Abstract
The nucleotide sequence diversity present among Hepatitis C Virus (HCV) isolates allows rapid adjustment to exterior forces including host immunity and drug therapy. This viral response reflects a combination of a high rate of replication together with an error-prone RNA dependent RNA polymerase providing for the selection and proliferation of the viruses with the highest fitness. We examined HCV subtype 1a whole genome sequences to identify positions contributing to genotypic and phenotypic diversity. Phylogenetic tree reconstructions showed two distinct clades existing within the 1a subtype with each clade having a star-like tree topology and lacking definite correlation between time or place of isolation and phylogeny. Identification of significant phylogenetically-informative sites at the nucleotide level revealed positions not only contributing to clade differentiation, but which are located at or proximal to codons associated with resistance to: protease inhibitors (NS3 Q41) or polymerase inhibitors (NS5B S368). Synonymous/nonsynonymous substitution mutation analyses revealed that the majority of nucleotide mutations yielded synonymous amino acids, indicating the presence of purifying selection pressure across the polyprotein with pockets of positive selection also being detected. Despite evidence for divergence at several loci, certain 1a characteristics were preserved including the length of the alternative reading frame/F protein (ARF/F) gene, and a subtype 1a-specific phosphorylation site in NS5A (S349). Our analysis suggests that there may be strain-specific differences in the development of antiviral resistance to viruses infecting patients that are dependent on the genetic variation separating these two clades.
Keywords: Bioinformatics, Drug resistance, Hepatitis C virus, Phylogeny, Selection Pressure
Introduction
Hepatitis C virus (HCV) infects nearly 3% of the world’s human population (more than 170 million people), resulting in chronic liver disease, hepatocellular carcinoma, and/or cirrhosis (1, 2). HCV is a member of the Hepacivirus genus of the Flaviviridae family with a genome that consists of a single-stranded, positive polarity RNA molecule that is approximately 9.6 kilobases (kb) in length. The RNA genome contains a single open reading frame which is translated into a polyprotein and cleaved co- and post-translationally by host and viral proteases into 3 structural (C, E1, E2) and 7 nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins which interact with each other and/or with host proteins to facilitate viral replication in the cell. Another protein, the alternative reading frame (ARF) or frameshift (F) protein, is produced following either a +1 to +2 ribosomal frameshift or by translation initiation at an alternative start site within the C gene (3, 4). The viral 5′ and 3′ untranslated regions (UTRs) consist of elaborate RNA structures including an internal ribosomal entry site (IRES) in the 5′ UTR, which interacts with multiple proteins (5, 6) to permit cap-independent translation initiation.
Due to polymerase fidelity and selection pressures (7), HCV exists as a quasispecies (8, 9) within its host. The virus has diverged over time into several different lineages, classified as six distinct genotypes, each with multiple subtypes. Only 75%–86% (10, 11) nucleotide identity exists between isolates of the same subtype while an average of 72% (12) nucleotide identity is present between isolates of different genotypes. Currently all genotypes are clinically treated with interferon and ribavirin, but the specific infecting genotype influences both the dose and success rate of therapy. For HCV subtype 1a, which is the most prevalent subtype in the United States (13), roughly half of the patients can be successfully treated (14). Nonresponse to antiviral therapy has been attributed to various host, environmental (15), and viral causes--including pre-existing mutations or mutations triggered by treatment (16, 17). Nonresponse due specifically to viral causes is especially interesting as it has previously been theorized that it is not merely one (or several) position(s) causing such resistance to treatment, but rather a synergism existing between multiple positions scattered across the genome (18). In this current study using complementary sequence analysis methods, we were able to demonstrate that HCV subtype 1a isolates can be further separated into two distinct sub-genotypes with potentially different phenotypic characteristics that may reflect differences in treatment response.
Materials and Methods
Alignments and Sequence Metadata
All available HCV subtype 1a whole genome sequences were obtained from the Viral Bioinformatics Resource Center (19) database (www.hcvdb.org) and GenBank in November 2008 (Supplemental Table 1). This represented 447 sequences. Nucleotide multiple sequence alignments were constructed with MUSCLE (20) and refined within the polyprotein coding region using ClustalW (21) as implemented in MEGA version 4.0 (22). Manual editing of the multiple sequence alignment ensured that the nucleotide alignment of the polyprotein coding region was codon-aligned so as to not disrupt the reading frame within the alignment itself. Highly conserved coding motifs such as those within the viral RNA-dependent RNA-polymerase and Helicase genes served as anchors points to ensure accuracy of the overall alignment. Outside of the polyprotein coding region, the 5′ and 3′ untranslated regions (UTRs) were well aligned overall, especially at more conserved anchor points such as the viral IRES element. Only a small subset of the sequences required the insertion of gaps to bring them into alignment with the rest of the sequences. As there were more sequences available from one of the two newly recognized clades identified during our analysis, a subset of the original 447 sequences were used to construct a separate alignment using 240 sequences consisting of 120 representative sequences from each clade so as to not bias the statistical results.
The 447 sequences used in this analysis were derived from samples taken from 397 individual patients with geographical data available for 322 of the sequences (72%), temporal data available for 307 sequences (69%), and treatment outcomes available for 20 patients (comprising 69, or 7% of the sequences). For these patients, multiple samples were obtained that represent pre- and post-treatment isolates. No other isolate metadata was available at the time this analysis was performed.
Phylogenic Analysis
Maximum Likelihood phylogenetic trees were reconstructed using GARLI version 0.951 (www.bio.utexas.edu/faculty/antisense/garli/Garli.html). The parameters were set to run until no topology improvement was observed for 10,000 consecutive generations using the substitution nucleotide general time-reversible model, using default settings for all other parameters. Geotemporal points of isolation were obtained from the respective GenBank sequence records.
Significant Informative Sites
Subtype 1a sequences were separated into 2 groups based on the results of the phylogenetic tree reconstructions. Perl scripts were used to perform chi-square tests of association on the list of 4205 parsimony-informative sites acquired from an alignment containing codon-aligned whole genome sequences from MEGA to identify all sites (including gaps), that significantly contributed to distinguishing the two clades. A Bonferroni correction was then performed on the dataset to yield the final list of 282 statistically significant sites which differentiated the two clades from each other. Informative sites were categorized as “hotspots” if there were 3 or more clade-informative sites within a contiguous string of 10 nucleotide bases.
Synonymous/Nonsynonymous Mutation Analysis
The synonymous nonsynonymous analysis program (SNAP) (23), (www.hiv.lanl.gov) was run locally to calculate nonsynonymous/synonymous (dN/dS) substitution ratios, from the codon-aligned nucleotide sequences. The calculation of comparative statistics, including standard deviation and variance, were not possible due to the large size of the results. Those regions having high nonsynonymous mutation values (defined as multiple adjacent high-scoring positions with at least one position having a SNAP score ≥ 1.0) in the SNAP average per-codon behavior (dN/dS) results were categorized as being under positive selection pressure.
Results
Phylogenetic Separation of Subtype 1a
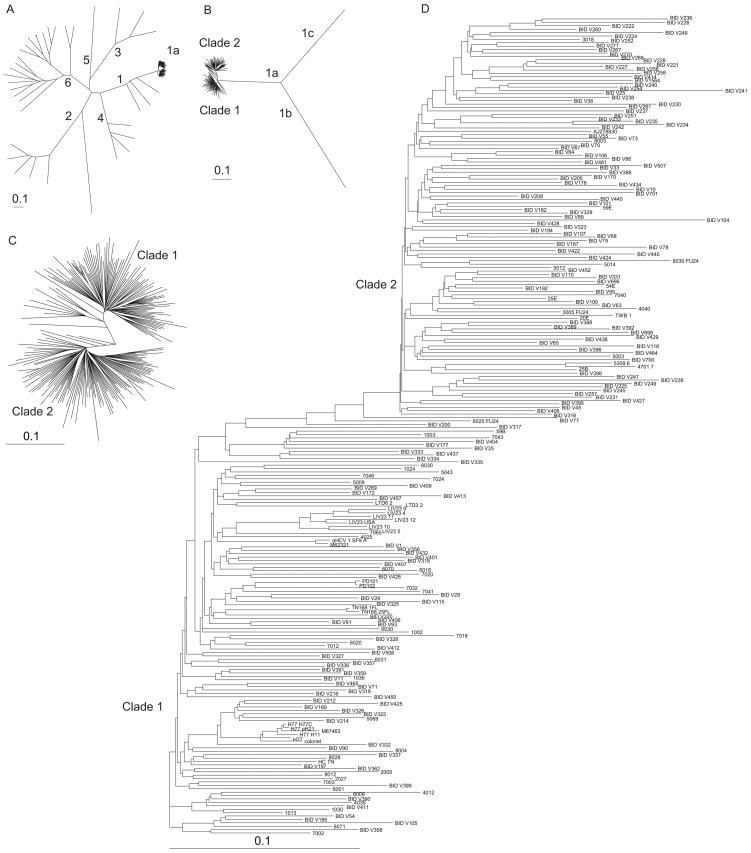
In order to determine the variation in HCV subtype 1a sequences at the nucleotide sequence level, we examined the maximum likelihood phylogenetic tree reconstruction for various representative HCV subtype 1a sequences with representatives from all other genotypes and subtypes (figure 1A). After noting that the HCV subtype 1a sequences were separated into two clades, we constructed a separate tree with the same subtype 1a sequences along with outgroups from subtypes 1b and 1c (figure 1B) for increased resolution. We confirmed these results by constructing separate maximum likelihood phylogenetic trees containing just the 240 subtype 1a sequences consisting of equal representatives from each clade (figures 1C and 1D). The various phylogenetic tree reconstructions showed that 2 distinct clades were present within the HCV subtype 1a branch. Isolate sequences present within each of these clades displayed a significant star-like topology. Such an observation would result from a genetic “bottleneck” among the ancestral sequences for each clade which was then followed by sequence divergence within the progeny sequences. Since separation among highly homologous sequences is commonly due to differences in either the time (24) or place (25) of isolation, we examined the isolation data associated with each sequence and found no correlation existing between either the time or location of isolation and the clade in which the sequence was placed (data not shown).
Figure 1. Phylogenetic trees showing evolutionary relationships between HCV isolates.
Maximum likelihood phylogenetic trees are presented that demonstrate the separation of the HCV 1a isolates into two distinct clades. An unrooted maximum likelihood phylogenetic tree of 94 representative HCV 1a whole genome sequences together with sequences from (A) all other genotypes and subtypes, or (B) the same representative subtype 1a sequences with outgroup sequences from subtypes 1b and 1c. Sequence names were removed for clarity. Maximum likelihood phylogenetic reconstructions containing 240 HCV 1a sequences and are shown as either (C) unrooted, without sequence names for clarity, or (D) rooted trees.
Inter-Clade Informative Sites
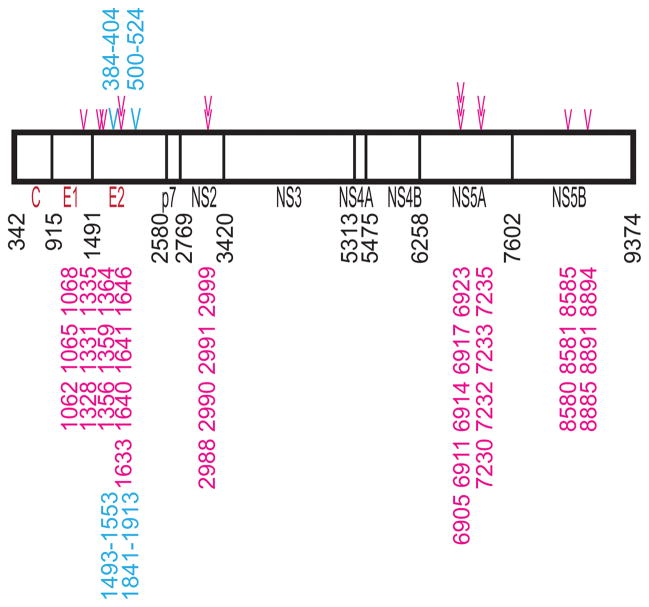
In order to identify the individual nucleotide positions that contribute most to the inter-clade differentiation, a list of all 4205 phylogenetically informative sites from the codon-aligned multiple genome alignment was assembled. A chi-square test of association was applied to identify all sites possessing a statistically significant non-random distribution of bases between the two clades. This analysis yielded a list of 282 informative sites that significantly differentiated the two clades (Supplemental Table 2) with p-values ranging from 3.53 × 10−24 to 1.15 × 10−5. The number of clade-informative sites that separate the two clades from each other were tallied within each genomic region as follows: 1 site in the 5′ UTR, 3 in C, 28 in E1, 55 in E2, 8 in p7, 24 in NS2, 49 in NS3, 4 in NS4A, 21 in NS4B, 47 in NS5A, 42 in NS5B, and 0 in the 3′ UTR. It is interesting that so many clade-informative sites exist within the NS3, NS5A, and NS5B nonstructural coding regions since such regions account for resistance against multiple antiviral compounds. It is unknown whether or not some of the sequences used in the present study included sequences from patients treated with protease inhibitors, though samples from patients identified as receiving treatment, received the standard combination of interferon and ribavirin. However, several clade-informative sites were identified within codons at the nucleotide level which are translated into amino acid residues that have been shown to be involved in protease inhibitor resistance (NS3 Q41) (26) and viral polymerase inhibitor resistance (NS5B H95 and S365) (27). Additional codons containing nucleotide informative sites located proximal to other functional amino acid residues were also identified (Table 1). Fourteen regions where clade-informative sites were clustered together were found scattered throughout the genome, forming clade-informative “hotspots” (Figure 2) at nucleotide positions: 1062, 1065, 1068, 1328, 1331, 1335, 1356, 1359, 1364, 1633, 1640, 1641, 1646, 2988, 2990, 2991, 2999, 6905, 6911, 6914, 6917, 6923, 7230, 7232, 7233, 7235, 8580, 8581, 8585, 8891, and 8894.
Table 1.
Clade-informative sites located within 3 residues of a functional site.
| Gene | Genome NT Position (strain H77) | Polyprotein Position (H77) | AA Seqa | Hit Gene Position | Known Gene Position (polyprotein) | Function (Resistance) | Position in Codon | SNAP Avgb | p-valuec |
|---|---|---|---|---|---|---|---|---|---|
| NS3 | 3461 | 1040 | QTRGLLGC | L14 | C16S (1042) | (ACH-806) | 3 | SN | 9.44E-14 |
| NS3 | 3464 | 1041 | TRGLLGC | G15 | C16S (1042) | (ACH-806) | 3 | S | 4.94E-15 |
| NS3 | 3518 | 1059 | GEVQIV | V33 | V36I (1062) | (Boceprevir) (Telaprevir) |
3 | SN | 7.46E-08 |
| NS3 | 3542 | 1067 | VSTATQ | A39 Q41 |
A39V (1065) Q41R (1067) |
(ACH-806) (Boceprevir)/(ITMN-191) |
3 | SN | 6.02E-17 |
| NS3 | 3572 | 1077 | NGVCWT | V51 | T54A (1080) | (Boceprevir)/(Telaprevir) | 3 | SN | 7.87E-14 |
| NS3 | 3657 | 1106 | YTNVDQ | Q80 | D79 (1105) | Near NS3 Catalytic Triad | 1 | NS | 5.42E-14 |
| NS3 | 3665 | 1108 | NVDQDL | L82 | D79 (1105) | Near NS3 Catalytic Triad | 3 | S | 7.66E-09 |
| NS3 | 3935 | 1198 | VDFIPV | V172 | V170A (1196) | (Boceprevir)/(Telaprevir) | 3 | S | 3.71E-20 |
| NS4a | 5390 | 1683 | VVIVGRI | V26 | V23A (1680) V29 (1686) |
(ITMN-191) NS3-NS4A allosteric activation |
3 | SN | 3.29E-12 |
| NS5a | 6990 | 2217 | KATCTA | A245 | P237-N276 (2209-2248) | Present in ISDR Region | 1 | NS | 4.82E-09 |
| NS5a | 6998 | 2219 | TCTANH | H247 | P237-N276 (2209-2248) | Present in ISDR Region | 3 | SN | 2.26E-11 |
| NS5a | 7189 | 2283 | SRRFAR | R311 | A310-K331 (2282-2303) | Cyclosporin Resistance Region | 2 | NS | 2.18E-07 |
| NS5a | 7208 | 2289 | ALPVWA | A317 | A310-K331 (2282-2303) | Cyclosporin Resistance Region | 3 | S | 1.05E-10 |
| NS5a | 7230 | 2297 | DYNPPL | L325 | A310-K331 (2282-2303) | Cyclosporin Resistance Region | 1 | SN | 3.47E-16 |
| NS5a | 7233 | 2298 | YNPPLV | V326 | A310-K331 (2282-2303) | Cyclosporin Resistance Region | 1 | NS | 3.30E-08 |
| NS5b | 7886 | 2515 | LTPPHS | H95 S96 |
H95Q (2515) S96T (2516) |
(A-782759) (R1479) |
3 | S | 1.34E-12 |
| NS5b | 8030 | 2563 | IMAKNE | E143 | N142T (2562) | (HCV-796) | 3 | S | 3.01E-14 |
| NS5b | 8696 | 2785 | ITSCSS | S365 | S365T/A (2785) S368A (2788) |
(HCV-796) (A-837093) |
3 | S | 1.03E-10 |
| NS5b | 8867 | 2842 | TLWARM | R422 | M423T/V/I (2843) | (AG-021541) | 3 | SN | 5.59E-18 |
| NS5b | 8939 | 2866 | LNCEIY | E446 | Y448H (2868) | (A-782759) (A-837093) |
3 | S | 1.57E-13 |
Underlined residues are the significant “hits” in the amino acid sequence which distinguish the two clades, bold residues are known functional residues.
Average nonsynonymous:synonymous ratio. S = synonymous, N = nonsynonymous, SN = synonymous > nonsynonymous, NS = nonsynonymous > synonymous.
P-value calculated from the chi-square score quantifying the probability of the nucleotide divergence at each column being attributable to random chance. Maximum p-value allowed after Bonferroni correction is 1.19 × 10−5.
Figure 2. Genomic Representation Showing Clade-informative Sites with Potential Public Health Impact.
The translated polyprotein is depicted with the protease cleavage sites marked by black vertical lines and are shown with nucleotide positions in black referencing the first nucleotide of each mature protein. The mature structural proteins are labeled in red while the nonstructural proteins are labeled in black letters. Cyan symbols indicate the two regions displaying positive selection pressures as measured by SNAP. The amino acid positions corresponding to the positive selection regions are specified in cyan letters above the polyprotein depiction. Magenta symbols represent the fourteen regions containing high concentrations (or “hotspots”) of phylogenetically-informative sites. Stacked symbols indicate overlapping “hotspots”. The individual nucleotide positions (in strain H77) are identified in magenta letters below the polyprotein representation.
Purifying Selection Pressures
SNAP was used to examine the synonymous and nonsynonymous substitution mutation ratios through a codon-by-codon pairwise comparison analysis using the method described by Nei and Gojobori (28). Overall there were approximately 3.8 times as many pairwise identical (67,816,832) as synonymous substitution classes (17,801,620) and 4.25 times as many synonymous as nonsynonymous (4,188,972) classes. There were also 2,138,124 small insertions/deletions present within the pairwise comparisons. When we examined the location of each informative site within their respective codons, 48 significant nucleotide clade-informative sites were present at the first codon position, 17 sites at the second position, 216 at the third position, and 1 site in the 5′ non-coding region. Previous quantitative analyses concluded that the majority of mutations in the 3rd position of the codon are synonymous, while the majority of mutations at either the 1st or 2nd codon positions are nonsynonymous (29). Our analysis showed that the majority of informative sites were synonymous (silent) mutations--complying with the known degeneracy (30) that exists within the genetic code and verifying that HCV 1a isolates contain relatively conserved amino acid sequences (16). There were also multiple cases of individual codons having a relatively equal nonsynonymous:synonymous ratio resulting from a distribution of both similar and dissimilar codons. In such cases SNAP reports an almost equal nonsynonymous:synonymous ratio which infers a lack of (or neutral) selection pressure.
To determine whether different selection pressures were present in either of the two clades separately or combined, we examined the average pairwise comparison output from the various SNAP tests. Each of three separate analyses was run with all sequences from either of the two clades, or with the combined set of sequences. Similar dS values, which measures the Jukes-Cantor corrected synonymous substitution rate, between the sequence sets (0.2150, 0.2314, and 0.2774 for clades 1, 2, and combined respectively) were found. Nearly identical dN values, which quantifies the Jukes-Cantor corrected nonsynonymous substitution rate, were observed (0.0217, 0.0221, and 0.0250). Neither the dN/dS, which measures the ratio of nonsynonymous to synonymous substitutions (0.0999, 0.09588, and 0.09002), nor the PN/PS value, which quantifies the uncorrected ratio of the proportion of observed nonsynonymous to synonymous substitutions (0.1133, 0.1098, and 0.1061), were noticeably different. The dN/dS and the PN/PS ratios, which are indicative of selection pressure(s), are each less than 1 for all analyses, indicating the presence of purifying selection across the entire coding region (31–34). Purifying selection involves the conservation of sequence regions throughout evolutionary history, presumably due to functional constraints. In comparing a set of viral isolates, a reduction in the rate of nonsynonymous mutations along with a stable (or increased) rate of synonymous mutations for a particular coding region in comparison with other coding regions is one indication of purifying selection pressure. Our SNAP results suggest that purifying selection may play a role in sequence conservation both at the codon and whole sequence levels within both clades. We also analyzed the SNAP statistics for the phylogenetically-informative sites from the combined sequence set. None of these sites showed strictly nonsynonymous changes, although 161 exhibited both nonsynonymous and synonymous mutation classes, and 120 were strictly synonymous. There were no clade-informative sites for insertions/deletions (indels), and only 1 that was located in a noncoding region.
Regions of Positive Selection
With an overall indication of purifying selection pressure, we hypothesized that there might be specific areas within the coding region that may possess a higher rate of nonsynonymous mutations as such cases have been previously reported (35–37). Upon examination of the SNAP results, we identified two regions as having elevated nonsynonymous:synonymous SNAP ratios--confirming positive selection (figure 2). Interestingly, several individual codons within these variable regions were found to maintain a preference for synonymous substitutions, or purifying selection.
The first region found to be under positive selection pressure was in the N-terminus of the E2 protein. This region, spanning the polyprotein codons 384–404, had 4 clade-informative sites and had been previously described as Hypervariable Region I (HVR1) (38, 39) in the E2 protein. Positive selection in this region has been ascribed to a dominant B cell epitope (40, 41), with increased variability associated with increased chance for viral persistence (39) and decreased variability being associated with viral clearance (42). The second positive selection region is found between codons 500 and 524 in the polyprotein, which is a known epitope target for both B cells and T cells (35, 43–46). This region has 4 clade-informative sites and also lies in the E2 coding region. Thus, although purifying selection pressures are dominant across the genome to maintain functionality, regions undergoing positive selection still exist, presumably as a mechanism to escape attack by the host’s immune response.
Antiviral Response by Clade
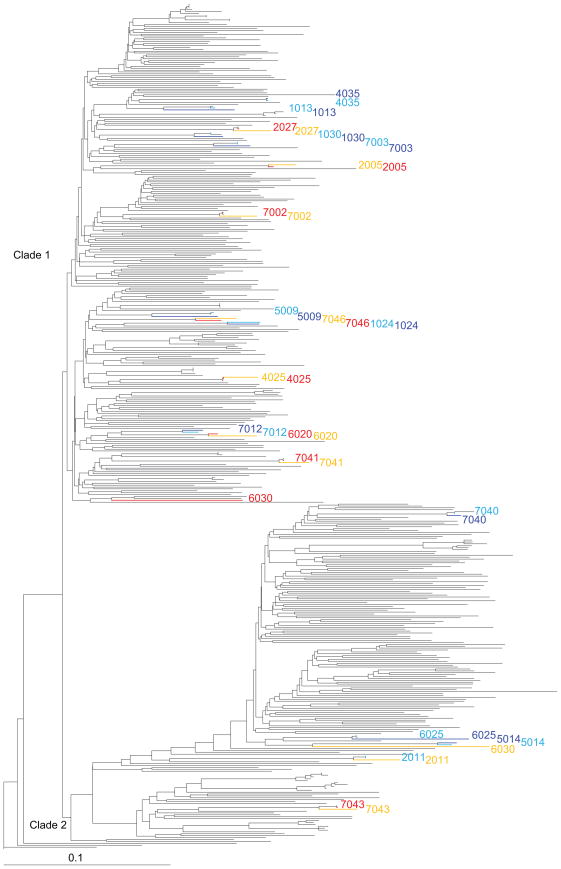
To ascertain whether the distinct subtype 1a clades respond to antiviral treatment differently, we reconstructed a maximum likelihood tree consisting of 447 whole genome sequences, which included the 240 sequences included in the original tree reconstruction. Clade separation and the star-like topology of the tree were both maintained when the additional sequence data was included (Figure 3). Clinical response data for 40 clinical isolates from 20 patients have been previously sequenced by the Virahep-C study group (16) and were categorized as either relapser (patient had no detectable viral RNA immediately after antiviral treatment, but viral RNA was detectable six months after therapy) or nonresponder (patient had detectable viral RNA after antiviral treatment). The treatment response data for these sequences were identified and overlaid onto the phylogenetic tree. This data revealed that for the 15 clade-1-infected patients having available clinical data 8 were categorized as relapsers and 7 were nonresponders. In the four patients that were infected with a clade 2 virus 1 was a relapser and 3 were nonresponders. For 19 of the 20 patients the sequence obtained before therapy and the sequence isolated after therapy was closely related. Curiously, one patient was observed to have a relapsing infection was initially infected with a clade 1 quasispecies which was then replaced by a dominant clade 2 quasispecies after antiviral treatment. While the one patient who cleared a clade one virus and relapsed with a clade two is noteworthy, it is unclear if this anomaly was due to clade 2 isolates being more resistant to treatment, due to the specifics of this patient’s immune system, or due to the random chance of isolating a co-infecting clade 2 viral RNA instead of clade 1 viral RNA from the patient, or a combination of these possibilities. Given the number of mutations that would be required for any genomic sequence to change from clade 1 to clade 2 (or vice versa), it is almost impossible that the introduction of random mutations into the genome would account for such a clade switch.
Figure 3. Phylogenetic Tree Showing 447 HCV 1a Sequences along with Virahep-C Clinical response data.
A maximum likelihood phylogenetic tree reconstruction containing all 447 non-chimeric HCV subtype 1a whole genome sequences that were available in November 2008. Both clades are labeled in black. Sequence names lacking clinical data were removed for clarity. Virahep-C Study Group clinical drug resistance data was also included with isolates being sequenced both before (pre) and after (post) treatment and classified as either responders (R) or nonresponders (N). Sequences were color-coded as follows: cyan = pre-N, blue = post-N, Red = pre-R, and Orange = post-R. Of particular note is the dominant quasispecies isolated from patient 6030 which changed from clade 1 to clade 2 either during or after treatment.
Discussion
In this work we use phylogenic analysis to show that subtype 1a HCV can be sub-categorized into at least two clades. Both clades are represented by isolates taken from various cities in the US and Europe at various times. The informative sites distinguishing clade 1 from clade 2 are distributed throughout the genome, but are particularly concentrated in the E1, E2, NS2, NS5A, and NS5B coding regions (Fig 2). Sequences in each clade form a star-like topology within the genotype 1a phylogenetic tree, which suggests the presence of two ideally-infectious or optimally-fit master genomes from which the viral sequence can drift to a limited degree within the clade-specific subgenotype. Although the two clades differ from one another, they each maintain at least three common subtype 1a functional features that distinguish the 1a subtype from all other subtypes. One such conserved features is a +1 to +2 reading frame shift during translation of the C protein that results in the production of the alternative reading frame (ARF) or F protein. There is very little divergence in the N terminus of core in either the standard reading frame (+1) or the alternative reading frame (ARF/F protein) (+2) reading frame and no informative sites that alter the length of the +2 reading frame within the N terminus of core, This ARF/F protein has the same termination codon in both 1a clades which makes it 18 amino acids shorter than its length in 80% of 1b isolates (47). Additionally, a subtype 1a-specific phosphorylation site is maintained in both clades at S349 of NS5A which is not a phospho-acceptor site in other HCV subtypes including 1b (48). A third defining 1a feature is in NS5B coding region in which both 1a clades sites were similar to each other but distinct from 1b codons that alter in vitro sensitivity to cyclosporine (49).
All but one (which was non-coding) of the clade-informative substitutions had a low nonsynonymous:synonymous ratio, suggesting that purifying selection pressure may be present either due to selection at the amino acid level to maintain protein structure or due to other RNA or protein functional constraints. But even within synonymous codons, the presence of one codon versus another may influence that rate at which particular phenotypes may arise. For example, depending on the codon being utilized at a particular amino acid position, a change in only one of the codon’s three bases may be needed for a virus to become resistant to antivirals. But in some cases, if a different synonymous codon is present, a change in two or more nucleotides may be necessary to achieve the same result. This raises the possibility that one subtype or clade may be more prone to development of drug-resistance mutation(s) than the other. For example, HCV genotype 1b genomes require two mutations to convert the NS3 155 codon from Arginine (CGN) to a protease-resistant Lysine (AAR) (50). An analysis of this codon within the HCV 1a clades demonstrates that both clades only require a single point mutation to switch from an Arginine (AGR) to the protease-resistant Lysine (AAR). As more drug resistant mutations are characterized, we anticipate that additional subtype and clade specific codon biases may be found. Analyses such as these are dependent on the accuracy of the genomic sequence of the virus isolates obtained for the study. In reality, the virus population present in any one patient represents a quasispecies population of related, but variable genomic sequences. Problems with the analysis of quasispecies populations of viruses is partially mitigated by the fact that the isolate sequences utilized in our analysis were in general obtained through PCR of the mixed quasispecies population and therefore represent a consensus sequence of the virus genome population within each host.
Altogether, 161 of the 282 clade-informative sites have a higher nonsynonymous ratio. Many of these changes result in conservative amino acid changes and may limited to sites with significant functional RNA structural phenotypes. Nevertheless, such sites could be important on a protein level for small molecule inhibitors. Nucleotide position 5390 (amino acid 1683) contains an informative site in which the consensus residue differs between clade 1 and clade 2 and is known to be near or within the portion of NS4A that binds and allosterically activates NS3 (51). Another site within this complex is either a Lysine (clade 1) or a Glutamine (clade 2) adjacent to the Histidine of the catalytic triad of the protease. Domain 3 of NS5A was found to contain clade informative sites that may influence response to cyclophilin inhibitors (52).
Both genotypes 1a and 1b respond poorly to current therapy when compared to other HCV genotypes. Despite multiple studies, no specific amino acid variants have unequivocally been shown to confer resistance clinically to either interferon or ribavirin. Correlations with response have been made when pretreatment sequence contains a number of mutants (usually >4) in defined regions (53, 54) although both the definition of this heterogeneity and the magnitude of the effect differ between studies. What is unclear is whether or not the accumulation of polymorphisms causing the deviation from the “ideal” consensus sequence is a result of a partial host immune response or of the antiviral therapy and whether such polymorphisms cripple the fitness of the virus. Regardless of which of these mechanisms is acting, this work suggests that clade-specific consensus sequences rather than subtype-specific consensus sequences maybe a useful comparator. We acknowledge that having clinical response data from a sample size of only 20 patients is insufficient for statistically-significant conclusions to be drawn as to the resistance profiles between the two clades; however, we anticipate that this scenario will change as more sequence data along with associated metadata is obtained in the future. Since either the 5′ UTR or a 222 base fragment of NS5B (10, 55) are the loci most commonly used to genotype HCV in the clinic, and given that there is one site within the 5′ UTR and several sites within the NS5B genotyping fragments that differ between clades 1 and 2, this sequence information could be combined with clinical data to begin to address the question of which clade, if either, is more resistant to therapy. Analysis of the data by examining variation between phylogenetic clades as opposed to analysis of individual amino acid differences between pairs of sequences may provide a more robust method for addressing this issue.
In conclusion, we were able to identify a statistically significant evolutionary divergence of subtype 1a HCV isolates into two distinct clades that cannot be attributed to either geography or time of isolation. In addition, there was a suggestion that response to therapy may be partially predicted by the clade membership of the infecting virus. Therapeutic studies of clade 1 and 2 HCV 1a viruses will be needed to definitively assess their response to treatment.
Supplementary Material
List of all whole genomes, and associated meta-data, used in this study.
Comprehensive list of all clade-informative nucleotide sites with associated statistics and other information.
Acknowledgments
We wish to thank Dr. John Tavis and the Virahep-C study group for providing the drug resistance data and Dr. Cecile Ané for statistical help and critical reading of the manuscript. This work was supported by PHS Training grant 5 T32 AI007150-29 to BP and NIH/NIAID contract number HHSN266200400036C to EJL. RS is supported by the American Cancer Society, a Veteran’s Administration Merit Award (5I01CX000117-02) and in part by the University of Wisconsin Institute for Clinical and Translational Research, and funded through an NIH Clinical and Translational Science Award (CTSA), grant number 1UL1RR025011.
Abbreviations
- ARF/F
alternative reading frame/frameshift protein
- C
Core protein
- E
envelope protein
- HCV
hepatitis C virus
- HVR1
hypervariable region 1
- IRES
internal ribosomal entry site
- NS
non-structural protein
- SNAP
synonymous nonsynonymous analysis program
- UTR
untranslated region
Footnotes
Disclosure:
The authors have no conflict(s) of interest.
1) Authors Declaration of personal interests:
None.
2) Declaration of funding interests:
(i) This study was funded [in part or in full] by: National Institutes of Health/National Institute for Allergies and Infectious Diseases, contract number HHSN266200400036C; Public Health Service, grant number 5 T32 AI007150-29; and National Institutes of Health, grant number 1UL1RR025011.
References
- 1.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13(17):2436–41. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C virus. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 3.Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA (New York, NY) 2001;7(5):710–21. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassilaki N, Mavromara P. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. The Journal of biological chemistry. 2003;278(42):40503–13. doi: 10.1074/jbc.M305504200. [DOI] [PubMed] [Google Scholar]
- 5.Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Archives of virology. 2006;151(3):509–24. doi: 10.1007/s00705-005-0642-6. [DOI] [PubMed] [Google Scholar]
- 6.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. The Journal of biological chemistry. 2000;275(36):27531–40. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. The Journal of general virology. 2004;85(Pt 11):3173–88. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 8.Kurosaki M, Enomoto N, Marumo F, Sato C. Evolution and selection of hepatitis C virus variants in patients with chronic hepatitis C. Virology. 1994;205(1):161–9. doi: 10.1006/viro.1994.1631. [DOI] [PubMed] [Google Scholar]
- 9.Zeuzem S. Hepatitis C virus: kinetics and quasispecies evolution during anti-viral therapy. Forum (Genoa, Italy) 2000;10(1):32–42. [PubMed] [Google Scholar]
- 10.Simmonds P, Holmes EC, Cha TA, et al. Classification of hepatitis C virusinto six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. The Journal of general virology. 1993;74 (Pt 11):2391–9. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 11.Bukh J, Miller RH, Purcell RH. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Seminars in liver disease. 1995;15(1):41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 12.Donlin MJ, Cannon NA, Yao E, et al. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. Journal of virology. 2007;81(15):8211–24. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. The New England journal of medicine. 2002;347(13):975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 14.Berg T, von Wagner M, Nasser S, et al. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology. 2006;130(4):1086–97. doi: 10.1053/j.gastro.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Conjeevaram HS, Fried MW, Jeffers LJ, et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131(2):470–7. doi: 10.1053/j.gastro.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Cannon NA, Donlin MJ, Fan X, Aurora R, Tavis JE. Hepatitis C virus diversity and evolution in the full open-reading frame during antiviral therapy. PLoS ONE. 2008;3(5):e2123. doi: 10.1371/journal.pone.0002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieffer TL, Sarrazin C, Miller JS, et al. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology (Baltimore, Md. 2007;46(3):631–9. doi: 10.1002/hep.21781. [DOI] [PubMed] [Google Scholar]
- 18.Torres-Puente M, Cuevas JM, Jimenez-Hernandez N, et al. Genetic variability in hepatitis C virus and its role in antiviral treatment response. Journal of viral hepatitis. 2008;15(3):188–99. doi: 10.1111/j.1365-2893.2007.00929.x. [DOI] [PubMed] [Google Scholar]
- 19.Greene JM, Collins F, Lefkowitz EJ, et al. National Institute of Allergy and Infectious Diseases bioinformatics resource centers: new assets for pathogen informatics. Infection and immunity. 2007;75(7):3212–9. doi: 10.1128/IAI.00105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular biology and evolution. 2007;24(8):1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Korber B. HIV Signature and Sequence Variation Analysis. In: Rodrigo AG, Learn GH, editors. Computational Analysis of HIV Molecular Sequences. Dordrecht, Netherlands: Kluwer Academic Publishers; 2000. pp. 55–72. [Google Scholar]
- 24.Auguste AJ, Pybus OG, Carrington CV. Evolution and dispersal of St. Louis encephalitis virus in the Americas. Infect Genet Evol. 2008 doi: 10.1016/j.meegid.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 25.Bragstad K, Nielsen LP, Fomsgaard A. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virology journal. 2008;5:40. doi: 10.1186/1743-422X-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curry S, Qiu P, Tong X. Analysis of HCV resistance mutations during combinationtherapy with protease inhibitor boceprevir and PEG-IFN alpha-2b using TaqMan mismatch amplification mutation assay. Journal of virological methods. 2008;153(2):156–62. doi: 10.1016/j.jviromet.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Kuntzen T, Timm J, Berical A, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology (Baltimore, Md. 2008;48(6):1769–78. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Molecular biology and evolution. 1986;3(5):418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 29.Lee MA, Keane OM, Glass BC, et al. Establishment of a pipeline to analyse non-synonymous SNPs in Bos taurus. BMC genomics. 2006;7:298. doi: 10.1186/1471-2164-7-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crick FH. Codon--anticodon pairing: the wobble hypothesis. Journal of molecular biology. 1966;19(2):548–55. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular biology and evolution. 1998;15(5):568–73. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 32.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. Journal of virology. 1997;71(1):663–77. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unemo M, Olcen P, Albert J, Fredlund H. Comparison of serologic and genetic porB-based typing of Neisseria gonorrhoeae: consequences for future characterization. Journal of clinical microbiology. 2003;41(9):4141–7. doi: 10.1128/JCM.41.9.4141-4147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mindell DP. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(8):3284–8. doi: 10.1073/pnas.93.8.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Hahn T, Yoon JC, Alter H, et al. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology. 2007;132(2):667–78. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Kuntzen T, Timm J, Berical A, et al. Viral sequence evolution in acute hepatitis C virus infection. Journal of virology. 2007;81(21):11658–68. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Booth JC, Kumar U, Webster D, Monjardino J, Thomas HC. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology (Baltimore, Md. 1998;27(1):223–7. doi: 10.1002/hep.510270134. [DOI] [PubMed] [Google Scholar]
- 38.Li H, McMahon BJ, McArdle S, et al. Hepatitis C virus envelope glycoprotein co-evolutionary dynamics during chronic hepatitis C. Virology. 2008;375(2):580–91. doi: 10.1016/j.virol.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheridan I, Pybus OG, Holmes EC, Klenerman P. High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression. Journal of virology. 2004;78(7):3447–54. doi: 10.1128/JVI.78.7.3447-3454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu YK, Igarashi H, Kiyohara T, et al. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223(2):409–12. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 41.Farci P, Shimoda A, Wong D, et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(26):15394–9. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farci P, Shimoda A, Coiana A, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science (New York, NY) 2000;288(5464):339–44. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 43.Mink MA, Benichou S, Madaule P, Tiollais P, Prince AM, Inchauspe G. Characterization and mapping of a B-cell immunogenic domain in hepatitis C virus E2 glycoprotein using a yeast peptide library. Virology. 1994;200(1):246–55. doi: 10.1006/viro.1994.1182. [DOI] [PubMed] [Google Scholar]
- 44.Zibert A, Kraas W, Ross RS, et al. Immunodominant B-cell domains of hepatitis C virus envelope proteins E1 and E2 identified during early and late time points of infection. Journal of hepatology. 1999;30(2):177–84. doi: 10.1016/s0168-8278(99)80059-2. [DOI] [PubMed] [Google Scholar]
- 45.Ching WM, Wychowski C, Beach MJ, et al. Interaction of immune sera with synthetic peptides corresponding to the structural protein region of hepatitis C virus. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(8):3190–4. doi: 10.1073/pnas.89.8.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze zur Wiesch J, Lauer GM, Day CL, et al. Broad repertoire of the CD4+Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J Immunol. 2005;175(6):3603–13. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 47.Xu Z, Choi J, Yen TS, et al. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. The EMBO journal. 2001;20(14):3840–8. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed KE, Rice CM. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. The Journal of biological chemistry. 1999;274(39):28011–8. doi: 10.1074/jbc.274.39.28011. [DOI] [PubMed] [Google Scholar]
- 49.Robida JM, Nelson HB, Liu Z, Tang H. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. Journal of virology. 2007;81(11):5829–40. doi: 10.1128/JVI.02524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Muh U, Hanzelka BL, et al. Phenotypic and structural analyses of hepatitis C virus NS3 protease Arg155 variants: sensitivity to telaprevir (VX-950) and interferon alpha. The Journal of biological chemistry. 2007;282(31):22619–28. doi: 10.1074/jbc.M610207200. [DOI] [PubMed] [Google Scholar]
- 51.Kwong AD, Kim JL, Rao G, Lipovsek D, Raybuck SA. Hepatitis C virus NS3/4A protease. Antiviral research. 1999;41(1):67–84. [PubMed] [Google Scholar]
- 52.Fernandes F, Poole DS, Hoover S, et al. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology (Baltimore, Md) 2007;46(4):1026–33. doi: 10.1002/hep.21809. [DOI] [PubMed] [Google Scholar]
- 53.Enomoto N, Sakuma I, Asahina Y, et al. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. The New England journal of medicine. 1996;334(2):77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 54.Torres-Puente M, Cuevas JM, Jimenez-Hernandez N, et al. Hepatitis C virus and the controversial role of the interferon sensitivity determining region in the response to interferon treatment. Journal of medical virology. 2008;80(2):247–53. doi: 10.1002/jmv.21060. [DOI] [PubMed] [Google Scholar]
- 55.Okamoto H, Tokita H, Sakamoto M, et al. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. The Journal of general virology. 1993;74 (Pt 11):2385–90. doi: 10.1099/0022-1317-74-11-2385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all whole genomes, and associated meta-data, used in this study.
Comprehensive list of all clade-informative nucleotide sites with associated statistics and other information.