Abstract
G protein-coupled receptors (GPCRs)1 constitute the largest family of integral membrane proteins present in all eukaryotic cells, yet relatively little information is known pertaining to their structure, folding, and stability. In this work, we describe several approaches to characterize conformational stability of the human adenosine A2a receptor (hA2aR). Thermal and chemical denaturation were not reversible, yet clear differences in the unfolding behavior were observed upon ligand binding via circular dichroism and fluorescence spectrometry. We found that the stability of hA2aR was increased upon incubation with the agonist N6-cyclohexyladenosine or the antagonist theophylline. When extracellular disulfide bonds were reduced with a chemical reducing agent, the ligand-binding activity decreased by ~40%, but reduction of these bonds did not compromise the unfolding transition observed via urea denaturation. Overall, these approaches offer a general strategy for characterizing the effect of surfactant and ligand effects on the stability of GPCRs.
Keywords: G protein-coupled receptor, membrane protein, adenosine, agonist, antagonist, stability, ligand-binding ability
The family of membrane proteins classified as G protein-coupled receptors (GPCRs) is among the most desirable targets for pharmaceutical development because of their predominant role in cellular signaling (1). All GPCRs are integral membrane proteins that are characterized by their seven α-helical transmembrane domains, and their ability to initiate intracellular signaling via trimeric G protein-activation following ligand binding.
GPCRs play critical roles in several diseases, yet the structural biology of these proteins remains poorly characterized, limiting progress towards rational design of pharmaceuticals (2). Currently, four high-resolution structures (rhodopsin, β1 adrenergic, β2 adrenergic, and adenosine A2a receptors) have been determined from this ~800-member GPCR superfamily, providing some insight into the conserved structural features within this complex protein family (3-6). Although these recent structures are significant steps toward an understanding of GPCR structure-function relationships, more information is needed before such data can be readily translated into the design of effective therapeutics, in part because only the inactive states of the receptors were resolved, with the exception of rhodopsin. In the case of the hA2aR crystal structure, a C-terminal truncation (Δ317) and the substitution of T4 lysozyme for intracellular loop three were used to facilitate crystallization (4); this variant lacks some key interactions, such as G protein-coupling and native agonist preferences, but has been of value in identifying antagonist leads in virtual screening (4, 7, 8). Structural analysis of GPCRs in more dynamic physiological conformations or with a variety of ligands will help address issues pertaining to ligand specificity, and reveal critical interactions that underlie receptor function for individual GPCRs.
In our lab, we have readily expressed and purified 6-10 mg/L of full-length hA2aR in an active ligand-binding form from a yeast expression system (9-13), facilitating our studies. Using this purified receptor, we have employed several biophysical techniques to elucidate the effects of agonist and antagonist molecules on the stability of human adenosine A2a receptor (hA2aR). Also, with the discovery of an unusually high number of disulfide bonds identified during crystallography (4), we hypothesized that hA2aR stability might be sensitive not only to ligand association but also to the presence of critical extracellular disulfide bonds.
Analysis of this receptor may serve as a basis to more broadly clarify critical aspects of receptor stability within the GPCR superfamily. In addition, identification of conditions that promote conformational stability of full-length hA2aR should facilitate crystallography and characterization of active state conformations.
EXPERIMENTAL PROCEDURES
Expression and Purification of hA2aR-His10
A vacuolar protease-deficient strain of S. cerevisiae, BJ5464 (MATα ura3-52 trp1 leu2Δ1 hisΔ200 pep::HIS3 prbΔ1.6R can1 GAL), containing the multi-integrating pITy-hA2aR-His10 plasmid was used for over-expression and purification of hA2aR-His10 as described previously (12). Specific radio-ligand binding is readily detectable for hA2aR-His10 localized within the yeast plasma membrane prior to solubilization (12, 13), and corresponds to the affinity preferences observed for native hA2aR (9). After purification and buffer exchange, purified hA2aR-His10 was reconstituted in mixed micelles consisting of 0.1% dodecyl-β-D-maltoside (DDM), 0.1% 3-(3-cholamidopropyl)-dimethylammoniopropane sulfonate (CHAPS), and 0.02% cholesteryl hemisuccinate (CHS) (Anatrace, Maumee, OH) in a 50 mM sodium phosphate buffer at a pH of 7.0, which has been shown to promote activity of reconstituted hA2aR in vitro (12).
Prior to biophysical measurements, UV-Vis spectra were taken of each protein sample (230 nm – 650 nm) in order to verify protein content (absorbance ~ 280 nm) and rule out any potential aggregation of the sample (absorbance ~ 600 nm). Protein purity of > 95% was also verified through Coomasie staining of SDS-PAGE for all samples. Radioligand-binding activity was verified for purified hA2aR-His10 utilized for biophysical experiments. Each batch of purified, micelle-reconstituted hA2aR-His10 was used within a 2-week time span, as receptor activity was found to decrease after approximately 20 days when stored at 4°C.
Reduction of Disulfide Bonds
Reduction of extracellular disulfide bonds within hA2aR was achieved via incubation with tris(2-carboxyethyl)phosphine (TCEP) (Thermo-Fisher, Chicago, IL). TCEP was the preferred reducing agent in these studies due to its general compatibility with spectroscopic and biophysical measurements. Surfactant solubilized hA2aR-His10 was incubated with 5mM dithiothreitol (DTT) or various concentrations of TCEP (from 0.5 mM to 10 mM), separated via electrophoresis on 12% SDS-PAGE, and blotted onto nitrocellulose for Western blot analysis as described previously (12). Mobility of TCEP-treated samples was compared to hA2aR-His10 without TCEP addition. Receptors with one or more reduced disulfide bonds are evident by a shift in mobility compared to untreated receptors, as has been previously reported for hA2aR heterologously expressed in yeast (9).
Ligand Binding
For experiments where free ligand was added to purified receptors, N6-cyclohexyladenosine (CHA) (Sigma, St. Louis, MO) and theophylline (Sigma) (14) served as representative agonist and antagonist molecules, respectively. Purified hA2aR-His10 was typically incubated with 100 μM of CHA or theophylline, well above the Ki values of 1.5-4 μM for CHA(15, 16) and 1.7-22 μM for theophylline (17, 18) in competition experiments with 3H-CGS21680 or 3H-NECA, respectively, for at least 15 minutes prior to biophysical analysis.
Radioligand binding was performed to assess the activity of hA2aR-His10 when incubated with increasing concentrations of urea. These studies were carried out at room temperature. Protein was purified as detailed above, but was not eluted from nickel resin particles. Concentrated solutions of urea (10 M) were prepared in 50 mM phosphate buffer with 0.1% DDM/0.1% CHAPS/0.02% CHS using ultra-pure urea (MP Biomedicals, Cleveland, OH), pH was adjusted to 7.0, and solutions were filter-sterilized. Urea solutions were prepared and used the same day to prevent the formation of cyanate and ammonium ions resulting from the decomposition of urea over time (19). Equal amounts of resin-protein slurry (at a dilution of ~1 – 2 μL settled resin/mL of solution) were combined with increasing amounts of ultra-pure urea ranging from 0 M urea to 8 M, and incubated on an end-over-end mixer for approximately 5-6 hours. After equilibrium was reached, samples were aliquoted into a 96-well Millipore Multi-screen Filter B 96-well glass fiber plate, and combined with 50 nM of agonist 3[H]-CGS-21,680 (Perkin Elmer, Wellesley, MA). Resin-bound protein was allowed to incubate on a rotating orbital mixer for 1.5 - 2 hours with radio-ligand prior to analysis with a MicroBeta Jet (Perkin Elmer) scintillation counter. Non-specific radio-ligand binding was calculated by incubation of radio-ligand/urea mixtures with empty resin slurry (without protein bound), which was subtracted from all samples. Previous data from our laboratory (Butz and Robinson, unpublished) showed that using cells or lysate that were not expressing protein yielded similar non-specific binding data as using a non-radioactive competitor ligand. Samples were run in triplicate, and error bars represent the standard deviation from the average.
Circular Dichroism
Protein secondary structure was characterized by CD spectroscopy measurements carried out on a Jasco J-810 spectropolarimeter with a Peltier temperature controller, as described previously (12). Spectra were measured from 260 nm to 196 nm, with 1 nm resolution, with at least 3 integrations obtained for each spectrum. Protein concentrations used in these experiments were typically 0.2 mg/mL per sample. Thermal scans were performed from 5°C to 95°C, in increments of 5°C with an incubation of 5 minutes at each temperature interval. Equilibrium times for unfolding experiments were determined empirically for thermal denaturation studies, and deemed sufficient when protein spectra no longer changed with time.
Representative data are reported from three or more independent trials, and error bars represent the standard deviation from the average. Appropriate reference spectra were subtracted in all cases, and mean residue ellipticity calculations were corrected for protein dilution incurred through ligand solution addition. Mean residue ellipticity was calculated according to:
| (1) |
where ellipticity is in millidegrees, MW is the protein molecular weight in g/mol, concentration is the sample concentration in mg/mL, l is the path length of the cuvette (0.1 cm), and aa is the number of amino acids per monomer. For hA2aR-His10, aa is 434 and MW was estimated from the primary sequence of the protein to be 47410 g/mol (20). Secondary structure thermal denaturation was fit to a single transition between two states when possible (19).
Fluorescence Spectroscopy
Fluorescence experiments were performed on an ISS-PC-1 (ISS, Champaign, IL) spectrofluorimeter. Measurements were made with the excitation polarizer set to 90° and the emission polarizer set to 0° to minimize the effects of light scattering on obtained spectra. Protein concentrations were chosen so that absorbance of the sample at 280 nm was less than or equal to 0.2 (typically 0.1 – 0.2 mg/mL protein). Ligand addition measurements were performed as detailed above, with ligand present in the reference cuvette, and any dilution of protein resulting from the volumetric addition of ligand was corrected for in the obtained spectra. For intrinsic tryptophan fluorescence measurements, an excitation wavelength of 290 nm was used and fluorescence was measured between 300 – 450 nm. When the full emission spectra was collected (280 nm – 440 nm), excitation was set at 265 nm to eliminate the presence of Rayleigh scattering in the emission spectra.
7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin (CPM) (Invitrogen, Carlsbad, CA) was used as a thiol-reactive fluorescent probe, as has been previously described for membrane proteins (21). In all cases, CPM was protected from light to minimize photobleaching effects. CPM was dissolved in DMSO at a concentration of 4 mg/mL, aliquoted, and stored at -80°C until use. Frozen stocks were thawed, diluted 1:40 in 50 mM sodium phosphate with 0.1% DDM/0.1% CHAPS/0.02% CHS buffer, and approximately 30 μL of this dye was added to a 370 μL sample containing purified protein, or protein with ligand. CPM was allowed to react with samples on an end-over-end mixer at 4°C for at least 70 – 80 minutes as this incubation time was necessary to achieve full reaction of CPM with accessible free thiols. Samples incubated with CPM were excited at 387 nm and emission spectra were collected from 400 – 550 nm.
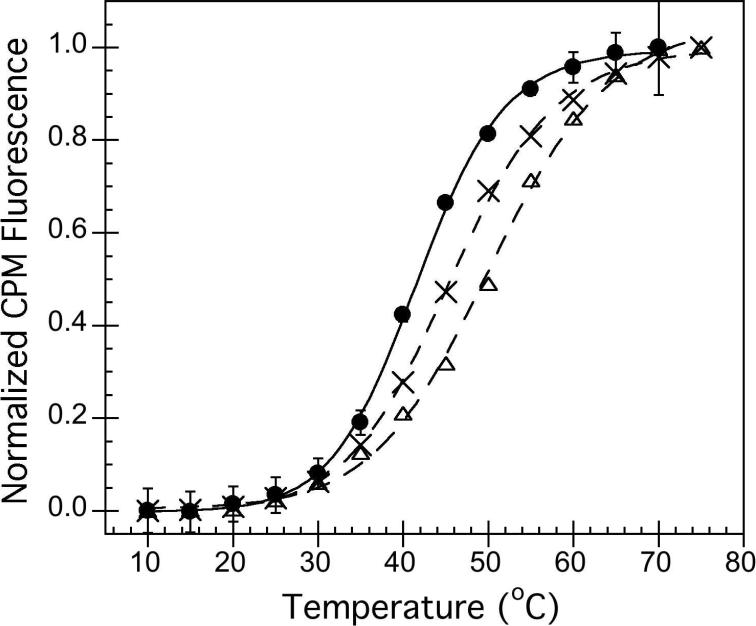
During thermal denaturation experiments, point emission measurements were taken where CPM fluorescence was measured at an emission wavelength of 463 nm. CPM fluorescence was normalized to the maximum intensity values in order to improve readability of the data shown in Figure 4B. However, the normalization did not alter the T1/2 values obtained using a model of a single-transition between two states and reported in Table 1. Thermal denaturation experiments for both intrinsic tryptophan and CPM fluorescence experiments were performed using a Peltier temperature controller, set to increase the temperature in 5°C increments. The sample chamber was held at each temperature for 5 minutes prior to fluorescence acquisition. Each trial was carried out in triplicate, and error bars represent the standard deviation from the average after fluorescence intensity normalization. Data for tertiary structure thermal denaturation were fit to a single transition between two states, where possible.
Figure 4.
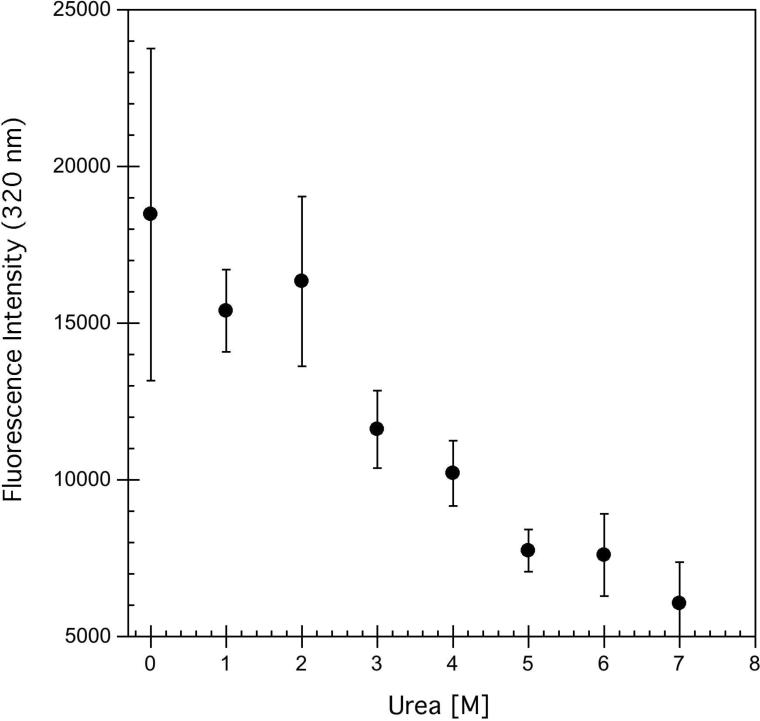
A) Representative thermal unfolding CD spectra obtained for hA2aR-His10, with temperatures increasing from 5°C to 95°C, in 15°C increments, where lighter greys are higher temperatures. B) Tertiary structure changes monitored through intrinsic tryptophan fluorescence where an excitation wavelength of 290 nm was used. Spectra represent temperatures from 11°C to 79°C, in 8°C increments, where lighter greys are higher temperatures. C) Chemical unfolding of hA2aR-His10 as a function of increasing urea, as determined by intrinsic fluorescence intensity changes at 320 nm. Error bars represent the deviation from the average of two or more biological replicates.
Table 1.
Midpoints of thermal unfolding (T1/2, °C) determined for hA2aR-His10.
| WT | +TCEP | +CHA | +Theophylline | |
|---|---|---|---|---|
| CD T1/2 [°C]i | 58.9 ± 0.4 | 55.9 ± 0.4 | 61.1 ± 0.6 | 58.4 ± 0.4 |
| CPM fluorescence T1/2 [°C] | 41.7 ± 0.2 | NDii | 46.3 ± 0.5 | 45.6 ± 0.2 |
T1/2 = midpoint of thermal unfolding; values were determined using a model for a single transition between two states
not determined: Addition of TCEP to CPM samples resulted in high levels of background fluorescence, preventing determination of thermal unfolding with CPM.
RESULTS
Conformational Changes in hA2aR-His10 Upon Ligand Binding
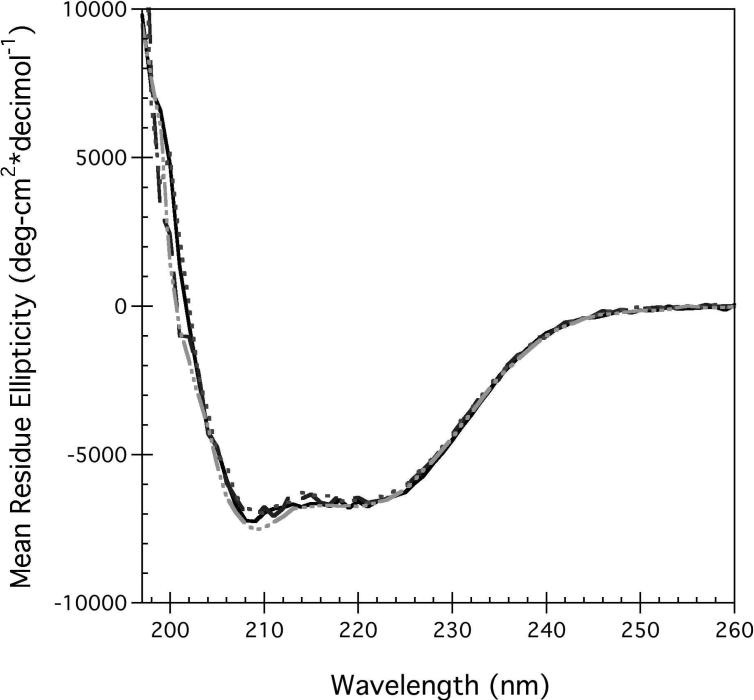
The secondary structure of purified, detergent-solubilized full-length hA2aR with a C-terminal decahistidine tag (hA2aR-His10) was predominantly α-helical as determined via circular dichroism (CD) measurements, as expected from the recently determined crystal structure (4). To evaluate the impact of bound ligands on receptor secondary and tertiary structure as well as overall thermodynamic stability, biophysical experiments were performed in the presence of agonist and antagonist for hA2aR. Upon incubation with N6-cyclohexyladenosine (CHA) (an A2aR agonist) and theophylline (an A2aR antagonist), no discernable change was observed in receptor secondary structure (Figure 2A).
Figure 2.
The impact of ligand binding and TCEP addition on hA2aR-His10 protein conformation observed via CD (A), and via intrinsic tryptophan fluorescence (Excitation wavelength, 256 nm) (B). Unliganded receptors (black), receptors with 100 μM of either CHA agonist (dark grey dashed) or theophylline antagonist (medium grey dotted), or reduced with 1 mM TCEP (light grey dash-dot). Lines are the interpolated spectra for measurements taken every 1 nm and averaged for three independent data sets.
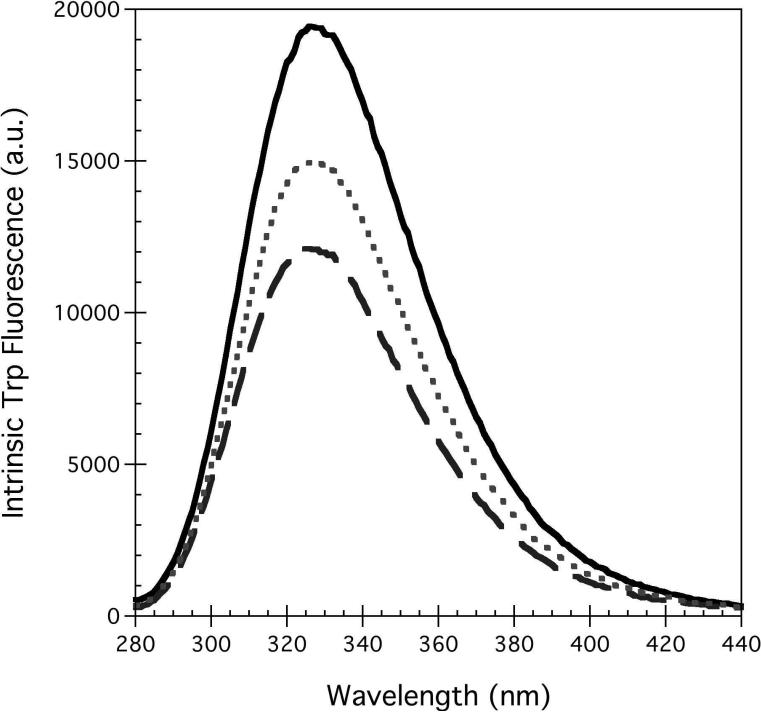
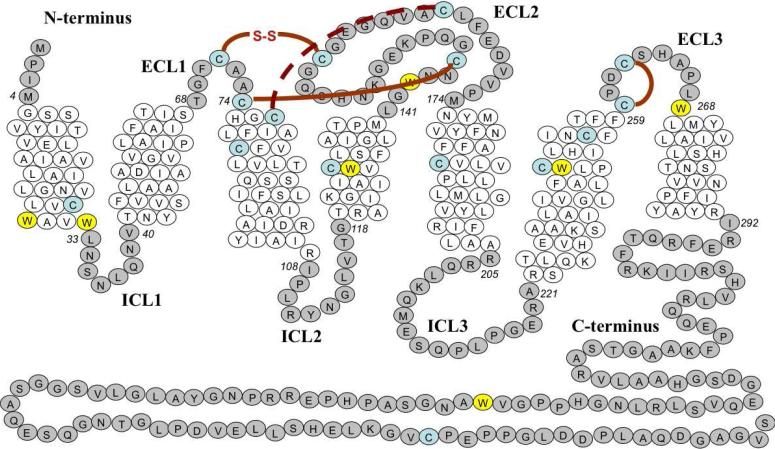
hA2aR contains seven Trp residues (Figure 1), most of which are located within transmembrane domains, one solvent-exposed on the C-terminus of the receptor, and Trp 246 at a critical site in the ligand binding pocket (7) (Figure 1), which suggested that intrinsic tryptophan fluorescence would serve as a good reporter of conformational changes to the tertiary structure in the presence of agonist and antagonist ligands. Native hA2aR shows an emission maximum at 328 nm, suggesting a relatively nonpolar environment for the Trp residues (Figure 2B). Upon binding of theophylline antagonist, the emission maximum was identical to hA2aR in the absence of ligand, but was blue-shifted to approximately 326 nm in the presence of CHA agonist (Figure 2B). Both ligands showed some absorption at 280 nm, due to the presence of aromatic rings. However, the inner filter effects contributing to the observed decrease in intensity are likely minimal, as shifting the excitation wavelength to minimize ligand absorption yielded similar behavior.
Figure 1.
Distribution of tryptophan (W) residues and cysteine residues (C) in full-length native hA2aR. Residues are numbered from the amino terminus, and are shown in italics for amino acids that flank the hydrophobic interface. Tryptophan residues are highlighted in yellow while cysteine residues are highlighted in blue. Intracellular loop regions (ICL) and extracellular loop regions (ECL) are also shown. Disulfide bonds are indicated between paired cysteines. The disulfide bond generally conserved among the GPCR superfamily is hatched red, while other bonds are displayed in solid red.
The Impact of Disulfide Bonds on hA2aR-His10 Conformation
The crystal structure for a C-terminal truncation of hA2aR fused to T4-lysozyme revealed an extensive disulfide bond network within the extracellular loop regions of the receptor (4). Disulfide bonds C71-C159, C74-C146, and C77-C166 tether extracellular loop 1 and 2, while a fourth disulfide bond (C259-C262) exists within extracellular loop 3 (Figure 1). Two of the disulfide bonds are thought to be unique to hA2aR, while the C77-C166 bond is generally conserved throughout most GPCRs (22). The CXXC motif within the third extracellular loop is also shared by the hA1 receptor, within the adenosine receptor family (23, 24).
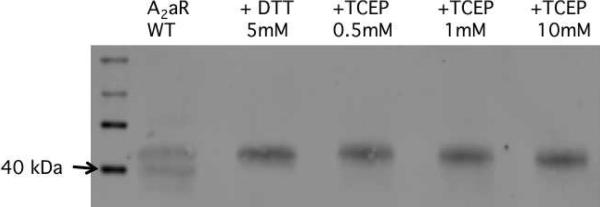
It has been postulated that these 4 disulfide bonds aid in ligand discrimination and/or ligand accessibility within the binding cavity of hA2aR (4). Here, disulfide bonds in hA2aR-His10 were reduced through incubation with the reducing agent tris(2-carboxyethyl)phosphine (TCEP) or dithiothreitol (DTT) and reduction was monitored via a reducing gel analysis (Figure 3), previously used to detect disulfide reduction in hA2aR (9). Upon incubation with 0.5 mM TCEP, the bands corresponding to hA2aR-His10 monomer was characterized by altered mobility through the gel (Figure 3A). Upon titration with increasing amounts of TCEP up to 10 mM, migration of hA2aR-His10 remained unchanged relative to the 0.5 mM addition suggesting that incubation with additional reducing agent did not bring about further disulfide bond reduction under these conditions.
Figure 3.
Effect of reducing agents on purified, solubilized hA2aR. Samples were incubated with 5 mM DTT or 0.5 mM – 10 mM TCEP, separated on 12% SDS-PAGE, and blotted onto nitrocellulose for detection with an anti-His primary antibody. Arrows denote molecular weight marker (40kDa) for MagicMark™Western protein standard (Life Technologies), close to the mobility of the monomeric band of wild-type hA2aR-His10.
hA2aR-His10 secondary structure remained highly α-helical in the presence of TCEP (Figure 2A), and the fluorescence spectra was relatively unchanged compared to that of native (non-reduced) receptors (data not shown), inferring that reduction of hA2aR's disulfide bonds had no direct effect on the conformation of unliganded receptor.
Thermal and Chemical Stability of Full-length hA2aR-His10
One of the barriers to determining stability of membrane proteins is that the detergent contributes to the stability, resulting in challenges to studying unfolding (25-28). Reversible folding of membrane proteins proves to be among the greatest difficulties associated with their biophysical characterization, and very few α-helical membrane proteins have been folded/unfolded in vitro (29, 30).
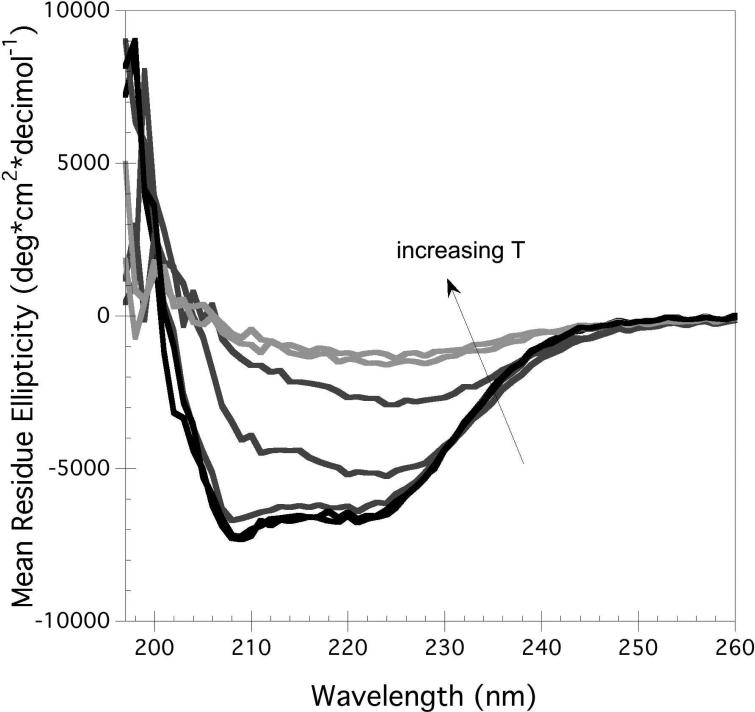
Thermal denaturation was found to be a useful approach to perturb hA2aR-His10 conformation. Figure 4 shows representative spectra generated for hA2aR-His10 as measured by CD (Figure 4A) and intrinsic fluorescence spectroscopy (Figure 4B) as temperatures were increased. Unfolding of the hA2aR-His10 secondary structure was characterized by a loss in α-helical structure, as seen through a loss of the characteristic trough at 208 and 222 nm (Figure 4A). Overall, an 80% loss in α-helical content was seen when hA2aR-His10 was heated to 90°C, and a midpoint of unfolding was fit based on a pseudo-two state model (Table 1). Upon lowering the temperature back to 10°C, we found that only ~10-20% α-helical content was recovered and these spectra did not overlap with initial measurements at 10°C (data not shown), indicating that the thermal unfolding transition was not reversible. It is likely this lack of reversibility results from aggregate formation at temperatures above 80 °C, as evidenced by increased absorbance in the visible range (~ 450 – 600 nm).
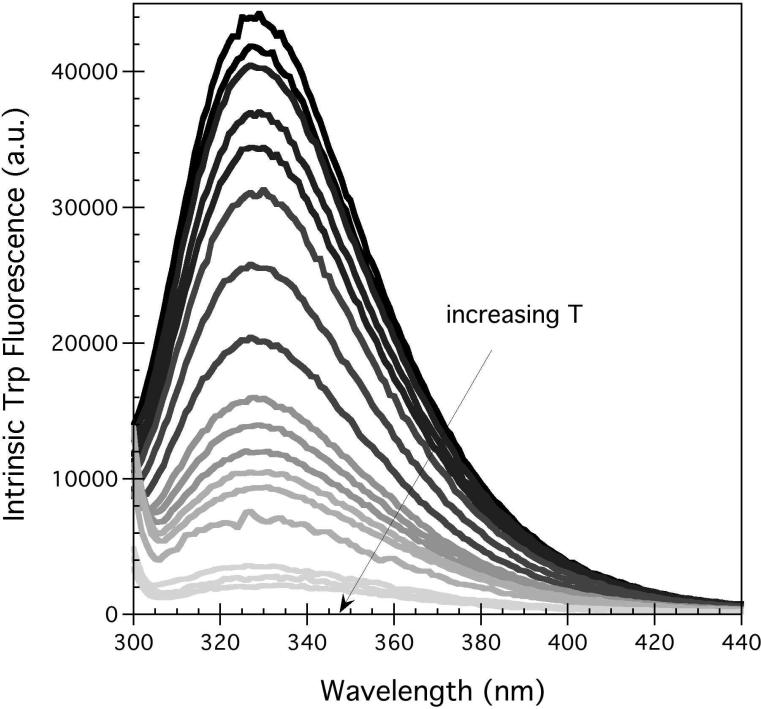
Increasing temperature from 5°C to 95°C led to a decrease in the tryptophan fluorescence emission intensity but little change in center of mass (Figure 4B). In the case of hA2aR, incubation with chemical chaotropes and SDS did not result in well-behaved denaturation, as intrinsic tryptophan fluorescence and CD did not decrease monotonically with increased concentration of SDS (data not shown) and urea (intrinsic tryptophan fluorescence, Figure 4C), consistent with the temperature denaturation, but unlike that of many small soluble proteins (31, 32). When the full-length, purified hA2aR-His10 was incubated with 0-2 M urea, the intrinsic tryptophan fluorescence intensity at 320 nm remained approximately the same, then decreased rapidly above 2 M urea (Figure 4C).
Thermal Unfolding of hA2aR-His10 Upon Ligand Addition
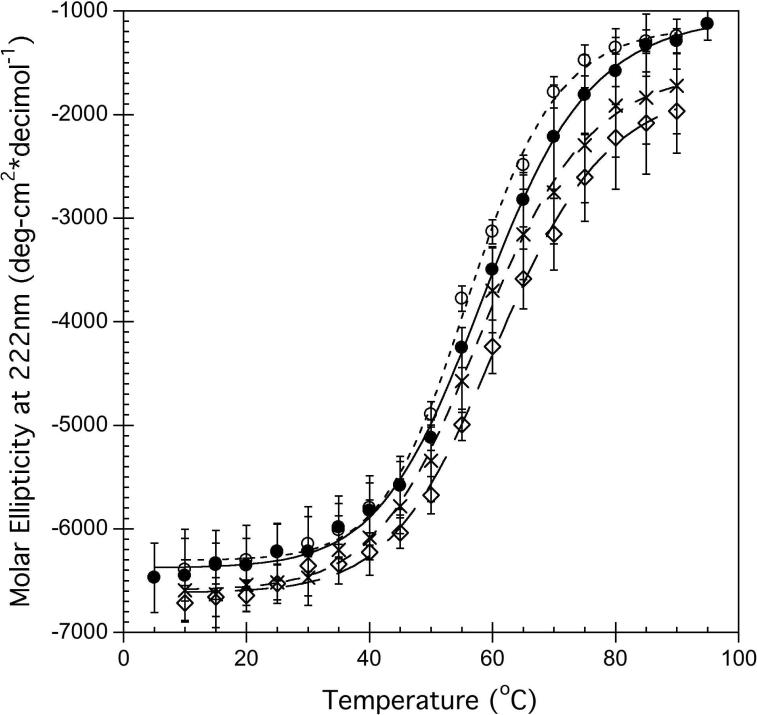
To determine the effect of ligands on hA2aR-His10 stability, molar ellipticity data at 222 nm as a function of temperature was determined (Figure 5A). Although the thermal denaturation was only partly reversible, a single transition between two states was observed in the CD data (Figure 5A). For hA2aRHis10, this unfolding transition occurred at 58.9 ± 0.4°C (Table 1). In the presence of the agonist CHA and the antagonist theophylline (Theo), changes to the thermal stability relative to hA2aR-His10 alone were observed (Figure 5A). In the agonist-bound state, the hA2aR-His10 secondary structure was slightly more resistant to thermal denaturation compared to the unliganded protein or protein bound to the antagonist, as a slight increase in the overall midpoint of unfolding was seen only in the presence of CHA (61.1 ± 0.6°C) (Table 1).
Figure 5.
Thermal unfolding of hA2aR-His10 monitored via CD at 222 nm (A) or via fluorescence intensity (463 nm) of a thiol-reactive fluorescent probe, CPM (B), for unliganded receptors (closed circles), receptors with 100 μM CHA agonist (open triangles), with 1000 μM theophylline antagonist (crosses), and reduced with 1 mM TCEP (open circles). Lines represent fit to a single-transition folding model for experimental data (points). Error bars represent standard deviation from the average of 3 or more trials. Note that CPM and TCEP are not compatible, as background fluorescence in the presence of TCEP was too large for detection of changes with temperature.
In these studies, we used an additional fluorescence-based reporter system as a complementary method to elucidate tertiary structure changes and folding transitions for hA2aR-His10. The thiol-reactive probe 7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin (CPM), which reacts with solvent-exposed cysteines, was utilized, as it has been useful for studying unfolding of membrane proteins (4, 21). Upon CPM addition to purified hA2aR-His10, observed fluorescence emission at 463 nm increased by nearly a factor of ten (data not shown). Note that for hA2aR, the reporter groups for CPM binding are six transmembrane cysteine residues, and one cysteine on the C-terminal end of the protein (Figure 1).
Addition of CHA (agonist) or theophylline (antagonist) to hA2aR-His10 yielded a distinct shift in the unfolding behavior from the unliganded hA2aR-His10 when fluorescence (due to CPM binding) as a function of temperature was examined (Figure 5B). When fit to an unfolding model that describes a single transition between two states, effective T1/2 values were 41.7 ± 0.2°C for unliganded hA2aRHis10, which increased to 46.3 ± 0.5 °C for hA2aR-His10 bound to CHA, and 45.6 ± 0.2°C when hA2aR-His10 was bound to theophylline (Table 1 and Figure 5B).
Thermal Unfolding of hA2aR-His10 Upon TCEP Addition
Due to the increased protein stability usually imparted to proteins via disulfide linkages (33-35) we explored whether these bonds also play a role in stabilization of hA2aR. The thermal denaturation of disulfide-reduced hA2aR-His10 monitored via CD led to a thermal midpoint of unfolding at 55.9 ± 0.4°C, or ~3°C less than untreated hA2aR-His10 (Figure 5A, Table 1). However, CPM fluorescence could not be determined, as TCEP addition (even in modest amounts) to CPM resulted in a pronounced increase in fluorescence intensity, which masked any folding transitions obtained.
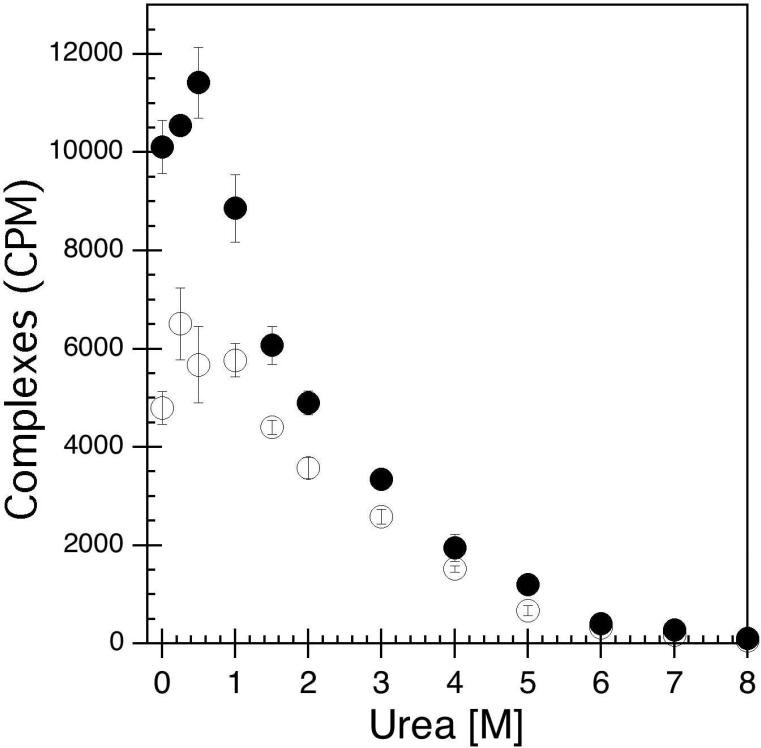
In order to determine whether the disulfide bond network in the extracellular loops of hA2aR impacted ligand binding as a metric for tertiary structure stability, surfactant-reconstituted receptors in the presence or absence of TCEP were incubated with urea to perturb protein structure. Receptor activity via radio-ligand binding was used as a sensitive metric to assess the folded state for disulfide bond-reduced hA2aR-His10. In this set of experiments, hA2aR-His10 or reduced hA2aR-His10 protein was bound to the same concentration of 3[H]-CGS-21,680 (a high-affinity A2aR agonist) over a range of urea concentrations (0 – 8M). As seen in Figure 6, in the absence of urea, reduced receptors exhibited diminished activity compared to native hA2aR-His10, by approximately 40%. At low urea concentration (< 1 M), the bound ligand obtained for hA2aR-His10 and reduced hA2aR-His10 were relatively unaffected compared to 0 M urea, as solubilized receptors largely maintained their maximal ligand-binding activity (Figure 6). Upon incubation with increasing amounts of urea, a sharp decrease in obtained radioactivity was observed, likely due to a decrease in receptor activity brought about by urea-associated unfolding. Both native and reduced hA2aR-His10 proteins lost approximately half of their binding affinity at ~2 M urea. When 6 M urea was reached, most ligand-binding activity for hA2aR-His10 was eliminated, both in the presence or absence of TCEP.
Figure 6.
Denaturation of hA2aR-His10 via urea addition leads to a loss of receptor ligand-binding activity (decrease in receptor-ligand complexes) as measured through radio-ligand binding with 3[H]-CGS-21,680 in the presence (open circles) or absence (closed circles) of the reducing agent TCEP. Error bars depict standard deviation from the average of three replicate samples at each urea concentration.
DISCUSSION
In this study, biophysical characterization of full-length human adenosine A2a receptor (hA2aR) was performed to understand receptor conformational changes and stability in the presence of extracellular ligands. We also investigated hA2aR conformational stability in the presence of reducing agent, since extensive disulfide bonding within the receptor's extracellular loop regions was identified within the crystal structure (4).
Slight Conformational Changes Accompany Ligand Binding
Biophysical methods were employed to examine changes in protein conformation upon binding of agonist and antagonist molecules to hA2aR-His10. From CD measurements, we were not able to detect appreciable changes in the receptor's α-helical secondary structure when bound to ligands (Figure 2A). One can infer from these data that receptor α-helical content does not change significantly during ligand binding. Therefore, it is more likely that transmembrane domain α-helices within hA2aR-His10 shift upon extracellular ligand binding, rather than incur any direct change to their α-helical conformation.
Although the fluorescence emission maximum remained unchanged for several conditions tested, a slight, reproducible shift was seen for hA2aR-His10 only in the presence of agonist (Figure 2B). Similar blue-shift behavior in the intrinsic fluorescence spectrum of agonist-bound GPCRs has also been reported for the BLT1 receptor upon binding LTB4 (36). Since there was a relatively small change in the emission maximum upon ligand binding (Figure 2B), we can infer that the average environment of the tryptophans in hA2aR-His10 does not undergo a major change.
A significant reduction of the fluorescence signal accompanied both agonist and antagonist binding relative to hA2aR-His10 in the absence of ligands, indicating that both ligands bury Trp residues of the receptor (Figure 2B). A larger decrease of fluorescence intensity in the presence of CHA compared to that of theophylline indicates that CHA binding affects the solvation state of more Trp residues of the receptor than theophylline does upon its binding.
Solvent-exposed regions of GPCRs have been hypothesized to play a role in accommodating ligand binding in both rhodopsin and β2AR (37), which likely also explains the Trp fluorescence decreases observed here, since many Trp residues within hA2aR are located at the interface of transmembrane helices and soluble loops, with Trp 246 at a critical site in the ligand binding pocket (7).
hA2aR Thermal and Chemical Denaturation
We have used thermal denaturation to characterize the unfolding pathway for solubilized hA2aR-His10 under a variety of conditions. Thermal unfolding of receptor secondary structure was well described as a single transition between two folded states (Figure 5A), as was thermal unfolding of tertiary structure when CPM was used to monitor cysteine accessibility (Figure 5B). Thermal unfolding monitored by CD spectroscopy was used to evaluate protein stability in the presence of ligand. As with many soluble (38) and membrane-bound proteins (39), the equilibrium thermodynamic analysis of the unfolding of hA2aRHis10 was hampered by irreversibility, due to aggregates formed to some degree with increased temperature. Therefore, it is important to clarify that folding transitions reported in this study are effective midpoints of unfolding, rather than equilibrium unfolding parameters.
The ellipticity was only slightly affected by temperature changes between 20 and 50 indicating that secondary structure is stable within this temperature range (Fig 4A). At higher temperatures the spectra change gradually with increasing temperature (Fig3A), as the minimum at 222 nm broadens and loses its intensity suggesting unfolding of the protein. Our data show that large heat-induced increases in the scattering (corresponding to greater than 300 V) that would be indicative of aggregation of partially unfolded protein take place only above ~ 80-85°C, which is much above calculated Tm values (Table 1).
It is important to note that a simple two-state folding/unfolding transition is characterized by two states only — the native state and the denatured state, with a single cooperative transition. However, here the midpoint of unfolding calculated via CD and CPM fluorescence for hA2aR-His10 did not occur at the same temperature: the secondary structure transition observed by CD occurred at 58.9°C while the tertiary structure transition (CPM) took place at 41.7 °C for unliganded receptors. This unfolding behavior is consistent with the unfolding model proposed by Popot and Engelman for α-helical membrane proteins, whereby α-helices act as stable, independent folding domains, and their interaction defines the tertiary structure which undergoes a separate folding transition (40). Such behavior has been observed previously for the well-studied GPCR rhodopsin, whereby pH-induced denaturation led to a loss in tertiary contacts between the receptor and retinal, yet secondary structure was largely intact as monitored via FTIR (41). Simulation of the thermal unfolding of rhodopsin has suggested that denaturation begins with a loss of hydrogen bonds and disulfide bridges within extracellular loops, which is directly coupled to the orientation of transmembrane helices, although they do not fully unfold (42). An alternate explanation is that a change of permeability or structure of the micelles at increasing temperature allows the CPM probe to enter the transmembrane domains before denaturation, or the probe itself may facilitate protein denaturation. Although dodecyl maltoside has a critical micelle concentration (CMC) of ~0.2 mM at 25°C (43, 44), we are working at ~16x the CMC of pure surfactant at 25°C, and the temperature dependence of micelle formation is expected to be small (~0.25 mM at 50°C) (43). However, we cannot rule out some changes in micelle structure or permeability with increasing temperature. A further consideration is that due to high hydrophobicity, α-helices in membrane proteins may not denature to the same extent as usually seen in soluble proteins (29).
In comparing unfolding of hA2aR-His10 using both conformational probes (tryptophan and CPM fluorescence), we have found that receptors bound to CHA and theophylline demonstrated enhanced stability compared to the unliganded receptor. Here we observe that wild-type hA2aR-His10 was ~3-4°C less stable than the engineered protein hA2aR-T4L-ΔC, which includes a C-terminal truncation and replacement of intracellular loop 3 with T4-lysozyme (4). Interestingly, Stevens and colleagues reported increased thermal stability in the presence of certain antagonists (4), consistent with our observations for theophylline binding to the full-length native receptor; however, they did not observe any changes in stability upon agonist binding. In contrast, we can observe a significant increase in GPCR stability in the presence of the agonist CHA for full-length receptor.
The Extracellular Disulfide Network is Critical to Ligand Binding
No changes were immediately observed within either the hA2aR-His10 secondary structure (Figure 2A) or tertiary structure (as measured through intrinsic Trp fluorescence) when disulfide bonds were reduced with TCEP. However, for reduced hA2aR-His10, the T1/2 calculated from thermal denaturation as observed by CD was 55.9°C, which is approximately 3°C less stable than unliganded hA2aR-His10 (Figure 5A and Table 1).
When hA2aR-His10 was incubated with the reducing agent TCEP, ~40% of the ligand-binding activity was lost (Figure 6). Although at least partial reduction of disulfide bonds in hA2aR-His10 was achieved in this study (Figure 3), it is important to note that the entire network may not have been fully disrupted. Other studies have shown that reducing agents, presumably acting to reduce disulfide bonds within extracellular loops, can diminish activity of hβ2AR (45, 46), the μ-opioid receptor (47), and a human thromboxane receptor (48). Loss of ligand-binding activity as a function of urea follows a similar trend for hA2aR-His10 and reduced hA2aR-His10, which implies that the tertiary structure unfolding pathways are likely analogous (Figure 6). Therefore, these experiments support the conclusion that while hA2aR activity is sensitive to disulfide bonding within extracellular loops, likely due to ligand accessibility within the agonist-binding site, disulfide bonding does not necessarily impart significant tertiary structure stability to the protein.
CONCLUSION
Biophysical studies conducted for full-length, solubilized hA2aR-His10 have shown important receptor behavior under a variety of conditions and have allowed for the identification of critical factors that influence stability. We have seen that while slight changes for hA2aR-His10 tertiary structure are evident, either agonist or antagonist binding significantly increased overall stability as observed in both receptor secondary and tertiary structure folding transitions. In addition to the insight drawn from these studies for hA2aR-His10, the approach developed here may be further applied towards other GPCRs to characterize whether these small changes in conformation upon ligand binding and stabilizing effects are commonly shared among other adenosine receptors, or even more broadly shared between members of the GPCR superfamily.
ACKNOWLEDGMENT
The authors thank Dr. K. Dane Wittrup (MIT) for the pITy plasmid and Dr. Marlene Jacobson (Merck) for the human adenosine A2a receptor gene.
Footnotes
List of abbreviations: N6-cyclohexyladenosine (CHA); 7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin (CPM); G protein-coupled receptor (GPCR); human adenosine A2a receptor (hA2aR); tris(2-carboxyethyl)phosphine (TCEP); dodecyl-β-D-maltoside (DDM), 3-(3-cholamidopropyl)-dimethylammoniopropane sulfonate (CHAPS); cholesteryl hemisuccinate (CHS); hA2aR with a C-terminal decahistidine tag (hA2aR-His10); circular dichroism (CD); critical micelle concentration (CMC)
REFERENCES
- 1.Kobilka B, Schertler GFX. New G-protein-coupled receptor crystal structures: insights and limitations. Trends in Pharmacological Sciences. 2008;29:79–83. doi: 10.1016/j.tips.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Chiu ML, Tsang C, Grihalde N, MacWilliams MP. Over-expression, solubilization, and purification of G protein-coupled receptors for structural biology. Comb. Chem. High Throughput Screen. 2008;11:439–462. doi: 10.2174/138620708784911456. [DOI] [PubMed] [Google Scholar]
- 3.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A(2A) Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–U482. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakola VP, Lane JR, Lin JY, Katritch V, Ijzerman AP, Stevens RC. Ligand Binding and Subtype Selectivity of the Human A(2A) Adenosine Receptor IDENTIFICATION AND CHARACTERIZATION OF ESSENTIAL AMINO ACID RESIDUES. Journal of Biological Chemistry. 2010;285:13032–13044. doi: 10.1074/jbc.M109.096974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, Yeager M, Kufareva I, Stevens RC, Abagyan R. Structure-Based Discovery of Novel Chemotypes for Adenosine A(2A) Receptor Antagonists. Journal of Medicinal Chemistry. 2010;53:1799–1809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butz JA, Niebauer RT, Robinson AS. Co-expression of molecular chaperones does not improve the heterologous expression of mammalian G-protein coupled receptor expression in yeast. Biotechnology and Bioengineering. 2003;84:292–304. doi: 10.1002/bit.10771. [DOI] [PubMed] [Google Scholar]
- 10.Niebauer RT, Robinson AS. Exceptional total and functional yields of the human adenosine (A2a) receptor expressed in the yeast Saccharomyces cerevisiae. Protein Expression and Purification. 2006;46:204–211. doi: 10.1016/j.pep.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 11.Niebauer RT, Wedekind A, Robinson AS. Decreases in yeast expression yields of the human adenosine A2a receptor are a result of translational or post-translational events. Protein Expression and Purification. 2004;37:134–143. doi: 10.1016/j.pep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 12.O'Malley MA, Lazarova T, Britton ZT, Robinson AS. High-level expression in Saccharomyces cerevisiae enables isolation and spectroscopic characterization of functional human adenosine A(2)a receptor. Journal of Structural Biology. 2007;159:166–178. doi: 10.1016/j.jsb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Malley MA, Mancini JD, Young CL, McCusker EC, Raden D, Robinson AS. Progress toward heterologous expression of active G-protein-coupled receptors in Saccharomyces cerevisiae: Linking cellular stress response with translocation and trafficking. Protein Science. 2009;18:2356–2370. doi: 10.1002/pro.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vangalen PJM, Stiles GL, Michaels G, Jacobson KA. Adenosine-a(1) and Adenosine-a(2) Receptors - Structure-Function-Relationships. Medicinal Research Reviews. 1992;12:423–471. doi: 10.1002/med.2610120502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piersen CE, True CD, Wells JN. A carboxyl-terminally truncated mutant and nonglycosylated A2a adenosine receptors retain ligand binding. Mol Pharmacol. 1994;45:861–870. [PubMed] [Google Scholar]
- 16.Price LA, Strnad J, Pausch MH, Hadcock JR. Pharmacological characterization of the rat A2a adenosine receptor functionally coupled to the yeast pheromone response pathway. Mol Pharmacol. 1996;50:829–837. [PubMed] [Google Scholar]
- 17.Muller CE. A(2A) adenosine receptor antagonists - future drugs for Parkinson's disease? Drug Future. 2000;25:1043–1052. [Google Scholar]
- 18.Jarvis MF, Schulz R, Hutchison AJ, Do UH, Sills MA, Williams M. [H-3] Cgs-21680, a Selective A2 Adenosine Receptor Agonist Directly Labels A2-Receptors in Rat-Brain. J. Pharmacol. Exp. Ther. 1989;251:888–893. [PubMed] [Google Scholar]
- 19.Hirs CHW, Timasheff SN. Methods in Enzymology. Vol. 131. Harcourt Brace Jovanovich; Orlando, FL: 1986. [Google Scholar]
- 20.Stoscheck CM. Quantitation of Protein. Methods in Enzymology. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 21.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 23.Fredholm BB, Abbracchio MP, Burnstock G, Daly JW, Harden TK, Jacobson KA, Leff P, Williams M. Nomenclature and Classification of Purinoceptors. Pharmacological Reviews. 1994;46:143–156. [PMC free article] [PubMed] [Google Scholar]
- 24.Cristalli G, Lambertucci C, Marucci G, Volpini R, Dal Ben D. A(2A) adenosine receptor and its modulators: Overview on a druggable GPCR and on structure-activity relationship analysis and binding requirements of agonists and antagonists. Current Pharmaceutical Design. 2008;14:1525–1552. doi: 10.2174/138161208784480081. [DOI] [PubMed] [Google Scholar]
- 25.Hong H, Joh NH, Bowie JU, Tamm LK. Methods in Enzymology: Biothermodynamics. Vol. 455. Elsevier Academic Press Inc; San Diego: 2009. Methods for Measuring the Thermodynamic Stability of Membrane Proteins; pp. 213–236. Part A. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbusch JP. Stability of membrane proteins: Relevance for the selection of appropriate methods for high-resolution structure determinations. Journal of Structural Biology. 2001;136:144–157. doi: 10.1006/jsbi.2001.4431. [DOI] [PubMed] [Google Scholar]
- 27.Stowell MHB, Rees DC. Advances in Protein Chemistry. Vol. 46. Academic Press Inc; San Diego: 1995. Structure and Stability of Membrane-Proteins; pp. 279–311. [DOI] [PubMed] [Google Scholar]
- 28.Haltia T, Freire E. Forces and Factors That Contribute to the Structural Stability of Membrane-Proteins. Biochimica Et Biophysica Acta-Bioenergetics. 1995;1228:1–27. doi: 10.1016/0005-2728(94)00161-w. [DOI] [PubMed] [Google Scholar]
- 29.Booth PJ, Curnow P. Membrane proteins shape up: understanding in vitro folding. Current Opinion in Structural Biology. 2006;16:480–488. doi: 10.1016/j.sbi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Bowie JU. Solving the membrane protein folding problem. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]
- 31.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annual Review of Biophysics. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni S, Ivarsson Y, Bah A, Bush-Pelc LA, Di Cera E. Mechanism of Na+ binding to thrombin resolved by ultra-rapid kinetics. Biophysical Chemistry. 2007;131:111–114. doi: 10.1016/j.bpc.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke J, Fersht AR. Engineered Disulfide Bonds as Probes of the Folding Pathway of Barnase - Increasing the Stability of Proteins against the Rate of Denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 34.Creighton TE, Goldenberg DP. Kinetic Role of a Metastable Native-Like 2-Disulfide Species in the Folding Transition of Bovine Pancreatic Trypsin-Inhibitor. Journal of Molecular Biology. 1984;179:497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- 35.Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. Disulfide bonds and protein folding. Biochemistry. 2000;39:4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- 36.Baneres JL, Martin A, Hullot P, Girard JP, Rossi JC, Parello J. Structure-based analysis of GPCR function: Conformational adaptation of both agonist and receptor upon leukotriene B-4 binding to recombinant BLT1. Journal of Molecular Biology. 2003;329:801–814. doi: 10.1016/s0022-2836(03)00438-8. [DOI] [PubMed] [Google Scholar]
- 37.Blumer KJ, Thorner J. An adrenaline (and gold?) rush for the GPCR community. Acs Chemical Biology. 2007;2:783–786. doi: 10.1021/cb7002412. [DOI] [PubMed] [Google Scholar]
- 38.Duy C, Fitter J. How aggregation and conformational scrambling of unfolded states govern fluorescence emission spectra. Biophysical Journal. 2006;90:3704–3711. doi: 10.1529/biophysj.105.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau FW, Bowie JU. A method for assessing the stability of a membrane protein. Biochemistry. 1997;36:5884–5892. doi: 10.1021/bi963095j. [DOI] [PubMed] [Google Scholar]
- 40.Popot JL, Engelman DM. Membrane-Protein Folding and Oligomerization - the 2-Stage Model. Biochemistry. 1990;29:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 41.Vogel R, Siebert F. Conformation and stability of alpha-helical membrane proteins. 2. Influence of pH and salts on stability and unfolding of rhodopsin. Biochemistry. 2002;41:3536–3545. doi: 10.1021/bi016024f. [DOI] [PubMed] [Google Scholar]
- 42.Rader AJ, Anderson G, Isin B, Khorana HG, Bahar I, Klein-Seetharaman J. Identification of core amino acids stabilizing rhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoudia M, Zana R. Aggregation Behavior of Sugar Surfactants in Aqueous Solutions: Effects of Temperature and the Addition of Nonionic Polymers. J. Colloid Interface Sci. 1998;206:158–167. doi: 10.1006/jcis.1998.5627. [DOI] [PubMed] [Google Scholar]
- 44.Drummond CJ, Warr GG, Grieser F, Ninham BW, Evans DF. Surface-properties and Micellar Interfacial Microenvironment of N-dodecyl-beta-D-Maltoside. Journal of Physical Chemistry. 1985;89:2103–2109. [Google Scholar]
- 45.Pedersen SE, Ross EM. Functional Activation of Beta-Adrenergic Receptors by Thiols in the Presence or Absence of Agonists. Journal of Biological Chemistry. 1985;260:4150–4157. [PubMed] [Google Scholar]
- 46.Prior TI, Patel V, Drummond GI. Inactivation of the Beta-Adrenergic-Receptor in Cardiac-Muscle by Dithiols. Can. J. Physiol. Pharmacol. 1985;63:932–936. doi: 10.1139/y85-154. [DOI] [PubMed] [Google Scholar]
- 47.Zhang PS, Johnson PS, Zollner C, Wang WF, Wang ZJ, Montes AE, Seidleck BK, Blaschak CJ, Surratt CK. Mutation of human mu opioid receptor extracellular “disulfide cysteine” residues alters ligand binding but does not prevent receptor targeting to the cell plasma membrane. Molecular Brain Research. 1999;72:195–204. doi: 10.1016/s0169-328x(99)00241-7. [DOI] [PubMed] [Google Scholar]
- 48.Dangelo DD, Eubank JJ, Davis MG, Dorn GW. Mutagenic analysis of platelet thromboxane receptor cysteines - Roles in ligand binding and receptor-effector coupling. Journal of Biological Chemistry. 1996;271:6233–6240. doi: 10.1074/jbc.271.11.6233. [DOI] [PubMed] [Google Scholar]