Southern analysis on miws and mirw DNA (the latter now molecularly analyzed in detail, Bharti et al., 2008) detected the absence of bands present in C57BL/6 mice (note, however, that the strain of origin for mirw is CBA/J, not C57BL/6). Although limited, this analysis confirmed the presence of a gene encoding a TCFE3-related transcription factor at the microphthalmia locus (Hughes et al., 1993).
Outlook
Progress in science, and potential benefits to mankind, often depend on chance events. In biology, the chance events of mutations, searched for in corresponding screens or appearing spontaneously, are striking examples of the serendipitous nature of science. Among them, the discovery of microphthalmia/Mitf is a case in point, and although Mitf research has now been pursued for close to seven decades, I am convinced the last chance event has not yet occurred in this field. Numerous are the opportunities to discover additional, unexpected mechanisms of how Mitf acts in the many cell types whose development and function it regulates. As we now recognize, the gene encodes not just one MITF protein but a large family of isoforms that differ in sequence, post-translational modifications, and expression patterns, and we have so far only glimpsed at the complexity this generates when one considers that MITF does not only form dimers with itself but also with the three related partners TCFE3, TCFEB and TCFEC that each come with their own isoform complexities. Also, our knowledge is still limited on other partner proteins or co-factors interacting with MITF and how such interactions might be influenced by post-translational modifications such as phosphorylation, sumoylation, ubiquitination, methylation, and acetylation. Moreover, answers to how MITF modifications are regulated by the activation of signaling pathways in the different cell types also require additional studies. Furthermore, we still know relatively little about the transcriptional regulation of Mitf promoters, in particular of those that operate outside the melanocyte lineage. Each of the above parameters controls the activity of MITF and determines in the end which sets of target genes are regulated, and this in turn determines whether cells proliferate, migrate, or differentiate during development and perform their proper functions in the adult. The multitude of unsolved questions will undoubtedly prompt many additional decades of Mitf research that will be punctuated by surprises, and I encourage everyone to join in this exciting field with an open mind for such surprises.
Acknowledgments
I would like to thank the many collaborators who have made this work possible, and my host institute, NINDS, for allowing us to pursue investigations on Mitf. I would also like to extend my thanks to Nancy Jenkins and Neal Copeland, Karen Steel, Harold Gainer, James Battey, Eirikur Steingrimsson, and Monique Dubois-Dalcq for invaluable comments on the manuscript. This work was supported by the intramural program of the NIH, NINDS.
References
- Arnheiter H, Hou L, Nguyen MTT, Bismuth K, Csermely T, Murakami H, Skuntz S, Liu W, Bharti K. Mitf—A matter of life and death for the developing melanocyte. In: Hearing V, Leong SPL, editors. From Melanocytes to Malignant Melanoma. Totowa, NJ: Humana Press; 2006. pp. 27–49. [Google Scholar]
- Bauer GL, Praetorius C, Bergsteinsdottir K, et al. The role of MITF phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue. Genetics. 2009;183:581–594. doi: 10.1534/genetics.109.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechey CV, Harrison MA. A new spontaneous W allele, W36H. Mouse Genome. 1994;92:502–502. [Google Scholar]
- Bharti K, Liu W, Csermely T, Bertuzzi S, Arnheiter H. Alternative promoter use in eye development: the complex role and regulation of the transcription factor MITF. Development (Cambridge, England) 2008;135:1169–1178. doi: 10.1242/dev.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2010;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- Diwakar G, Zhang D, Jiang S, Hornyak TJ. Neurofibromin as a regulator of melanocyte development and differentiation. J Cell Sci. 2008;121:167–177. doi: 10.1242/jcs.013912. [DOI] [PubMed] [Google Scholar]
- Ebi Y, Kasugai T, Seino Y, Onoue H, Kanemoto T, Kitamura Y. Mechanism of mast cell deficiency in mutant mice of mi/mi genotype: an analysis by co-culture of mast cells and fibroblasts. Blood. 1990;75:1247–1251. [PubMed] [Google Scholar]
- Gerstengarbe S. … kenntnisreich, überlegt, kritisch gut veranlagt und von guter Darstellungsgabe – die Genetikerin Paula Hertwig. BIOspektrum. 2003;4/2003:378–380. [Google Scholar]
- Grüneberg H. Some observations on the microphthalmia gene in the mouse. J Genet. 1948;49:1–13. doi: 10.1007/BF02986377. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Stivers C, Odenwald W, Arnheiter H, Pignoni F, Steingrimsson E. The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics. 2004;167:233–241. doi: 10.1534/genetics.167.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hertwig P. Neue Mutationen und Kopplungsgruppen bei der Hausmaus. Z. Indukt. Abstammungs- u. Vererbungsl. 1942;80:220–246. [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Nakayama A, Li H, Swenson LB, Opdecamp K, Asher JH, Jr, Arnheiter H, Glaser T. Mutation at the anophthalmic white locus in Syrian hamsters: haploinsufficiency in the Mitf gene mimics human Waardenburg syndrome type 2. Hum Mol Genet. 1998;7:703–708. doi: 10.1093/hmg/7.4.703. [DOI] [PubMed] [Google Scholar]
- Hollander WF. Complementary alleles at the mi-locus in the mouse. Genetics. 1968;60:189. [Google Scholar]
- Hou L, Panthier JJ, Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development (Cambridge, England) 2000;127:5379–5389. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- Hughes MJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J Biol Chem. 1993;268:20687–20690. [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Konyukhov BV, Osipov VV. Interallelic complementation of microphthalm ia and white genes in mice. Genetika. 1968;4:65–76. [Google Scholar]
- Kozmik Z, Ruzickova J, Jonasova K, et al. Assembly of the cnidarian camera-type eye from vertebrate-like components. Proc Natl Acad Sci U S A. 2008;105:8989–8993. doi: 10.1073/pnas.0800388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development (Cambridge, England) 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Mankoo BS, Skuntz S, Harrigan I, Grigorieva E, Candia A, Wright CV, Arnheiter H, Pachnis V. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development (Cambridge, England) 2003;130:4655–4664. doi: 10.1242/dev.00687. [DOI] [PubMed] [Google Scholar]
- Mochii M, Ono T, Matsubara Y, Eguchi G. Spontaneous transdifferentiation of quail pigmented epithelial cell is accompanied by a mutation in the Mitf gene. Dev Biol. 1998;196:145–159. doi: 10.1006/dbio.1998.8864. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development (Cambridge, England) 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development (Cambridge, England) 1997;124:2377–2386. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Vanvooren P, Riviere M, Arnheiter H, Motta R, Szpirer J, Szpirer C. The rat microphthalmia-associated transcription factor gene (Mitf) maps at 4q34-q41 and is mutated in the mib rats. Mamm Genome. 1998;9:617–621. doi: 10.1007/s003359900832. [DOI] [PubMed] [Google Scholar]
- Rehli M, Den Elzen N, Cassady AI, Ostrowski MC, Hume DA. Cloning and characterization of the murine genes for bHLH-ZIP transcription factors TFEC and TFEB reveal a common gene organization for all MiT subfamily members. Genomics. 1999;56:111–120. doi: 10.1006/geno.1998.5588. [DOI] [PubMed] [Google Scholar]
- Skuntz S, Mankoo B, Nguyen MT, Hustert E, Nakayama A, Tournier-Lasserve E, Wright CV, Pachnis V, Bharti K, Arnheiter H. Lack of the mesodermal homeodomain protein MEOX1 disrupts sclerotome polarity and leads to a remodeling of the cranio-cervical joints of the axial skeleton. Dev Biol. 2009;332:383–395. doi: 10.1016/j.ydbio.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Barkway C, Bock GR. Strial dysfunction in mice with cochleo-saccular abnormalities. Hear Res. 1987;27:11–26. doi: 10.1016/0378-5955(87)90022-0. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Arnheiter H, Hallsson JH, Lamoreux ML, Copeland NG, Jenkins NA. Interallelic complementation at the mouse Mitf locus. Genetics. 2003;163:267–276. doi: 10.1093/genetics/163.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Moore KJ, Lamoreux ML, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences [see comments] Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Hara Y, Vyas D, Hodgkinson C, Fex J, Grundfast K, Arnheiter H. Cochlear disorder associated with melanocyte anomaly in mice with a transgenic insertional mutation. Mol Cell Neurosci. 1992;3:433–445. doi: 10.1016/1044-7431(92)90055-7. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Perez-Jurado LA, Nakayama A, Hodgkinson CA, Li X, Schneider M, Miki T, Fex J, Francke U, Arnheiter H. Cloning of MITF, the human homolog of the mouse microphthalmia gene and assignment to chromosome 3p14.1-p12.3. Hum Mol Genet. 1994;3:553–557. doi: 10.1093/hmg/3.4.553. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- Tomita M, Itoh H, Ishikawa N, Higa A, Ide H, Murakumo Y, Maruyama H, Koga Y, Nawa Y. Molecular cloning of mouse intestinal trefoil factor and its expression during goblet cell changes. Biochem J. 1995;311 (Pt 1):293–297. doi: 10.1042/bj3110293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimura T, Hashimoto K, Morii E, Tunio GM, Tsujino K, Kondo T, Kanakura Y, Kitamura Y. Involvement of transcription factor encoded by the mouse mi locus (MITF) in apoptosis of cultured mast cells induced by removal of interleukin-3. Am J Pathol. 1997;151:1043–1051. [PMC free article] [PubMed] [Google Scholar]
- Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J Exp Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Chen Y, Li H, et al. Allele-specific genetic interactions between Mitf and Kit affect melanocyte development. Pigment Cell Melanoma Res. 2010;23:441–447. doi: 10.1111/j.1755-148X.2010.00699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe HG, Coleman DL. Mi-spotted:a mutation in the mouse. Genet Res Camb. 1964;5:432–440. [Google Scholar]