Abstract
The folate receptor α (FRα) is critical for normal embryonic and fetal development. The receptor has a relatively narrow tissue specificity which includes the visceral endoderm and the placenta and mediates delivery of folate, inadequacy of which results in termination of pregnancy or developmental defects. We have previously reported that the FRα gene is negatively and directly regulated by estrogen and positively but indirectly by progesterone and glucocorticoid. To further investigate hormonal control of this gene and in view of the growing evidence for the importance of the androgen receptor (AR) in endometrial and placental functions, we examined the response of the FRα gene to androgen. Here we demonstrate that the FRα gene is directly activated by androgen. The P4 promoter of the FRα gene is the target of hormone-dependent activation by the androgen receptor (AR) in a manner that is co-activator-dependent. The site of functional association of AR in the FRα gene maps to a 35bp region occurring ~1500bp upstream of the target promoter. The functional elements within this region are an androgen response element (ARE) half-site and a non-canonical C/EBP element that cooperate to recruit AR in a manner that is dependent on the DNA-bound C/EBPα. Since the placenta is rich in C/EBPα, the findings underscore the multiplicity of mechanisms by which the FRα gene is under the exquisite control of steroid hormones.
Keywords: Folate receptor, Folate, C/EBPα, Androgen receptor
1 INTRODUCTION
The folate receptor type α (FRα) is a glycopolypeptide that is anchored to the cell surface by a glycosyl phosphatidylinositol (GPI) membrane anchor [1-4] and is expressed with a rather narrow tissue specificity [5]. FRα binds with a relatively high affinity (Kd < 10-9M) to the circulating form of folate, 5-methyl tetrahydrofolate, and mediates its transport across the membrane by an endocytic process [6, 7]. The maintenance of pregnancy as well as normal fetal development is critically dependent on the maintenance of an adequate folate status which requires a regular supply of the vitamin from a dietary source [8]. Elegant genetic studies in mice by Finnell and co-workers have established a critical role for FRα in development that is principally related to ensuring adequate folate delivery to the developing embryo and fetus [9, 10]. It has also been demonstrated that FRα is required for transplacental transport of folate resulting in several-fold higher folate levels in the fetal vs. the maternal circulation [11]. FRα is highly expressed in the visceral endoderm which nourishes the embryo [12] and also in the placenta, peaking in term placental tissue of embryonic origin [13]. Mutations in FRα have been implicated in neurodegenerative effects [14]. Physiological mechanisms that control FRα expression may therefore be expected to be critical during development.
Previous studies from this laboratory have demonstrated that the FRα gene (FOLR1) is a non-classical target for regulation by the steroid hormones estrogen, progesterone and glucocorticoids. The hormone-bound estrogen receptor (ER) directly represses the FRα gene by associating as a co-repressor complex with TAFII30 within its core promoter [15, 16]. The progesterone receptor (PR) [17] and the glucocorticoid receptor (GR) [18] activate the FRα gene in response to hormone, but indirectly through the products of upstream target genes.
There is growing evidence for a role for the androgen receptor (AR) in endometrial tissue proliferation and differentiation in the normal menstrual cycle which appears to involve a complex interplay with progestogens and estrogens [19]. AR is strongly expressed in the nuclei of decidual tissue during pregnancy [20]. It has also been recently demonstrated that AR and PR target distinct gene networks in decidual tissue with a primary function for AR in cytoskeletal organization and cell motility [21]. It was therefore undertaken in this study to investigate a potential role for androgen/AR in regulating FRα expression and to elucidate the mechanism of this regulation. Studying the endocrine mechanisms controlling FRα levels and the various factors involved may be expected to provide a better understanding of certain abnormalities of pregnancy and development.
2 Materials and Methods
2.1 Chemicals and reagents
Dulbecco's minimum essential medium (DMEM), and penicillin/streptomycin/L-glutamine stock mix were purchased from Life Technologies, Inc. (Carlsbad, CA); HAM's F-12 medium from Lonza (Walkersville, MD) Fetal bovine serum (FBS) and charcoal stripped FBS (CS-FBS) were from from Invitrogen (Carlsbad, CA). FUGENE 6 was purchased from Roche Diagnostics (Indianapolis, IN). Luciferase assay reagents were from Promega (Madison, WI). Affinity purified rabbit anti-human AR (sc-816), C/EBPα(sc-61), His-probe(sc-803), mouse anti-GAPDH (sc-47724), and normal rabbit IgG control (sc-2027) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA). Vent DNA polymerase was purchased from New England Biolabs (Beverly, MA). Custom oligonucleotide primers and biotinylated oligonucleotide probes were from Integrated DNA Technologies (Coralville, IA). Protein A-sepharose beads and streptavidin sepharose beads were purchased from Amersham (Uppsala, Sweden). The reagents for RT-PCR and real-time PCR were purchased from Applied Biosystems (Branchburg, NJ).
2.2 DNA constructs and expression plasmids
Construct design used either natural restriction sites or restriction sites created by PCR using Vent DNA polymerase (New England Biolabs) and custom oligonucleotides (Integrated DNA Technologies). The PCR products were first digested with the appropriate restriction enzymes and cloned into the pGL3-basic plasmid (Promega).The 5’ deletion constructs of the FRα promoter, i.e., FRα (-3394nt to +33nt), FRα ΔP1 (-3113nt to +33nt), FRα ΔP4 (-3394nt to +33nt with the deletion from -146 nt to -34nt), FRα P4 (-176nt to +33 nt), -1565nt to +33nt, and -1555nt to +33nt and the 3’ mutation constructs, i.e., mut(-1549nt to -1546nt), mut (-1545nt to -1542nt), mut (-1541nt to -1538nt), mut (-1537nt to – 1534nt), mut (-1533nt to -1530nt) were all constructed by PCR using the appropriate primers and subcloned at MluI (upstream) and XhoI (downstream) sites in the pGL3 basic plamid. For the 3’ mutation constructs, the sequential 4-base mutations were obtained by changing A-T pairs to G-C pairs and vice versa. GAL4-TATA-Luc plasmid (pG5luc) was purchased from Promega. ARE-TATA-Luc, FRα (-1565nt to -1536nt), (-1565nt to -1533nt), (-1549nt to -1536nt), or (-1565nt to -1548nt)-TATA-Luc, (C/EBP)3-TATA-luc, (C/EBP)3-ARE-TATA-luc were made by cloning appropriate annealed oligos with the addition of KpnI(5’) and NheI(3’) terminal restriction sites into the large segment of GAL4-TATA-Luc digested by KpnI and NheI. To generate ARE-TATA-luc, a high affinity ARE, AGTACGTGATGTTCT, [22] was inserted upstream of the TATA box. The Renilla luciferase transfection control was the pRL-null plasmid from Promega (Madison, WI). The recombinant plasmids were amplified in E. coli strain XL1 Blue and purified using the Qiagen plasmid kit (Qiagen, Chatsworth, CA). The entire cloned DNA sequence in each construct was verified by automated DNA sequence analysis. The co-regulator expression plasmids were provided by Dr. Brian Rowan at Tulane University.
2.3 Cell culture and transfection
HeLa (American Type Culture Collection) cells were cultured in Dulbecco's minimum essential medium (DMEM) supplemented with FBS (10%), penicillin (100 units/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). ACH-3P immortalized human placental trophoblast cells [23] (kindly provided by Dr. Ursula Hiden at Medical University Graz, Austria) were cultured in HAM's F-12 medium supplemented with FBS (10%), penicillin (100 units/ml), streptomycin (100 μg/ml), and L-glutamine (2 mM). To obtain hormone depletion, HeLa cells were grown in phenol red-free media supplemented with charcoal-stripped FBS (5% v/v), L-glutamine (2 mM), insulin (2 μg/ml), and transferrin (40 μg/ml) for 72h. Hormone depletion in ACH-3P cells was achieved by growing them in HAM's F-12 medium supplemented with charcoal stripped FBS (5%). HeLa cells were transfected with DNA constructs in 6-well plates (Corning, New York, NY) using FuGENE 6 (Roche Diagnostics), according to manufacturer's protocol. Normally, 500 ng reporter and 25-100 ng of each expression plasmid were used where not specially indicated. Transfection efficiency and promoter specificity were controlled using the pRL-null plasmid expressing Renilla luciferase and measurement of Renilla luciferase activity in the cell lysates.
2.4 Assay of cell surface FRα
In the fluorimetric assay, cells were washed with ice-cold acid buffer (10mM sodium acetate, pH 3.5, 150 mM NaCl) to remove FRbound endogenous folate. The cells were then washed with ice-cold PBS [10mM sodium phosphate (pH 7.5), 150 mM NaCl] and incubated with 5 nM fluorescein-conjugated folate [24] at 4°C for 30 min. The cells were washed twice with PBS and the bound folate was measured by flow cytometry as described [24]. Non-specific background fluorescence was measured by blocking with a 100-fold excess of unlabeled folic acid and the values were subtracted. In the radiolabeling assay, cells were incubated with 27nM of [3H] folic acid in serum free FFRPMI for 1h at 37°C. At the end of the incubation the cells were washed with ice cold PBS to remove unbound radioactivity. The cells were washed with 1ml of acid buffer (10mM sodium acetate, pH 3.5/150mM NaCl) for 1 min on ice. The acid wash eluate was counted by liquid scintillation and represents the amount of [3H] folic acid bound. The cells were also incubated with a 20-fold excess of unlabeled folic acid, relative to the amount of [3H]folic acid added, to ensure the specificity of binding via FR. All samples were measured in triplicate.
2.5 Luciferase assay
After incubation as indicated, transfected cells were washed with PBS and harvested in 500 μl of renilla luciferase assay lysis buffer provided with the renilla luciferase assay system (Promega).The culture plates were placed on an orbital shaker with gentle shaking at room temperature for 15 min. The cell lysates were centrifuged at top speed for 30 seconds in a refrigerated microcentrifuge and the supernatant was measured for firefly or renilla luciferase activity using the appropriate luciferase substrate from Promega in a luminometer (Lumat LB 9501; Berthold; Wildbad, Germany). All luciferase assays were performed at least in triplicate.
2.6 Biotin-DNA pull-down assay
HeLa cells were transfected with AR expression plasmid or plasmid vector. 48h after transfection, cells were treated with vehicle or testosterone (10 nM) for 60 min, and then washed twice with PBS and harvested in PBS. Cell pellets were lysed with lysis buffer (400 mM NaCl; 10 mM Tris, pH 8.0; 1 mM EDTA; 1 mM EGTA; 0.1% Triton X-100; 1 mM PMSF; and 5 μg/mL each of aprotinin, leupeptin, and pepstatin A) supplemented with vehicle or testosterone (10nM). The lysates were centrifuged at 16, 000g for 10 min and the resultant supernatants were diluted 1: 4 with dilution buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.5 mM EGTA; 10% glycerol; 0.25% nonidet P-40) in the presence of ligand as indicated. Then 300 μg of lysate was incubated with 1 μg of the appropriate biotinylated DNA probe and 10 μg poly (dI-dC) with or without 200 μg of proper unlabeled probe at 4°C on a rotary shaker for 1 h, and then incubated with 30 μl of 50% streptavidin-sepharose A beads overnight. The samples were centrifuged at 600g for 5 minutes, and the pellets were washed four times with washing buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 0.5 mM EGTA; 100 mM NaCl; 10% glycerol; 0.25% nonidet P-40) in the presence of ligand for 5 minutes with rotation. The proteins were released by boiling in 100 μl SDS loading buffer (62.5 mM Tris-HCl, pH 6.8; 10% glycerol; 2% SDS; 5% 2-mercaptoethanol; and 0.05% bromophenol blue) and analyzed by western blots to probe for AR and C/EBPα. The 5’ biotin-labeled probes were: FRα (5’-biotin-AGGGTTTGTTCCCGCAGGAACTGAACCCAAAGGA TCAC), (C/EBP)3 [5’-biotin-(TGCAGATTGCGCCAATCTGCA)3]. The unlabeled probess were: wt (wild type unlabeled probe for FRα; the sequence was the same as the biotin labled FRα probe), mA (FRα element with mutated ARE, AGGGTTTTCTCGCGCAGGAACTG AACCCAAAGGATCAC), dm (FRα element with double mutations in both ARE and CCAAT element, AGGGTTTTCTCGCGCAGGAACGTCTCTTCGAGGATCAC).
2.7 RNA isolation, RT-PCR and real-time PCR
Total RNA from HeLa cells, T47D and ACH-3P cells was prepared using the RNeasy Mini kit (Qiagen). Reverse transcription PCR (RT-PCR) followed by real-time PCR was used to measure mRNAs for luciferase, FRα and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For the reverse transcription reaction, 200 ng of total RNA was reverse transcribed with random primers by using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA) The resulting cDNA was measured by quantitative real-time PCR using the Real-time PCR master mix (Applied Biosystems) in the 7500 StepOne Plus Real Time PCR System (Applied Biosystems, Foster). The primers and TaqMan probe for FRα, Luciferase and GAPDH were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). All samples were assayed in triplicate and normalized to GAPDH values in the same samples.
2.8 Chromatin immunoprecipitation (ChIP) assay
HeLa cells were transfected with His- tagged AR expression plasmid or vector. 48h after transfection, cells were treated with vehicle or testosterone (10 nM) for 1h, washed with cold PBS and subjected to ChIP analysis using anti-His antibody (sc-803) and negative control IgG (sc-2027) following the procedure described previously [16]. The recruitment of His-AR to the FRα gene was measured by real time PCR. Real-Time PCR analysis of chromatin-immunoprecipitated products was performed using the following FRα promoter primers and TaqMan probe: FRα promoter probe (-1913nt to -1935nt), 5'-6 FAM-TCGGGACAGGTTGAACGGGAACC-3'; sense primer (-1876nt to – 1893nt), 5'-CCCCAAGGCCAAGGAGAA-3'; and antisense primer (-1966nt to -1945nt), 5'-CGGGAACAAACCCTAACTGTTT -3'. The samples were tested in triplicate. To perform the ChIP assay in ACH-3P cells, the cells were first grown in stripped serum media for 48 hours and treated with 10nM R1881 or testosterone for 2 hours. The cells were washed with cold PBS and subjected to ChIP analysis with or without anti-AR antibody (sc-816X from Santa Cruz) following the procedure described. The recruitment of AR to the FRα gene was measured by real time PCR as described above.
2.9 Co-Immunoprecipitation Assay
Total protein from ACH-3P cells were prepared using RIPA buffer. Co-Immunoprecipitation assay was performed by using AR antibody (sc-816) or C/EBPα antibody (sc-61) and the proteins detected by Western blot analysis as described [30].
3.0 Statistical analyses
All experimental values are presented as mean ± SE. The statistical significance of differences (P value) between values being compared was determined using ANOVA. In all cases, differences noted in the text are reflected by a P value < 0.001.
3 RESULTS
3.1 The folate receptor type α gene is directly regulated by androgen through activation of its basal P4 promoter
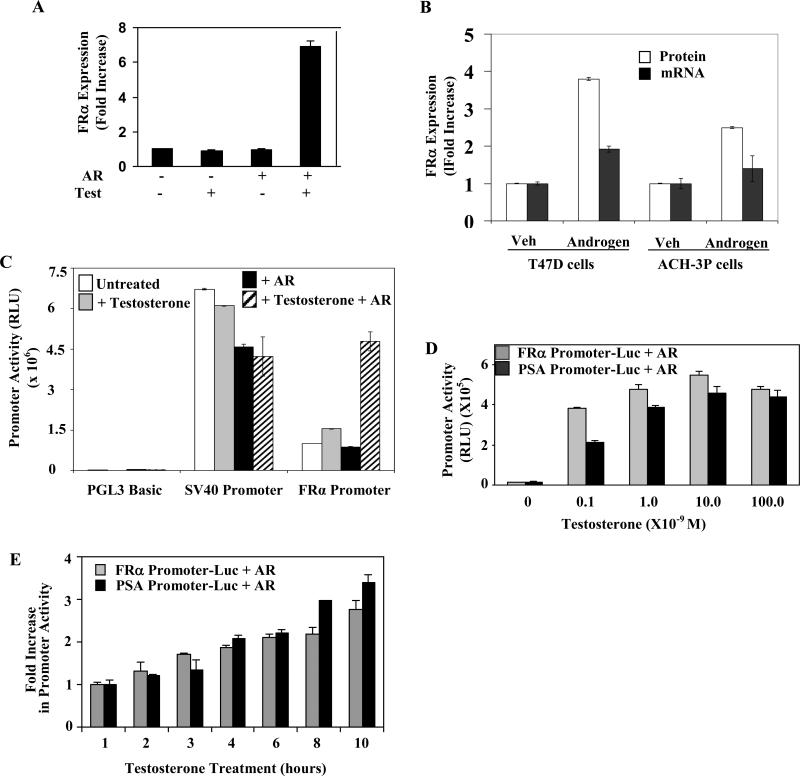
When AR was ectopically expressed in HeLa cells which are folate receptor (FR) type α-positive and AR-negative, treatment with androgen increased the cell surface expression of FR in the transfected population of cells (Fig 1A). In the AR and FRα-positive T47D cells, a similar treatment also increased FRα mRNA and FRα protein expression (Fig. 1B). Similarly in the AR and FR positive ACH-3P immortalized placental trophoblasts, androgen treatment increased cell surface FR at both the mRNA and protein levels (Fig. 1B). In T47D and ACH-3P cells, the fold increase in FRα protein was higher than that of the mRNA; this difference could reflect the possibility that the turnover of the de novo synthesized protein was slower than that of the mRNA or that additional mechanisms are involved at the translational level. Further mechanistic experiments in this study focused on transcription and were conducted in HeLa cells because these cells are amenable to transfection and AR expression in these cells may be controlled by ectopic expression. As described below, the results were then extended to ACH-3P cells.
Figure 1. Specific regulation of FRα by androgen.
(A) HeLa cells transfected with AR expression plasmid were treated with 10 nM testosterone (Test) or vehicle for 3 days. FRα cell surface expression was quantified. (B) T47D cells or ACH-3P cells were treated with 10 nM R1881 or vehicle for 72h. FRα cell surface expression and the FRα mRNA were measured. (C) Hormone deplete HeLa cells were transfected with one of the following constructs: the FRα promoter Luc, SV40 promoter Luc or PGL3 Basic Luc (empty vector control plasmid). The cells were co-transfected with either the expression plasmid for AR or the vector control as well as the Renilla luciferase plasmid (transfection control) and either untreated or treated for 48h with testosterone (10 nM). The cells were then harvested to measure luciferase activity (D) Hormone deplete HeLa cells were transfected with the FRα promoter Luc or the PSA promoter Luc. The cells were co-transfected with either the expression plasmid for AR as well as the Renilla luciferase plasmid (transfection control) and either untreated or treated for 48h with different doses of testosterone. The cells were then harvested to measure luciferase activity. (E) Hormone deplete HeLa cells were transfected with the FRα promoter Luc or the PSA promoter Luc. The cells were co-transfected with either the expression plasmid for AR as well as the Renilla luciferase plasmid (transfection control). 48h post-transfection, the cells were either untreated or treated for different periods with testosterone (10 nM). The cells were then harvested to measure luciferase activity. P values for the differences noted in the text were < 0.001.
The FRα gene organization includes seven exons and six introns and its transcription is driven by two TATA-less promoters, P1 and P4. The proximal P4 promoter (genomic coordinate for +1nt: 71,903,499 hg19 on chr 11), which generates a single transcript is generally predominant in malignant cell lines including HeLa cells [25]. In transfected HeLa cells, testosterone activated an FRα promoter-reporter construct spanning both the P1 and P4 regions (-3394nt to +33nt) in an AR-dependent manner but did not activate the SV40 promoter, indicating the specificity of its action on the FRα promoter (Fig. 1C). The testosterone dose response (Fig. 1D) as well as the time course of activation by testosterone (Fig. 1E) of the FRα promoter paralleled those of a prostate-specific antigen (PSA) promoter (-642nt to +12nt)-reporter construct, which is activated through its proximal androgen response elements (AREs). Those observations suggested that the FRα promoter could be a direct target of AR action.
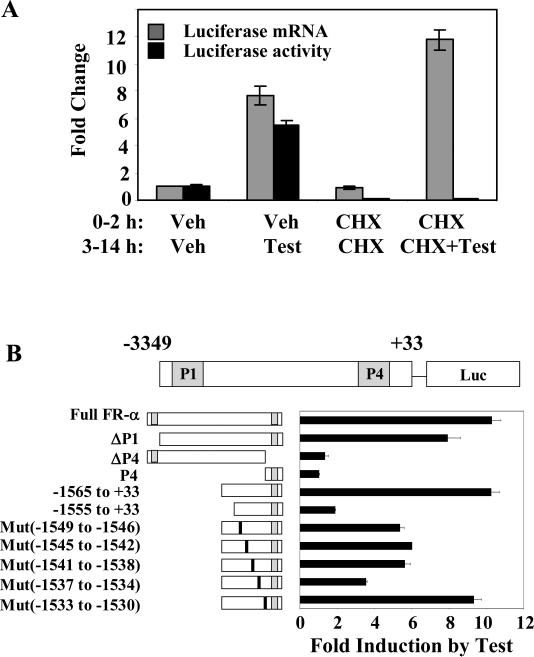
Further evidence for the absence of an intermediate gene target in androgen stimulation of the FRα promoter, i.e., the absence of a need for new protein synthesis, was provided by the observation that treatment with cycloheximide did not alter the promoter activation by testosterone/AR as measured by luciferase mRNA expression under conditions in which it abrogated the reporter enzyme activity (Fig. 2A).
Figure 2. Direct regulation of FRα by androgen and mapping of the regulatory elements.
(A) HeLa cells were transfected with FRα (-1565 to +33) promoter-luciferase reporter and co-transfected with AR; 24h later, the cells were then treated with either 10 μM cycloheximide (CHX) or vehicle for 2 h followed by the introduction of testosterone (10 nM) or vehicle for further 12h. Total RNA was extracted for quantitation of luciferase mRNA by real-time reverse transcription-PCR. In a parallel experiment, the cells were harvested to measure the luciferase protein expression by measuring luciferase activity. (B) HeLa cells were transfected with the following promoter-luciferase reporter constructs: the full FRα promoter [FRα(-3394nt to +33nt)-luc]; the full FRα promoter with deletion of either the P1 promoter (ΔP1) [FRα(-3113nt to +33nt)-luc] or the P4 promoter (ΔP4) [FRα(-3394nt to +33nt, Δ-146 nt to -34nt)-luc]; the P4 promoter [FRα (-176 nt to +33nt)-luc]; the FRα promoter with 5’ deletions (-1565nt to +33nt and -1555nt to +33nt); the FRα promoter constructs with sequential 4 base pair mutations from -1549nt through -1530nt. The positions of the mutated nucleotides are indicated by the vertical black bars (not to scale). The cells were co-tranfected with AR followed by treatment with either testosterone (10 nM) or vehicle and harvested for luciferase assay 48h post-transfection. P values for the differences noted in the text were < 0.001.
The testosterone/AR response of FRα promoter-luc did not diminish upon deletion of the P1 promoter but was abrogated when the P4 promoter was silenced by deleting its essential Sp1 binding elements (Fig. 2B), indicating that the P4 promoter is the target of AR. Further, the species of endogenous FRα mRNA that is up-regulated by testosterone/AR in HeLa cells was identified by RNase protection assays as the P4 promoter driven transcript (results not shown). However, the minimal P4 promoter alone could not respond to androgen and required the inclusion of an upstream region (Fig. 2B).
Based on the effect of progressive 5′ deletions on the testosterone/AR response, the target site of AR action was mapped downstream of -1565nt in the FRα promoter (Fig. 2B). To bracket the 3′ boundary of the target site, sequential 4-base mutations were introduced beginning at -1549nt and their effects on the promoter activity tested (Fig. 2B). In this manner, the site of AR action was mapped to -1565nt to -1534nt (genomic coordinates: 71,901,933 - 71,901,964 hg19 on chromosome 11).
3.2 A composite ARE half-site and non-canonical C/EBP element is the functional target site of AR action in the FRα gene
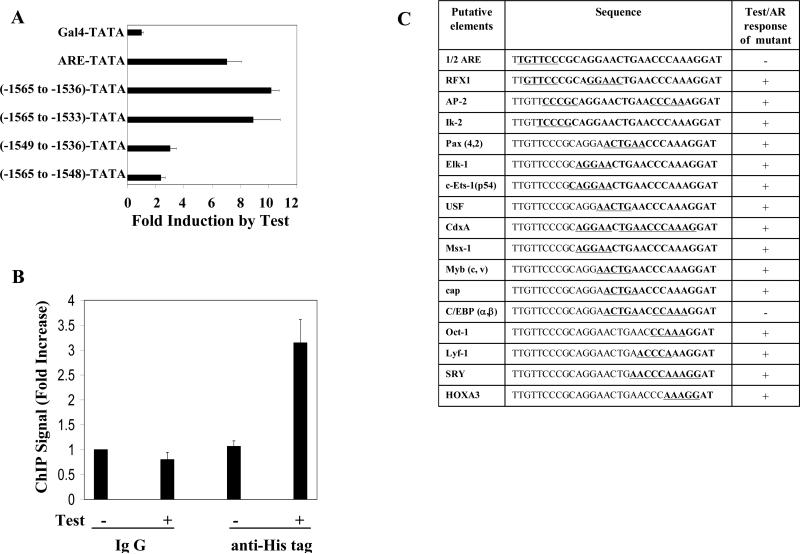
When the FRα gene sequence element -1565nt to -1536nt was transposed to the 5′ end of a heterologous minimal promoter, the promoter became responsive to testosterone/AR to an extent comparable to transposing a canonical ARE (Fig.3A). A 3′ extension of the transposed FRα promoter sequence to -1533nt did not increase the androgen response and further, within the transposed element, both the 5′ and 3′ regions were required similar to the observations in the FRα promoter (Fig.3A) indicating that the sequence -1565nt to -1536nt in the FRα promoter was necessary and sufficient to mediate the androgen response.
Figure 3. Identification of the site of AR recruitment in the FRα promoter.
(A) HeLa cells were transfected with the following constructs: a TATA-box dependent minimal promoter-luciferase construct containing an upstream GAL4 element (GAL4-TATA) (pG5luc); pG5luc in which the GAL4 element was replaced by an ARE element (ARE-TATA) or by FRα 5’ upstream sequences (-1565nt to -1536nt, -1565nt to -1533nt, -1549nt to -1536nt or -1565nt to -1548nt). The cells were co-transfected with AR expression plasmid and treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h); the cells were then harvested to measure luciferase activity. (B) Hela cells were transiently transfected with His-tag AR. 48h later, cells were treated with either testosterone (10 nM) or vehicle for 1h and subjected to ChIP assay using either an His-tag specific rabbit antibody or negative control rabbit IgG. The target sequence mapped in Fig. 2B and 3A was quantified in the immunoprecipitate by real-time PCR. (C) The FRα sequence -1565nt to -1536nt was inserted in place of the GAL4 element in GAL4-TATA-luc. Putative cis elements identified using the TRANSFAC database and the MATCH program were individually mutated within this construct. HeLa cells were transfected with each construct and co-transfected with either AR expression plasmid or a vector control. Testosteone (10 nM) treatment and measurement of luciferase assay were carried out as described for Fig. 2B and 3A. The results are summarized in terms of either a positive response of the promoter to testosterone (activation, +) or the absence of promoter activation by testosterone (-). P values for the differences noted in the text were < 0.001.
The ability of AR to directly associate with the endogenous FRα gene in the region spanning -1565nt to -1536nt was demonstrated by a quantitative chromatin immunoprecipitation (ChIP) assay using His-tagged AR; the association required a brief treatment with testosterone (Fig. 3B).
All of the known cis-elements for the binding of transcription factors within the FRα -1565nt to -1536nt sequence identified using the TRANSFAC database and the MATCH program [26, 27] are listed in Fig. 3C. Using the minimal promoter construct containing this sequence, the effects of a series of systematic mutations in this region were tested on promoter activation by testosterone/AR. Key functional nucleotides within each putative cis-element (underlined in Fig. 3C) were mutated by changing A-T pairs to G-C pairs and vice versa. In Fig. 3C, in order to mutate the RFX1 element without disrupting the overlapping ARE ½ site, RFX1 binding was disrupted by mutating the downstream ½ site which does not overlap with the ARE ½ site. Additional experiments including the use of promoters with a strong consensus RFX1 element demonstrated the inability of RFX1 to recruit AR (results not shown).In this manner it was determined that all of the putative cis-elements could be eliminated without loss of testosterone/AR responsiveness except a 5′ ARE half-site and a 3″ non-canonical C/EBPα/β binding element (Fig. 3C). Mutation of either of these two elements also abrogated the testosterone response of the full length FRα promoter (data not shown).
Taken together, the preceding results indicate that the site of functional association of AR in the FRα gene is spanned by the sequence -1565nt to -1536nt and identify the putative functional elements within this sequence as an upstream ARE half-site and a downstream non-canonical C/EBPα/β element.
3.3 AR and C/EBPα physically interact at the composite FRα element
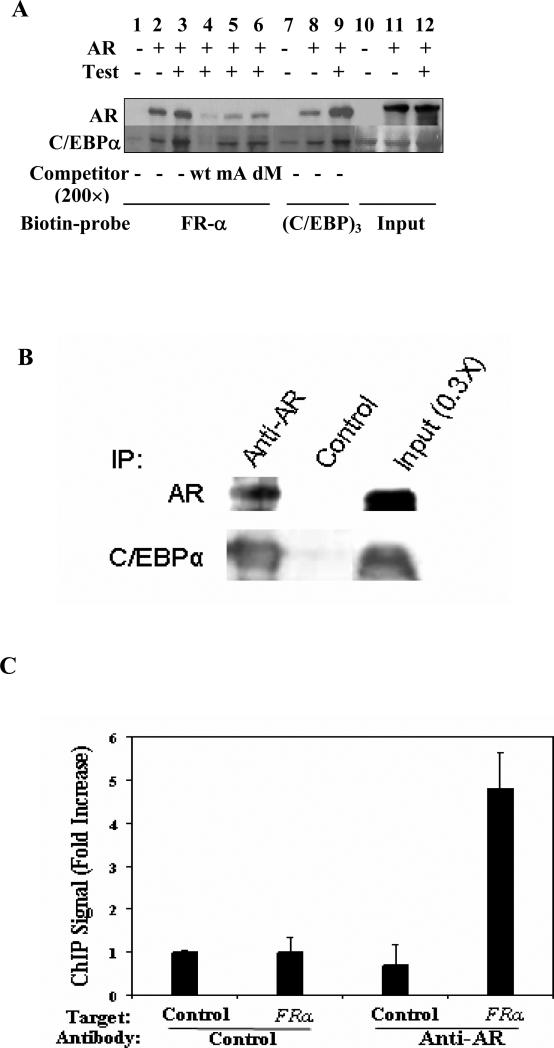
Interaction of AR and C/EBPα at the androgen target site in the FRα gene was tested by an in vitro biotinylated DNA pull down assay using the probe sequence FRα -1565nt to -1536nt in the presence of total HeLa cell lysates with absent or ectopic AR expression and with endogenous C/EBPα (Fig. 4). The FRα probe pulled down both AR and C/EBPα in a manner that was favored by treatment of the cells with testosterone (Fig. 4). However, in the absence of AR, the pull down of C/EBPα was greatly diminished (Fig.4). An excess amount of the unlabeled wild-type probe effectively competed for the binding of both AR and C/EBPα with the biotinylated probe; an unlabeled probe with mutations in the ARE half-site was less effective as a competitor and the competition further decreased when the C/EBP element was also mutated (Fig.4). These results demonstrate cooperative association of C/EBPα and AR with the composite element in the FRα gene sequence -1565nt to -1536nt.
Figure 4. Physical interaction between AR and C/EBPα and recruitment of endogenous AR to the FRα gene.
(A) DNA pull down assays for biotinylated forms of a synthetic FRα promoter element (-1570nt to-1533nt) or 3-tandem repeat C/EBP elements [(C/EBP)3]: Hela cells were tranfected with AR plasmid for 48h and treated with testosterone (10 nM) or vehicle for 1h before being harvested to prepare total cell lysates. The lysates were incubated with each biotinylated probe in the continued presence of either testosterone or vehicle. Control assays included incubation with a 200-fold excess of unlabeled synthetic DNA corresponding to FRα, -1570nt to-1533nt (wild type probe, wt); the wt probe in which the ARE half-site was mutated (mA); the wt probe in which both the ARE half-site and the C/EBP element were mutated (dM). The biotinylated DNA probe and associated proteins were pulled down using streptavidin sepharose beads. The proteins were eluted from the beads and probed by western blot analysis using antibody to AR or C/EBPα. (B) Co-immunoprecipitation of AR and C/EBPα in ACH-3P cells: Endogenous AR in the cell lysates was immonoprecipitated and probed by western blot using antibodies to either AR or C/EBPα. Bands were detected using either antibody probe in the immunoprecipitate obtained using anti-AR antibody but not in the negative control. (C) Chromatin immunoprecipitation of AR in R1881 treated ACH-3P cells. The control target sequence (irrelevant target) corresponds to the estrogen receptor gene. The FRα target sequence corresponds to the -1565nt to -1534nt region.
The ability of endogenous AR and endogenous C/EBPα to physically associate with each other in ACH-3P cells was demonstrated by their co-immunoprecipitation using an antibody specific for AR (Fig. 4B). Further, recruitment of endogenous AR in ACH-3P cells to the composite element in the FRα gene in situ was demonstrated by ChIP assay using the antibody to AR (Fig. 4C). These results extend the above mechanistic findings in HeLa cells to the physiologically relevant conditions in ACH-3P cells.
3.4 Trans-activation of the FRα promoter by AR is supported by AR co-activators
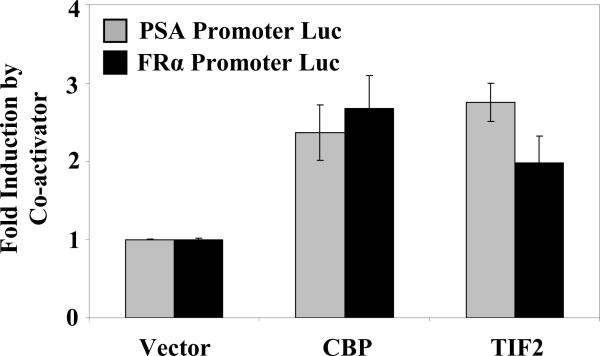
The effect of representative AR co-activators on the C/EBPα-dependent trans-activation of the FRα promoter by AR was examined (Fig. 5). The PSA promoter (control) and the FRα(-1565nt to +33nt) promoter were tested; in both cases, CBP and TIF2 acted as activators of the testosterone/AR response, indicating co-activator dependence of the transcriptional activity of AR mediated through recruitment by C/EBPα.
Figure 5. Effect of AR co-activators on the activation of the FRα promoter by androgen.
HeLa cells were separately transfected with two different promoter-luciferase constructs (PSA Promoter-Luc or FRα Promoter-Luc) as well as the Renilla luciferase control plasmid and were co-transfected with an expression plasmid for AR together with the co-activator CBP or TIF2 or with the vector control. The cells were treated with testosterone(10 nM) for the duration of the transfection (48 h) and harvested for luciferase assays. The luciferase values were normalized by the corresponding basal values in the absence of transfection with co-activators. The control experiment (PSA promoter Luc) has been previously reported [28]. P values for the differences noted in the text were < 0.001.
3.5 Trans-activation per se of the FRα promoter by AR-C/EBPα is partially androgen-independent
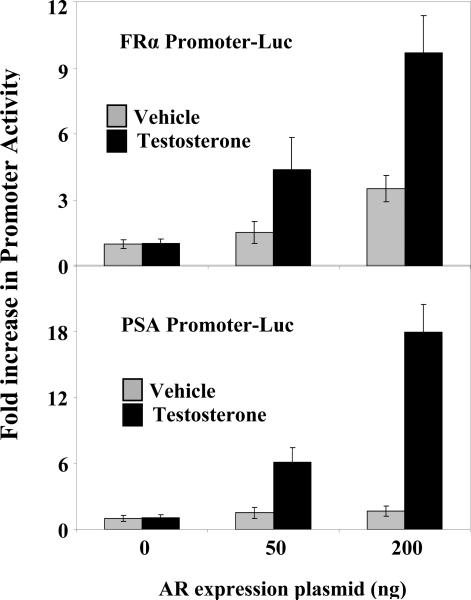
This laboratory [28] and others [29] have reported that in HeLa cells a substantial amount of ectopic AR can enter the nucleus independent of ligand and that the hormone-independent localization of AR in the nucleus increases with the dose of ectopic AR. The forced localization of apo-AR in the nuclear compartment of HeLa cells by over-expression of ectopic AR has been used to test hormone-dependence of trans-activation by AR, i.e., independent of its hormone-dependence for nuclear import [28]; in this manner, it has been established that for a purely tethered association of AR at a C/EBP element in its target promoter, trans-activation per se by AR is entirely hormone-independent. Therefore, it was of interest to use this approach to test the hormone-dependence of transactivation of the FRα promoter by AR since in this promoter, AR is only partially tethered by C/EBPα. The PSA promoter whose activation by AR is entirely androgen-dependent was used as the control.
As expected, in HeLa cells, despite increasing expression levels of ectopic AR, the AR activation of the PSA promoter was entirely androgen-dependent (Fig.6). Activation of the FRα promoter by AR on the other hand, did occur without the need for hormone, although androgen was required for optimal activation (Fig. 6). The results demonstrate that gene activation through partial tethering of AR at the composite ARE-C/EBP element of the FRα promoter is partially androgen-independent.
Figure 6. Partial ligand-independence of FRα promoter activation upon overexpression of AR.
HeLa cells were transfected with either the FRα promoter (top) or the PSA-promoter (bottom) luciferase reporter construct together with different amounts of AR expression plasmid (5-200 ng per 3 × 105 cells). The cells were treated with testosterone (10 nM) or vehicle for the duration of the transfection (48 h) and harvested for luciferase assays. P values for the differences noted in the text were < 0.001.
4 DISCUSSION
The results demonstrate the mechanism of a profound regulation of the FRα gene by androgen in different cell lines including human placental trophoblasts that is distinct from the modes of action of other steroid hormone receptors on this gene. AR transcriptionally activates the FRα gene by acting on its proximal P4 promoter from a site of physical association with the gene that is ~1500bp upstream of the promoter. This site comprises a composite element that recruits AR through the cooperative function of an ARE half-site and C/EBPα bound to DNA at a non-canonical site. FRα gene activation by AR is partially ligand-independent and is supported by steroid receptor co-activators.
C/EBPα has recently been shown to act as a tether that enables AR to associate with and regulate many of its target genes [28]. C/EBPα acts as a co-repressor of the classical androgen response element (ARE)-mediated gene activation by AR [28, 30] but also redirects the transcriptional activity of AR to C/EBP elements [28]. The association of AR with DNA bound C/EBPα and gene activation by this mechanism are independent of hormone and insensitive to the classical androgen antagonist, flutamide [28]. Further, C/EBPα-tethered gene activation by AR is insensitive to disruption of AR dimerization [28]. The foregoing studies of the regulation by androgen of the folate receptor type α (FRα) gene which exemplify a variant mechanism of AR tethering in which the tethering protein (C/EBPα) functions in concert with an ARE half-site. It is noteworthy that in the context of this partial tethering by C/EBPα, transcriptional activation by AR is largely hormone-dependent, suggesting that the C/EBP element essentially substitutes for the second ARE half-site to largely recruit the ligand-induced AR dimer.
Although most tissues do not require FRα for cellular folate uptake[31], hormonal control of the FRα gene is particularly significant in specific tissues during pregnancy since the receptor is critical for normal development. The placenta, which is one of the few tissues in which the physiological role of FRα has been established with clarity, co-expresses FRα, ER, PR, GR and AR as well as C/EBPα [12, 13, 19, 20, 32]. The negative and direct regulation of the FRα gene by estrogen is opposed by the indirect actions of progesterone as well as glucocorticoids. In this context, the C/EBPα-dependent activation of the FRα gene by androgen, elucidated in the current study, may offer an additional and important mechanism to ensure adequate expression of FRα in the appropriate tissues. The relative levels of steroid hormones, including androgen, as well as the molecular mediators of their actions may be critical factors that determine whether an adequate supply of folate is available for the developing embryo.
Role of funding source
This work was funded by NIH R01 grants CA 103964 and CA 80183 and by the Harold and Helen McMaster endowment to M.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supported by NIH R01 grants CA 103964 and CA 80183 and by the Harold and Helen McMaster endowment to M.R.
5 REFERENCES
- 1.Elwood PC. Molecular cloning and characterization of the human folate-binding protein cDNA from placenta and malignant tissue culture (KB) cells. J Biol Chem. 1989;264(25):14893–14901. [PubMed] [Google Scholar]
- 2.Lacey SW, Sanders JM, Rothberg KG, Anderson RG, Kamen BA. Complementary DNA for the folate binding protein correctly predicts anchoring to the membrane by glycosylphosphatidylinositol. J Clin Invest. 1989;84(2):715–720. doi: 10.1172/JCI114220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratnam M, Marquardt H, Duhring JL, Freisheim JH. Homologous membrane folate binding proteins in human placenta: cloning and sequence of a cDNA. Biochemistry. 1989;28(20):8249–8254. doi: 10.1021/bi00446a042. [DOI] [PubMed] [Google Scholar]
- 4.Yan W, Ratnam M. Preferred sites of glycosylphosphatidylinositol modification in folate receptors and constraints in the primary structure of the hydrophobic portion of the signal. Biochemistry. 1995;34(44):14594–14600. doi: 10.1021/bi00044a039. [DOI] [PubMed] [Google Scholar]
- 5.Elnakat H, Ratnam M. Distribution, functionality and gene regulation of folate receptor isoforms: implications in targeted therapy. Adv Drug Deliv Rev. 2004;56(8):1067–1084. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Kamen BA, Smith AK. A review of folate receptor alpha cycling and 5-methyltetrahydrofolate accumulation with an emphasis on cell models in vitro. Adv Drug Deliv Rev. 2004;56(8):1085–1097. doi: 10.1016/j.addr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell. 2002;2(4):411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 8.Molloy AM, Scott JM. Folates and prevention of disease. Public Health Nutr. 2001;4(2B):601–609. doi: 10.1079/phn2001144. [DOI] [PubMed] [Google Scholar]
- 9.Piedrahita JA, Oetama B, Bennett GD, van Waes J, Kamen BA, Richardson J, Lacey SW, Anderson RG, Finnell RH. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat Genet. 1999;23(2):228–232. doi: 10.1038/13861. [DOI] [PubMed] [Google Scholar]
- 10.Finnell RH, Wlodarczyk B, Spiegelstein O, Triplett A, GelineauvanWaes J. Folate transport abnormalities and congenital defects. Kluwer Academic publishers; Norwich, MA: 2001. [Google Scholar]
- 11.Antony AC. The biological chemistry of folate receptors. Blood. 1992;79(11):2807–2820. [PubMed] [Google Scholar]
- 12.Salbaum JM, Finnell RH, Kappen C. Regulation of folate receptor 1 gene expression in the visceral endoderm. Birth Defects Res A Clin Mol Teratol. 2009;85(4):303–313. doi: 10.1002/bdra.20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barber RC, Bennett GD, Greer KA, Finnell RH. Expression patterns of folate binding proteins one and two in the developing mouse embryo. Mol Genet Metab. 1999;66(1):31–39. doi: 10.1006/mgme.1998.2772. [DOI] [PubMed] [Google Scholar]
- 14.Steinfeld R, Grapp M, Kraetzner R, Dreha-Kulaczewski S, Helms G, Dechent P, Wevers R, Grosso S, Gartner J. Folate receptor alpha defect causes cerebral folate transport deficiency: a treatable neurodegenerative disorder associated with disturbed myelin metabolism. Am J Hum Genet. 2009;85(3):354–363. doi: 10.1016/j.ajhg.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelley KM, Rowan BG, Ratnam M. Modulation of the folate receptor alpha gene by the estrogen receptor: mechanism and implications in tumor targeting. Cancer Res. 2003;63(11):2820–2828. [PubMed] [Google Scholar]
- 16.Hao H, d'Alincourt-Salazar M, Kelley KM, Shatnawi A, Mukherjee S, Shah YM, Ratnam M. Estrogen-induced and TAFII30-mediated gene repression by direct recruitment of the estrogen receptor and co-repressors to the core promoter and its reversal by tamoxifen. Oncogene. 2007;26(57):7872–7884. doi: 10.1038/sj.onc.1210592. [DOI] [PubMed] [Google Scholar]
- 17.Shatnawi A, Tran T, Ratnam M. R5020 and RU486 act as progesterone receptor agonists to enhance Sp1/Sp4-dependent gene transcription by an indirect mechanism. Mol Endocrinol. 2007;21(3):635–650. doi: 10.1210/me.2006-0274. [DOI] [PubMed] [Google Scholar]
- 18.Tran T, Shatnawi A, Zheng X, Kelley KM, Ratnam M. Enhancement of folate receptor alpha expression in tumor cells through the glucocorticoid receptor: a promising means to improved tumor detection and targeting. Cancer Res. 2005;65(10):4431–4441. doi: 10.1158/0008-5472.CAN-04-2890. [DOI] [PubMed] [Google Scholar]
- 19.Critchley HO, Saunders PT. Hormone receptor dynamics in a receptive human endometrium. Reprod Sci. 2009;16(2):191–199. doi: 10.1177/1933719108331121. [DOI] [PubMed] [Google Scholar]
- 20.Milne SA, Henderson TA, Kelly RW, Saunders PT, Baird DT, Critchley HO. Leukocyte populations and steroid receptor expression in human first-trimester decidua; regulation by antiprogestin and prostaglandin E analog. J Clin Endocrinol Metab. 2005;90(7):4315–4321. doi: 10.1210/jc.2004-2338. [DOI] [PubMed] [Google Scholar]
- 21.Cloke B, Huhtinen K, Fusi L, Kajihara T, Yliheikkila M, Ho KK, Teklenburg G, Lavery S, Jones MC, Trew G, Kim JJ, Lam EW, Cartwright JE, Poutanen M, Brosens JJ. The androgen and progesterone receptors regulate distinct gene networks and cellular functions in decidualizing endometrium. Endocrinology. 2008;149(9):4462–4474. doi: 10.1210/en.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoenmakers E, Verrijdt G, Peeters B, Verhoeven G, Rombauts W, Claessens F. Differences in DNA binding characteristics of the androgen and glucocorticoid receptors can determine hormone-specific responses. J Biol Chem. 2000;275(16):12290–12297. doi: 10.1074/jbc.275.16.12290. [DOI] [PubMed] [Google Scholar]
- 23.Hiden U, Wadsack C, Prutsch N, Gauster M, Weiss U, Frank HG, Schmitz U, Fast-Hirsch C, Hengstschlager M, Potgens A, Ruben A, Knofler M, Haslinger P, Huppertz B, Bilban M, Kaufmann P, Desoye G. The first trimester human trophoblast cell line ACH-3P: a novel tool to study autocrine/paracrine regulatory loops of human trophoblast subpopulations--TNF-alpha stimulates MMP15 expression. BMC Dev Biol. 2007;7:137. doi: 10.1186/1471-213X-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McAlinden TP, Hynes JB, Patil SA, Westerhof GR, Jansen G, Schornagel JH, Kerwar SS, Freisheim JH. Synthesis and biological evaluation of a fluorescent analogue of folic acid. Biochemistry. 1991;30(23):5674–5681. doi: 10.1021/bi00237a006. [DOI] [PubMed] [Google Scholar]
- 25.Zheng X, Kelley K, Elnakat H, Yan W, Dorn T, Ratnam M. mRNA instability in the nucleus due to a novel open reading frame element is a major determinant of the narrow tissue specificity of folate receptor alpha. Mol Cell Biol. 2003;23(6):2202–2212. doi: 10.1128/MCB.23.6.2202-2212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31(13):3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, Voss N, Stegmaier P, Lewicki-Potapov B, Saxel H, Kel AE, Wingender E. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Gonit M, Salazar MD, Shatnawi A, Shemshedini L, Trumbly R, Ratnam M. C/EBPalpha redirects androgen receptor signaling through a unique bimodal interaction. Oncogene. 2010:723–738. doi: 10.1038/onc.2009.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenster G, Trapman J, Brinkmann AO. Nuclear import of the human androgen receptor. Biochem J. 1993;293(Pt 3):761–768. doi: 10.1042/bj2930761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chattopadhyay S, Gong EY, Hwang M, Park E, Lee HJ, Hong CY, Choi HS, Cheong JH, Kwon HB, Lee K. The CCAAT enhancer-binding protein-alpha negatively regulates the transactivation of androgen receptor in prostate cancer cells. Mol Endocrinol. 2006;20(5):984–995. doi: 10.1210/me.2005-0240. [DOI] [PubMed] [Google Scholar]
- 31.Zhao R, Matherly LH, Goldman ID. Membrane transporters and folate homeostasis: intestinal absorption and transport into systemic compartments and tissues. Expert Rev Mol Med. 2009;11:e4. doi: 10.1017/S1462399409000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(Pt 3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]