Abstract
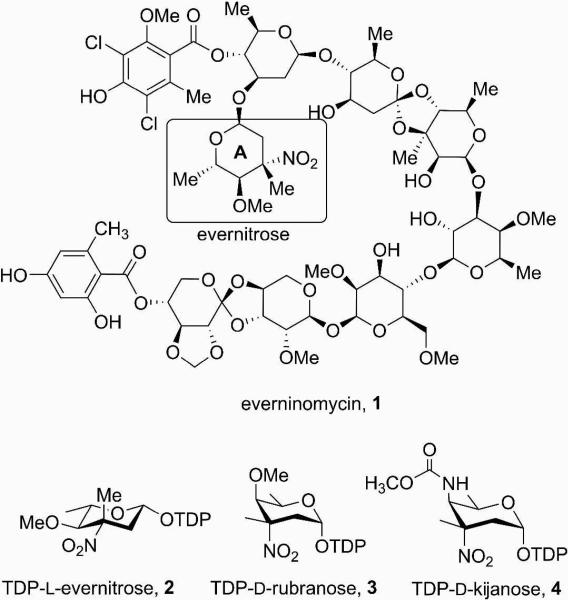
Everninomicin is a highly modified octasaccharide that belongs to the orthosomycin family of antibiotics and possesses potent gram-positive antibiotic activity, including broad-spectrum efficacy against multidrug resistant enterococci and Staphylococcus aureus. Among its distinctive structural features is a nitrosugar, l-evernitrose, analogs of which decorate a variety of natural products. Recently, we identified a nitrososynthase enzyme encoded by orf36 from Micromonospora carbonacea var. africana that mediates the flavin-dependent double oxidation of synthetically-generated thymidine diphosphate (TDP)-l-epi-vancosamine to the corresponding nitroso sugar. Herein, we utilize a five enzyme in vitro pathway both to verify that ORF36 catalyzes oxidation of biogenic TDP-l-epi-vancosamine and to determine whether ORF36 exhibits catalytic competence for any of its biosynthetic progenitors, which are candidate substrates for nitrososynthases in vivo. Progenitors solely undergo single oxidation reactions and terminate in the hydroxylamine oxidation state. Performing the in vitro reactions in the presence of 18O2 establishes that molecular oxygen, rather than oxygen from water, is incorporated into ORF36-generated intermediates and products, and identifies an off-pathway product that correlates with the oxidation product of a progenitor substrate. The 3.15 Å resolution x-ray crystal structure of ORF36 reveals a tetrameric enzyme that shares a fold with acyl-coA dehydrogenases and class D flavin-containing monooxygenases, including the nitrososynthase KijD3. However, ORF36 and KijD3 have unusually open active sites in comparison to these related enzymes. Taken together, these studies map substrate determinants and allow the proposal of a minimal monooxygenase mechanism for amino sugar oxidation by ORF36.
Keywords: Everninomicin, natural product biosynthesis, antibiotic biosynthesis, amine oxidation, nitrososynthase, flavin-containing monooxygenase, acyl-CoA dehydrogenase, induced fit
The orthosomycins are oligosaccharide antibiotics that possess potent broad spectrum antibacterial activity (1-3). These compounds are thought to target protein translation in bacteria by binding to a single site on the 50S ribosomal subunit (4). One such orthosomycin is everninomicin (1, Ziracin®, Figure 1), which harbors an unusual nitrosugar moiety that may be key for its activity (5, 6). Everninomicin was developed through Phase III clinical trials that were eventually discontinued due to apparent pharmacological complications (7). Since it is not uncommon for related antibiotics to exhibit similar efficacy but have altered pharmacological properties, the exploration of compounds related to everninomicin may identify molecules suitable for clinical use. Only limited chemical derivatization studies of everninomicin have been performed (5, 6), likely since the complexity of orthosomycins render the chemical synthesis required for structure-activity and optimization studies challenging. Enzymes in the natural biosynthetic pathway of everninomicin could be used to complement chemical synthesis during rational design of the antibiotic's scaffold.
Figure 1.
Everninomicin, its precursor TDP-l-evernitrose and related, proposed TDP-nitrosugar precursors to rubradirin and kijanimicin.
Using a comparative genomics approach, we recently identified and performed preliminary characterization of an enzyme from the everninomicin producer Micromonospora carbonacea var. africana (8). This enzyme, encoded by orf36 (also called EvdC) catalyzes the 2-step oxidation of the close substrate analog, TDP-l-epi-vancosamine 10, to its nitroso congener 14 (8), and as a result has been referred to as a nitrososynthase. The nitrososythases are an expanding family enzymes that catalyze nitroso sugar formation on oligosaccharide antibiotics, and include RubN8 of the rubradirin biosynthetic pathway (9), whose product is TDP-d-rubranose (3, Figure 1), KijD3 (10, 11) of the kijanimicin biosynthetic pathway, which yields TDP-d-kijanose (4, Figure 1), and isobutylamine-N-oxidase (12) of the valanimicin pathway. Formation of the nitroso sugar by nitrososynthases is consistent with previous studies that suggest that the oxidation to the nitro oxidation state is the result of a spontaneous photochemical reaction (9).
N-oxidation of primary amines has been observed in a variety of biological systems. For example, oxidation of amines to nitroso and nitro groups via a hydroxylamine intermediate can be catalyzed by Rieske N-oxygenases (13) and non-heme diiron monooxygenases (14, 15), while oxidation of substrate nitrogen atoms to the corresponding hydroxylamine or oxide can be accomplished by P450 enzymes (16, 17) and several single-component flavin monooxygenases (18, 19). Based on preliminary biochemical data (8), ORF36 and related nitrososynthases are likely flavin-containing monooxygenases. The flavin monooxygenases are a diverse set of enzymes that catalyze various oxidation reactions, including hydroxylation, halogenation, sulfoxidation, and Baeyer-Villiger oxidations (20). The double oxidation of an amino sugar represents a new addition to this set of reactivities. Sequence and structural similarities divide flavin-containing monooxygenases into subclasses (20), and nitrososynthases may be best classified as Class D flavin-containing monooxygenases, which are related in sequence and structure to the acyl-CoA dehydrogenases (11, 21). The protein scaffold of this superfamily can house distinct reaction types, with acyl-CoA dehydrogenases catalyzing dehydrogenation, and flavin-containing monooxygenases performing monooxygenation (22, 23). Interestingly, ORF36 and other nitrososynthases are more similar in sequence to the dehydrogenases (~25% identity) than to flavin-containing monooxygenases (~15% identity), seemingly at odds with the functional evidence suggesting that ORF36 performs a monooxygenase reaction.
To date, the characterized Class D flavin-containing monooxygenases (24-27) participate in two-component flavin-dependent monooxygenase systems (20, 28, 29) in which a first enzyme, a flavin reductase, reduces oxidized flavin with NAD(P)H, and a second enzyme, the monooxygenase, preferentially binds reduced flavin (24, 26, 29), activates molecular oxygen, and oxygenates the substrate (28). Consistent with the classification as a Class D flavin-containing monooxygenase, previous biochemical characterization of ORF36 confirmed that activity depends upon flavin and NADPH, and is dramatically enhanced by the addition of flavin reductase, but displays relatively weak, reversible binding for oxidized flavin (8). Curiously, these in vitro assays of ORF36 activity suggested that the enzyme exhibits flexible flavin cofactor utilization, with efficient aminosugar oxidation observed using either reduced FAD or FMN as the cofactor (8). Similar promiscuous cofactor usage was observed in the Class D flavin monooxygenase 4-hydroxyphenylacetate monooxygenase from Acinetobacter baumannii (24).
Application of classical flavoenzyme methodologies to our studies of ORF36 was impeded by several factors, including the instability of the putative substrate (TDP-l-evernosamine), the challenges entailed in its synthesis, and the low affinity of the flavin cofactor, which normally provides a convenient readout for monitoring the redox state of flavoenzymes throughout the reaction coordinate. To address these issues, we have implemented a modified procedure to generate the close substrate analog TDP-l-epi-vancosamine (des-4-O-Me evernitrosamine) 10 via an in vitro enzymatic synthetic pathway in yields sufficient to study enzyme turnover and to confirm the activity of this substrate analog. Fortuitously, this synthetic strategy provides access to progenitor substrates, which allows the assessment of enzymatic competence on biological precursors on the presumed substrate. We next performed 18O2-incorporation studies with biogenic TDP-l-epi-vancosamine, which provided evidence for a monooxygenase mechanism, and identified a new activity of ORF36 with the substrate analog TDP-l-epi-vancosamine. Finally, we determined the x-ray crystal structure of ORF36. Along with the previously determined structure of the homolog KijD3 (11), this structure reveals the chemical and physical constraints of the protein scaffold that houses the double oxidation of an amino sugar. To the best of our knowledge, ORF36 is the first structurally characterized enzyme with demonstrated nitrososynthase activity. Moreover, these studies comprise the first detailed structural and biochemical insights into the biosynthesis of the stage III clinical candidate everninomicin and its precursor TDP-evernosamine. The assay of progenitor substrates delineates the most likely timing of N-oxidation in biosynthesis of TDP-evernitrose, and isotopic incorporation studies reveal an additional reaction product and permit the assignment of this new class of flavoenzyme as a monooxygenase.
EXPERIMENTAL PROCEDURES
Overexpression and purification of enzymes
ORF36 from M. carbonacea var. africana (8), EvaA–E from Amycolatopsis orientalis (30), and RfbB from Salmonella enterica (strain LT2) (31, 32) were purified from freshly transformed E. coli BL21(DE3) by nickel affinity chromatography as previously described with the exception of EvaE, which was isolated as an insoluble preparation. EvaE preparations were generated from 3 L induced cultures, which were disrupted via a French pressure cell. The insoluble fraction was isolated by centrifugation and washed with 20 mM Tris-HCl, pH 7.5, resuspended in 10 mL, 20 mM Tris-HCl, pH 7.5, 5% glycerol, and stored at -80 °C prior to use.
Preparation of TDP-6-deoxy-4-keto-d-glucose 6
Reactions (1 mL) containing 8 mM thymidine 5’-diphosphate-glucose disodium salt (4.5 mg), 50 mM Tris-HCl (pH 8.0), 4 mM NADP+, and 160 μM RfbB were incubated at 24 °C for two hours. Proteins were removed by 10K molecular weight cutoff filtration (Centricon, Millipore Corp.) and the product was purified via SAX-HPLC (see below) followed by lyophilization. The final yield was 4.0 mg of TDP-6-deoxy-4-keto-d-glucose 6 (88% yield). Compound 6 was analyzed by ESI-LC MS, which determined the m/z to be 563.20 ([M-H + H2O]-, calculated for C16H25N2O16P2 is 563.07).
Preparation of other TDP-sugars substrates and intermediates
Other TDP-sugars were biochemically synthesized from 6 using purified enzyme preparations and the general procedures outlined by Chen et. al. (30) with the following modifications. Briefly, thimidine-5’-diphosphate-3-amino-2,3,6-trideoxy-3-C-methyl-d-erythrohexopyranos-4-ulose 8 was synthesized via tandem reaction with EvaA, EvaB and EvaC and purified as described below in a 36% overall yield for three steps. ESILS mass spectra yielded the m/z of 560.27 ([M-H + H2O]− calculated for C17H28N3O14P2 is 560.10). Thymidine-5’-diphospho-3-amino-2,3,6-trideoxy-3-C-methyl-l-arabinohexose (TDP-l-epi-vancosamine 10) was generated from 8 (1 mM) in a 1-mL reaction containing 100 mM Tris-HCl (pH 7.5), 5 mM NADPH, 20 μM EvaD and 50 μL of the resuspended EvaE pellet. The reaction was incubated at 24 °C for 2 h, proteins and cell debris were removed using a 10K centrifugal filter (Centricon), and the product was purified by SAX-HPLC (described below) followed by lyophilization which resulted in 300 μg (55% yield) of 10. ESI-LC MS confirmed the mass of 10; [M-H]− calculated for C17H28N3O13P2 is 544.11; found 544.27.
Purification of TDP-sugars
Preparative enzymatic reaction progress was followed via HPLC using a series 600 Waters HPLC system with a Waters 2996 diode array detector. Analytical separations were performed on an Adsorbosphere strong anion exchange (SAX) column (5 mm, 4.6 × 20 mm, Alltech Associates) and a linear gradient of 50 to 500 mM NH4HCO2, pH 3.5, at 1 mL/min for 45 min. Preparative HPLC separations were performed using a semi-preparative Adsorbosphere SAX column (5 μm, 22 × 250 mm) using a similar protocol, but at 10 mL/min. The pH of the fractions containing TDP-chromophores (267 nm) were immediately adjusted to ~7 with 1 M NH4OH and fractions containing reaction product were pooled, lyophilized and stored at -80 °C until assayed. Resuspended compound concentrations were determined by measuring the absorbance at 267 nm using a Nanodrop spectrophotometer (Thermo, Inc.) and comparison to a standard curve of dTDP.
ORF36 enzymatic reactions with 8, 9, and 10
Reactions of ORF36 with 8 and 10 were performed in a total volume of 50 μL containing 30 μM of the corresponding substrates with 30 μM FAD, 1 U/mL catalase, 0.05 U flavin reductase, and 2.0 mM NADPH. Compound 9 was generated in situ by including 20 μM EvaD in the reaction with 8. All reactions were initiated by addition of 12 μM ORF36 and 10 μL samples were withdrawn at increasing time points (0 - 120 min) and were quenched with 10 μL of acetone and stored at -80 °C until analyzed by LC-ESI/MS. Control assays were performed omitting ORF36, FAD, or NADPH.
18O2-incorporation assays
Reactions with 18O2 gas were performed in a 5 mL round bottomed flask fitted with a rubber septum and a balloon (~5 mL, connected via a syringe needle). The reaction vessel at 4 °C was connected via stainless steel cannula to a closed gas cylinder (Cambridge Isotope Laboratories) containing 1 L, 22 psi of 18O2 that was also fitted with a rubber septum. A freshly prepared solution (200 μL) containing 20 mM Tris-HCl, pH 7.5, 30 μM FAD, 1 U/mL catalase, 0.05 U/mL flavin reductase, 2.0 mM NADPH, and 0.5 mg/mL ORF36 was introduced into the flask and the entire reaction system was degassed in vacuo (25 - 50 mm Hg) for 20 minutes. The 18O2 cylinder was opened to release sufficient 18O2 gas to fill the flask and the small balloon with ~5-10 mL of gas. Reactions were initiated by the addition of 30 μM of vacuum degassed TDP-l-epi-vancosamine 10 via a gas-tight syringe. Reactions were followed for 2 h at 30 °C by withdrawing 25 μL aliquots via syringe at increasing time points, quenching via addition of 25 μL acetone and storing samples at -80 °C until analyzed by LC-ESI/MS as described below.
LC-ESI/MS method for ORF36 Substrates
The oxidation of ORF36 substrates was analyzed using Agilent 1100 HPLC system (Agilent, Palo Alto, CA) with a LCQ Quantum Decca XP ion trap mass analyzer (ThermoFinnigan, San Jose, CA) equipped with an API electrospray ionization source fitted with a 50 μm I.D. deactivated fused silica capillary. Injections of 10 μl were separated using a Hypercarb column (3 × 50 mm, Thermo Inc). Mobile phases were: (A) H2O with 50 mM NH4CH3COO and 0.1% (v/v) diethylamine and (B) H2O/acetonitrile (5:95) with 50 mM NH4CH3COO and 0.1% (v/v) diethylamine. Gradient conditions were as follows: 0-5 min, B = 15%; 5-15 min, linear gradient to 35% B; 15 to 16 min, linear gradient to 100% B; 16 to 21 min, B = 100%; 21 to 22 min, linear gradient to 15% B; 22-30 min, B = 15%. The flow rate was maintained at 0.3 ml/min. The mass spectrometer was operated in both the negative ion and full scan profile modes and the electrospray needle was maintained at 3,400 V. The ion transfer tube was operated at -47.50 V and 275°C. The tube lens voltage was set to -46 V. The collision energy for all product ion scans was set at 30%. For TDP-l-epi-vancosamine 10, full product and ion scans were set as follows: 544→155-548; 558→155-560; 560→155-562. For the assays with 8 and 9, the scans were set as follows: 542→155-548; 560→155-562; 558→155-560.
Data were acquired in profile mode. The following optimized parameters were used for the detection: N2 sheath gas 46 psi; N2 auxiliary gas 13 psi; spray voltage 3.4 kV.
Crystallization of ORF36
Initial crystallization conditions for ORF36 were identified by sparse matrix screening and were improved using diffraction-based feedback on the Life Sciences Collaborative Access Team (LS-CAT) beamlines 21-ID-D, 21-ID-F, and 21-ID-G at the Advanced Photon Source (APS), and beamline 9-1 at Stanford Synchrotron Radiation Lightsource (SSRL). Optimized crystals of ORF36 were grown using the hanging drop vapor diffusion technique at 277 K from drops containing 1 μL protein solution (7 mg/mL ORF36 in 20 mM Tris.HCl, 5% glycerol, 1 mM DTT, pH 7.5) and 1 μL crystallization solution (0.1 M Tris-HCl pH 8.5, 0.2 M MgCl2, 10% polyethylene glycol 4000), equilibrated against 1 mL crystallization solution. Crystals belonging to the hexagonal space group P65 (Table 1) reached a maximal size of 0.1 mm × 0.1 mm × 0.4 mm in 5-6 days. The volume of the unit cell was consistent with a tetramer in the asymmetric unit and 53% solvent content. Crystals were cryoprotected by equilibration in crystallization solution supplemented with 20% glycerol or 20% ethylene glycol and cryocooled in liquid nitrogen prior to data collection.
Table 1.
Crystallographic Data Collection Statistics
| Dataset | Native 1 | Native 2 |
|---|---|---|
| Resolution (Å)a | 50.0-3.60 (3.73-3.60) | 50-3.15 (3.26-3.15) |
| Space group | P65 | P65 |
| Unit cell dimensions (Ǻ) | a=b=104.01 c=294.36 | a=b=103.81 c=295.95 |
| Rsymb | 6.3 (43.8) | 11.6 (47.4) |
| Completeness (%) | 99.8 (100.0) | 99.6 (99.5) |
| Imean/σ | 29.2 (5.4) | 8.93 (2.50) |
| # total reflections | 159943 | 127962 |
| # unique reflections | 20697 (2046) | 31158 (3113) |
| Redundancy | 7.7 (7.7) | 4.1 (3.5) |
| wavelength (Å) | 1.12713 | 1.20000 |
| Beamline | APS 21-ID-D | APS 21-ID-D |
Values in parenthesis refer to the high-resolution bin.
Rsym = Σhkl Σi |Ii(hkl) - I(hkl)| (100) / ΣhklΣi Ii(hkl), where Ii(hkl) and I(hkl) are the ith and mean measurements of the intensity of reflection hkl.
Native data set 1 was collected at 100 K at the beamline 21-ID-D of the Advanced Photon Source using a Rayonix MX 225 detector. Native data set 2 was collected at 100 K on beamline 9-2 at SSRL using a MarUSA MarMosaic-325 CCD detector. Data integration and scaling were performed with the HKL2000 suite of programs (Table 1) (33).
Structure determination and refinement
The structure of ORF36 was determined by molecular replacement in Phaser (34) using a polyalanine version of the human short-branched chain acyl-CoA dehydrogenase tetramer as the search model (PDB entry 2JIF (35), 25% sequence identity). Molecular replacement was only successful with the Native data set 1. The molecular replacement solution was transferred into the higher-resolution Native data set 2 (Table 1) and refined using alternating rounds of manual model building in XtalView (36) and refinement in CNS (37) with strict non-crystallographic symmetry (NCS) constraints. Once the Rcryst reached 26%, each chain was examined for main chain differences, which were observed within the central β domain between the ORF36 monomers. As a result, during the final four cycles of refinement, each monomer was refined with NCS restraints applied to the N- and C-terminal domains, but deviations from NCS were allowed in the central β domain.
The final model of ORF36 comprises residues 1 to 395 (of 412 total residues), with several breaks observed in chains B, C, and D. In chain B, electron density for residues 143-151, 177-184, and 229-235 is not observed; in chain C, electron density for residues 144-156, 176-182, 218-221 and 235 is not observed; in chain D electron density for residues 226-227 and 316 is not observed. The test set of reflections for the Rfree consisted of 9.1% of the data, totaling 2816 reflections. The final model has an Rcryst of 24.2%, an Rfree of 28.1%, 88.5% of residues in the most favored regions of the Ramachandran diagram, 10.9% in the additionally allowed regions, 0.6% in the generously allowed regions, and 0.0% in the disallowed regions (Table 2). Figures 5 and 8 and Supplementary Figures S2, S6, and S7 were prepared with PyMOL (38). Figure 7 was prepared using MOLSCRIPT (39) and Raster3D (40).
Table 2.
Crystallographic refinement statistics
| Dataset | Native 2 |
|---|---|
| Resolution Limits (Å)a | 50 – 3.15 (3.26 – 3.15) |
| Rcrystb | 0.242 |
| Rfreeb | 0.281 |
| Bond length deviation (Å) | 0.009 |
| Bond angle deviation (°) | 1.3 |
Values in parenthesis refer to the high-resolution bin.
Rcryst and Rfree = Σhkl||Fo| - k|Fc||/Σhkl|Fo| where Fo and Fc are the observed and calculated structure factors for reflection hkl and k is a weighting factor. Rfree is calculated on only test reflections.
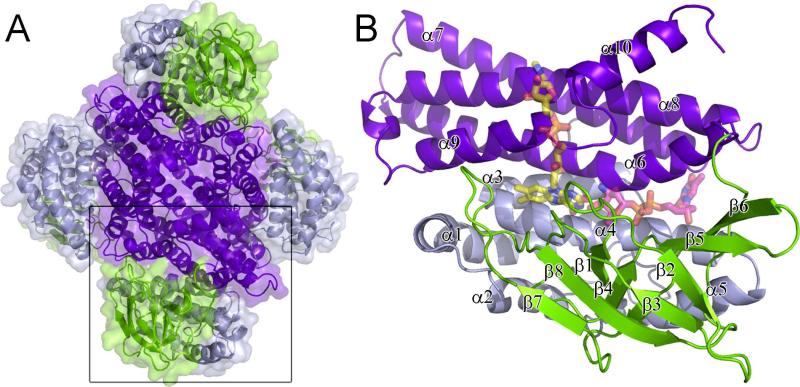
Figure 5.
Structure of ORF36. (A) The ORF36 tetramer is shown as a cartoon representation with a transparent surface, colored by domain. The N-terminal domains (residues 1-129) are colored grey, the central β-sheet domains (residues 130-235) green and the C-terminal domains (residues 235-398) purple. (B) The ORF36 monomer shown in cartoon representation with modeled FAD and TDP-l-evernosamine shown in transparent sticks. Colors are as follows: FAD carbons, yellow; TDP-l-evernosamine carbons, magenta; oxygen, red; nitrogen, blue; phosphate, orange. The view in panel (B) is rotated 45° about a vertical axis from the bottom monomer shown boxed in A.
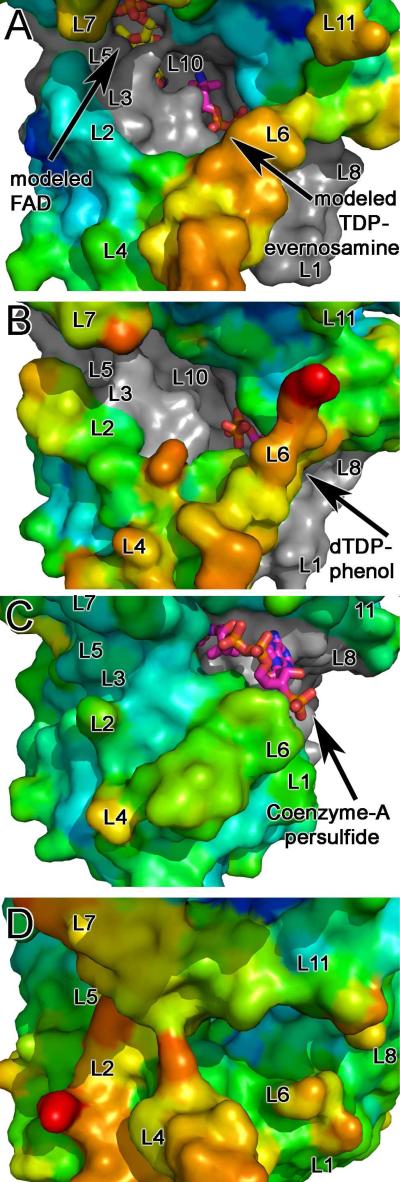
Figure 8.
Surface representation of the active site clefts of nitrososynthases, acyl-CoA dehydrogenases, and flavin-containing monooxygenases are shown colored by B-factor. Active site loops are labeled L1-L11. Surfaces of each enzyme are shown colored using a rainbow color ramp in which red corresponds to the highest B-factor and blue corresponds to the low B-factor. Areas of the surface that form the base of the active site cleft are shown in grey. Substrates and co-factors are shown as sticks with atoms colored as follows: FAD: carbons, yellow, oxygen, red; nitrogen, blue; phosphate, orange; TDP-l-evernosamine, dTDP, CoenzymeA persulfide: carbons, magenta; oxygen, red; nitrogen, blue; phosphate, orange. All panels are shown in an orientation rotated by 45° about a horizontal axis with respect to Figure 3A. (A) ORF36. Modeled FAD and TDP-l-evernosamine are displayed as sticks. In this orientation, loop L9 is concealed. The B factor color ramp gradient minimum and maximum values are 70 and 180 Å2, respectively, and for this structure, the Wilson B is 111.2 Å2 with an average protein B factor of 115.3 Å2. (B) KijD3 (PDB entry 3M9V)(11) with the dTDP of dTDP-phenol displayed as sticks. As in (A), loop L9 is concealed. The B factor color ramp minimum and maximum values for this panel are 10 and 80 Å2, and the average protein B value is 27.6 Å2. The Wilson B factor was not reported. (C) Human short/branched-chain acyl-CoA dehydrogenase (PDB entry 2JIF)(35). The FAD molecule is concealed by the protein surface in this orientation, and coenzymeA persulfide is shown in stick representation. Loops L9 and 10 are concealed in this orientation. The B factor color ramp gradient has minimum and maximum values of 14 and 60 Å2, the Wilson B is 25.84 Å2 and the average protein B factor is 22.76 Å2. (D) A. baumannii 4-hydroxyphenylacetate monooxygenase (PDB entry 2JBR)(24). FMN, 4-hydroxyphenylacetate, and loops L3, L9 and L10 are concealed in this orientation. The B factor color ramp gradient minimum and maximum values are 40 and 90 Å2, and the average protein B value is 56.77 Å2. The Wilson B factor was not reported.
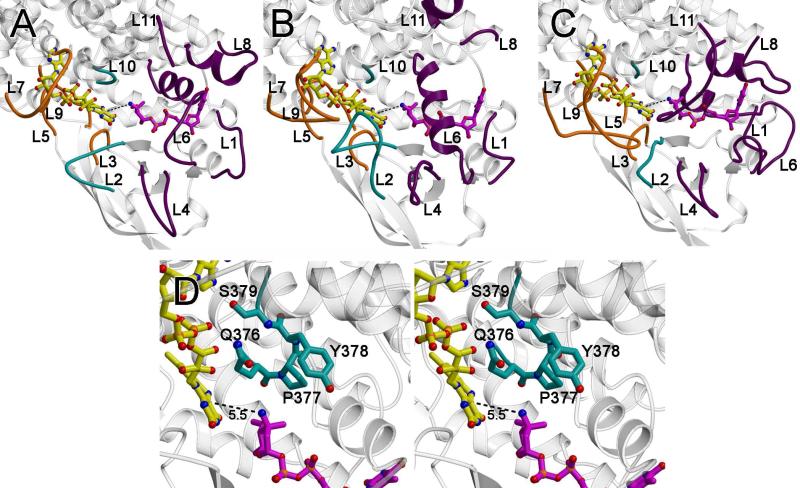
Figure 7.
Active site loops. The loops that form the active site are as follows: the loops between α3 and α4 (loop L1; residues 106 – 109), β1 and β2 (loop L2; residues 134-142), β3 and β4 (loop L3, residues 158-164), β4 and β5 (loop L4; residues 178-180), β6 and β7 (loop L5; residues 201-213), β9 and α6 (loop L6; residues 248-253), α6’ and α7’ (where the ’ indicates this is from an adjacent monomer; loop L7; residues 272-280), α7 and α8 (loop L8; residues 310-317), α8’ and α9’ (loop L9, residues 351-353), α9 and α10, (loop L10; residues 375-379) and at the C-terminus (loop L11; residues 390-412). (A) A ribbon diagram of ORF36 shown with modeled flavin (yellow carbons) and TDP-l-evernosamine (magenta carbons) as sticks. Active site loops L3, L5, L7, and L9, are predicted to interact with flavin and are colored orange, active site loops L1, L4, L6, L8, and L11 are predicted to interact with substrate and are colored purple, and active site loops L2 and L10 are predicted to interact with both substrate and cofactor and are colored teal. (B) A ribbon diagram of human short branched-chain acyl Co-A dehydrogenase (PDB entry 2JIF) (35), highlighting loops L1 to L11 colored are colored as in (A). (C) A ribbon diagram of A. baumannii 4-hydroxyphenylacetate monooxygenase (PDB entry 2JBT) (24) highlighting loops L1 to L11 colored as in (A). (D) Stereoview of the ORF36 loop L10 containing a tandem cis-peptide. Loop L10 is shown in sticks with teal carbons. The Q376-P377 and P377-Y378 bonds both adopt a cis conformation. Modeled flavin and substrate are displayed as in (A).
Modeling of substrate and cofactor
FAD was modeled into the active site by superpositioning the ORF36 structure with that of human short-branched chain acyl-CoA dehydrogenase (PDB entry 2JIF(35)) and using the position of bound FAD from 2JIF as a starting point. Attempts to improve the position of FAD using the molecular docking program Molecular Operating Environment (MOE, Chemical Computing Group, Montreal, Canada) did not result in a solution that both alleviated steric clashes between the FAD and protein and retained a reasonable position for the isoalloxazine ring.
A model for TDP-l-evernosamine was constructed by manual combination of deoxythymidine diphosphate (ligand name TYD) and the epi-vancosaminyl derivative of vancomycin (ligand name VAX), both available via the HIC-Up server (41). The TDP-l-evernosamine was manually positioned within the active site such that the amino sugar was within 5.5 Å of the expected position of the C4a of the isoalloxazine ring, but the location of the TDP varied. The docking algorithm in MOE was used to improve each manual starting point. Docking was performed using a radius of 10 Å around the initial position of the ligand, the triangle matcher algorithm for placement, London dG rescoring, and force field refinement. Between 30 and 100 positions were retained from each trial and validated manually by evaluating the orientation of the sugar moiety in the active site, and the distance of the amino group from the expected position of C4a of the isoalloxazine ring. The optimized position for TDP-l-evernosamine had a location of the TDP similar to that experimentally observed to that of dTDP-phenol crystallized in complex with the nitrososynthase KijD3 (11).
Phylogenetic analysis
The evolutionary history was inferred using the Neighbor-Joining method (42). The bootstrap consensus tree inferred from 1000 replicates (43) is taken to represent the evolutionary history of the taxa analyzed (43). The evolutionary distances were computed using the Poisson correction method (44) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There was a total of 333 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (45).
RESULTS
Substrate Determinants of Activity
NCBI BLAST analysis of sequenced microbial genomes contained in GenBank (46) reveals an ever-increasing number of nitrososynthase homologs, which corresponds with the observation that nitrosugars decorate a wide variety of known bioactive natural products with distinct scaffolds. The hypothetical TDP-sugar precursors of the known nitrosugar-containing natural products (Figure 1) possess conserved structural features, with notable functional variability evident at the C-4 position (found as a hydroxyl, O-methyl or N-carbamoyl group) and C-5 position (which can posses either the d or l- configuration). This structural diversity in the substrates is in striking contrast with the 60-65% sequence identity between proposed nitrososynthases (11), prompting us to investigate the possibility that the reported nitrososynthases act on a common intermediate biochemically upstream of their amino congeners in the corresponding pathways (47).
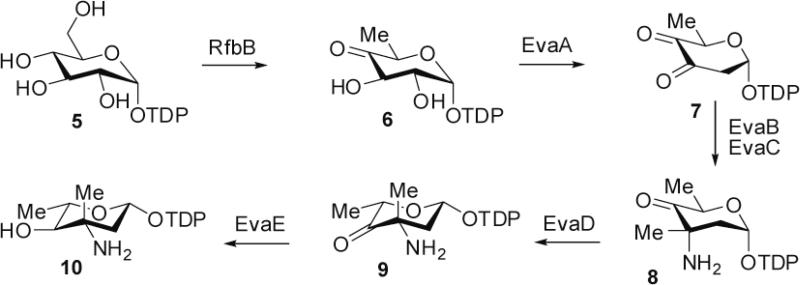
To address this possibility and to further delineate the timing of amine oxidation in broader nitrosugar biosynthesis, we reconstituted the biosynthetic pathway for TDP-l-epi-vancosamine 10 using enzymes obtained by the Walsh group for the chloroereomycin pathway (Figure 2) (30), and assayed for the biochemical competence of amino-functional ketosugar precursors 8 and 9 (Figure 3). These TDP-sugars were enzymatically biosynthesized and purified using ion exchange chromatography. While we were unable to isolate sufficient quantities of the substrate to perform Michaelis-Menten kinetics (and the inherent storage instability of these glycosyl-1-phosphates likely precludes their accurate quantitation), we were able to compare turnover of substrates at identical, titrated substrate concentrations (30 μM).
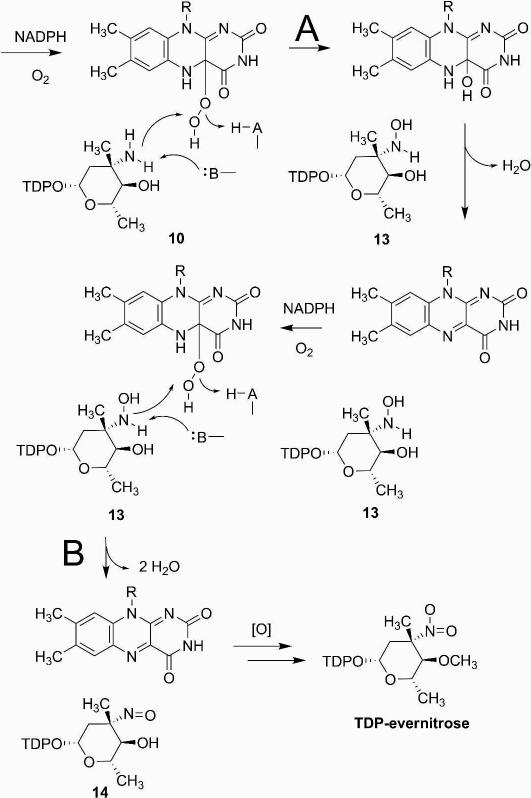
Figure 2.
Pathway utilized for biochemical synthesis of TDP-l-epi-vancosamine 10 and keto amine precursors 8 and 9.
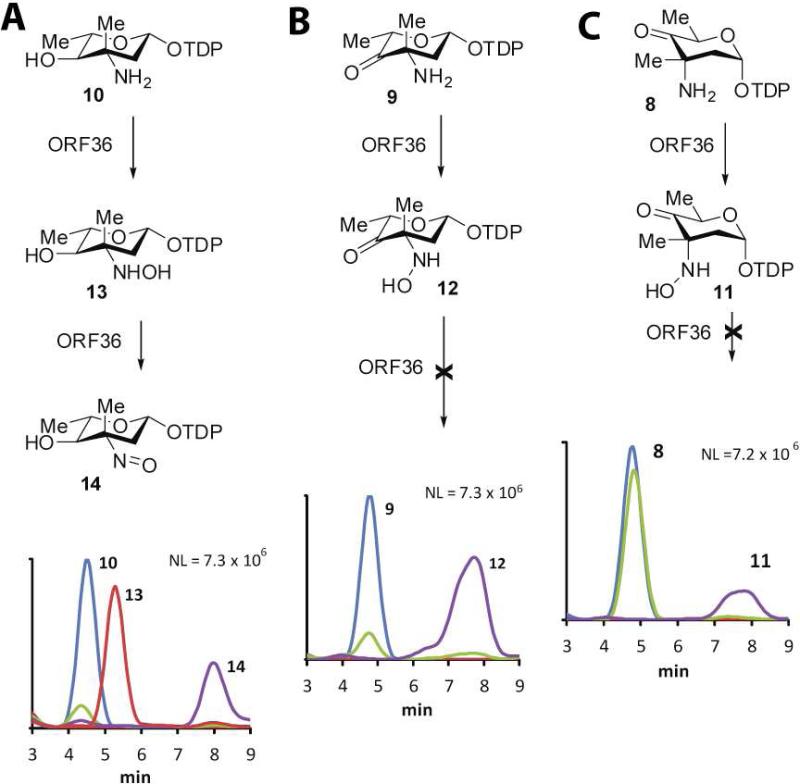
Figure 3.
ORF36 catalysis with potential substrates. HPLC/ESI-MS traces of ORF36 reactions with three potential substrates. (A) TDP-l-epi-vancosamine 10 (m/z = 544), 0 min (blue), 100 min (green); hydroxylamine intermediate 13 (m/z = 560), 15 min (red); nitroso product 14 (m/z = 558), 100 min (purple). (B) keto amine 9 (m/z = 542), t = 0 min (blue), 100 min (green); hydroxylamine intermediate 12 (m/z = 558), 100 min (purple). (C) keto amine 8 (m/z = 542), 0 min (blue), 100 min (green); hydroxylamine intermediate 11 (m/z = 558), 100 min (purple).
As analyzed by ESI-LC MS, all potential substrates displayed a degree of chemical competence for single oxidation to hydroxylamine (Figure 3), but only TDP-l-epi-vancosamine 10 was competent for the double oxidation that results in the nitroso congener 14 (Figure 3). The precursor of TDP-l-epi-vancosamine 10, ketosugar 9, was assayed by including a catalytic excess of the C-5 epimerase (EvaD) in the nitrososynthase oxidation reaction. Compound 9 was rapidly oxidized to the hydroxylamino congener 12 but did not further oxidize to other species under these conditions. The penultimate precursor to TDP-l-epi-vancosamine 10 is the C-5 epimeric ketosugar 8, which was also oxidized to the hydroxylamino oxidation state 11, but at a substantially slower apparent rate than when the epimerase was added to reactions. This is in accord with the recently reported partial oxidation of 8 by KijD3 (11), which likewise only evidenced the formation of hydroxylamine. Unlike these studies, however, we did not observe an additional peak at m/z = 527. While the formation of m/z = 527 was rationalized by the authors as a decomposition product of 11 (proposed as dTDP-2,3,6-trideoxy-4-keto-3-methyl-d-glucose), this structure corresponds to that of a reduction product of the nitroso sugar, not a fragment or likely decomposition product, and the identity of this compound remains undetermined. In comparison, ORF36 assays with compounds 8 and 9 produced species corresponding only to the hydroxylamino compounds 11 and 12 within the expected mass ranges, and no other detectable TDP-sugar decomposition products were apparent by MS or MS/MS analysis. The oxidation of biogenic TDP-l-epi-vancosamine 10 confirms our previous results using the chemically synthesized compound (8) and, importantly, also validates the successful biocatalytic generation of its biosynthetic progenitors 8 and 9. Taken together, these data indicate that catalytic competence of ORF36 improves in substrates more closely related to TDP-l-epi-vancosamine 10, and that an abortive reaction occurs with the biochemical progenitors. While the effect of C-4 O-methylation remains to be determined, the observation of both O-methyl and hydroxyl functionality in everninomicin congeners isolated from M. carbonacea var. africana suggests that O-methyl substitution at this position is not essential for nitrososynthase reactivity and may occur prior or subsequently to amine oxidation reactions and/or glycosylation (3).
18O2 incorporation experiments
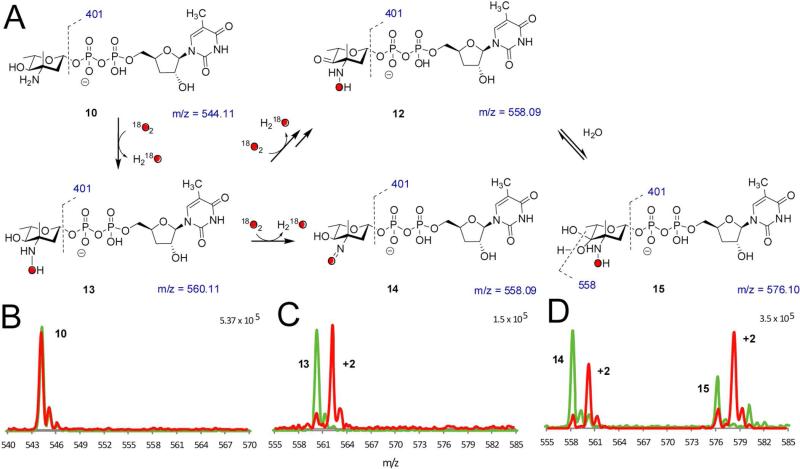
We next performed the reactions under an 18O2 atmosphere to determine whether molecular oxygen, as opposed to oxygen from H2O, was incorporated into the product. Reactions containing NADPH, FAD, flavin reductase, and ORF36 were incubated with a catalytic excess of catalase under an atmosphere of either 18O2 or 16O2 (Figure 4). We reproducibly observed a high level of incorporation of 18O into the hydroxylamine 13 (m/z = 560) and nitroso congeners 14 (m/z = 558), which demonstrated enrichment of the +2 m/z shift at 86% and 87% incorporation, respectively.
Figure 4.
18O2 incorporation studies. (A) Summary of proposed species observed in 18O2 incorporation experiments. (B – D) 18O2 incorporation with ORF36 and TDP-l-epi-vancosamine 10 incubated with 16O2 (green) and 18O2 (red). Masses shown under structures correspond to unlabeled species. HPLC/MS and MS/MS data were collected at a 15 minutes reaction time, when the hydroxylamine intermediate was most abundant. (B) TDP-l-epi-vancosamine 10 (tR = 4.5 min). (C) hydroxylamine 13 (m/z = 560, tR = 5.3 min). (D) Peaks at tR = 7 – 9 min are proposed to correspond to the nitroso compound 14 (tR = 8.8 min) and an additional oxidation product with m/z = 576 (tR = 7.8 min). This new mass has a distinct retention time from the nitrososugar 14 and, based on MS/MS analysis, the additional mass in the m/z = 576 ion is constrained to the pyranose ring. One possible structure is the 4-keto-3-hydroxylamino sugar 12, which is detected as a hydrate 15 under ESI conditions. MS/MS analysis supports this hypothesis in that the labeled ion m/z = 578 readily fragments to m/z = 560.09 and this fragmentation pattern is identical to the reaction product of 9 shifted by 2 mass units.
As well as confirming the incorporation of molecular oxygen into the expected products, these experiments identified a new reaction product. In addition to the labeling of the nitroso sugar 14, we also observed a +2 m/z shift in what we have previously assigned to be the electrospray induced hydrate of the nitroso moiety (14 + H2O m/z = 576 (8)). However, the single incorporation of oxygen into this species and the lack of demonstrable back-exchange of labeled nitroso 14 with H2O rule out the proposed dihydroxylamino species. Moreover, we discovered that subsequent to isolation of 14 by ion exchange HPLC and re-analysis, the nitroso group did not form the hydrate under our standard HPLC/MS conditions (data not shown). Indeed, closer examination of the chromatograms indicated that within the broad peak encompassing this region, the nitrososugar 14, and the m/z = 576 species possess different retention times (8.8 and 7.8 min respectively). Both species contain fragmentation peaks diagnostic of a TDP-sugar including the loss of TDP (m/z = 401) and TMP (m/z = 321) but the unknown m/z 576 compound demonstrated an additional major fragment at 558, indicating loss of water. We have thus far been unable to isolate the m/z 576 species from reactions generating the nitroso sugar for further characterization. However, based on MS/MS analysis, which demonstrated that oxidation is constrained to the pyranose ring, and considering the likelihood that oxidation is occurring on the C-3/4 heteroatoms, we propose that the m/z = 576 peak corresponds to a product oxidized at C-4, likely the C-4 ketone 12. Notably, this putative structure is advantageously identical to 12 generated as a reaction product of substrate analog 9 (Figure 3), which possesses an identical fragmentation pattern and retention time to the m/z = 576 peak. It should also be noted, however, that other structures are possible, including a possible electrocyclic rearrangement of the α−hydroxyketone (not shown) (48).
X-ray crystal structure of ORF36
The structure of ORF36 (Figure 5) was determined to 3.15 Å resolution by molecular replacement using a polyalanine model of human short-branched chain acyl-CoA dehydrogenase (PDB entry 2JIF) (35) as the search model. ORF36 forms a tetramer (Figure 5A) both in solution (Figure S1 of the Supporting Information) and in the crystal with 3,500 Å2 of surface area buried per monomer. Each ORF36 monomer adopts a three-domain α/β/α fold (Figure 5B) with the N- and C-terminal α−helical domains separated by a β-sheet domain. Interactions between the adjacent C-terminal helical domains predominates the contacts mediating tetramerization.
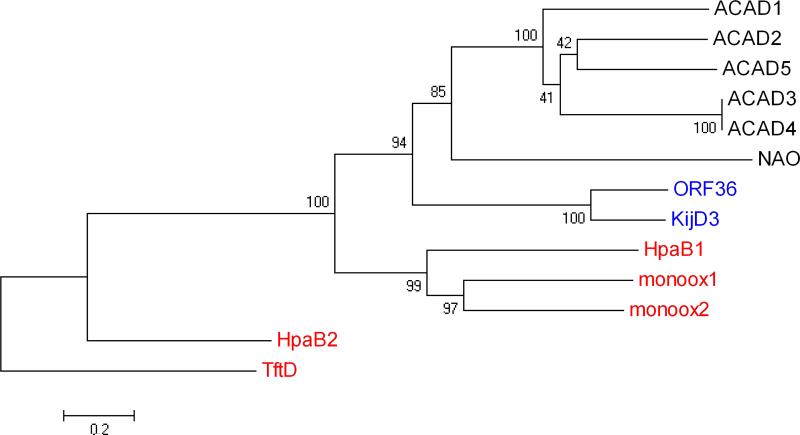
A search for structural homologs of ORF36 using the EMBL Dali server (49) identified the recently-released nitrososynthase KijD3 (11) as the most similar structural homolog. This pair of nitrososynthases shares structural similarity with members of the acyl-CoA dehydrogenase superfamily of proteins (21), which contains dehydrogenases, oxidases and flavin-containing monooxygenases (Supporting Information Table S1). Intriguingly, within this superfamily, ORF36 and KijD3 are markedly more similar in both sequence and structure to the dehydrogenases than to the nitrososynthase functional homologs, the monooxygenases. From this, we hypothesize that nitrososynthases independently evolved monooxygenase activity from acyl-CoA dehydrogenases, while most other Class D flavin monooxygenases and acyl-CoA dehydrogenases underwent a separate evolutionary divergence. This proposal is corroborated by phylogenic analysis of these structural relatives (Figure 6).
Figure 6.
Evolutionary relationships of 13 taxa including nitrososynthases, acyl-CoA dehydrogenases, and flavin-containing monooxygenases. The evolutionary history was inferred using the Neighbor-Joining method (42). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed (43). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (43). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (44) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). There were a total of 333 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (45). The nitrososynthases ORF36 and KijD3 are highlighted in blue text, the Class D flavin-containing monooxygenases in red, and the acyl-CoA dehydrogenases and nitroalkane oxidase are in black text. Proteins included here (which are also included in Supporting Information, Table S1) are named as follows: ACAD1, short branched-chain acyl-CoA dehydrogenase (H. sapiens), 2JIF (35); ACAD2, butyryl-CoA dehydrogenase (M. elsedenii), PDB entry 1BUC (58); ACAD3 medium-chain acyl-CoA dehydrogenase (S. scrofa), PDB entry 3MDE (59); ACAD4, medium-chain acyl-CoA dehydrogenase (S. scrofa), PDB entry 1UDY (60); ACAD5, short-chain acyl-CoA dehydrogenase (R. norvegicus), PDB entry 1JQI (61); HpaB1, 4-hydroxyphenylacetate monooxygenase (A. baumannii), PDB entry 2JBT (24); monoox1, 3-hydroxy-9,10-seco-nandrost-1,3,5(10)-triene-9,17-dione hydroxylase (Rhodococcus sp Rha1), PDB entry 2RFQ (62); monoox2, putative hydroxylase (Rhodococcus sp Rha1), PDB entry 2OR0 (63); NAO, nitroalkane oxidase (F. oxysporum), PDB entry 2C0U (64); HpaB2, 4-hydroxyphenylacetate monooxygenase (T. thermophilus), PDB entry 2YYI (25); TftD, chlorophenol-4-monooxygenase (B. cepacia), PDB entry 3HWC (26).
Cofactor and substrate modeling
Extensive co-crystallization and crystal soaking experiments of ORF36 with various concentrations and combinations of FAD, FMN, the isolated isoalloxazine riboflavin (each in oxidized and reduced states), and the substrate TDP-l-epi-vancosamine were performed. None of these resulted in the appearance of new electron density for cofactor or substrate within the active site. Instead, FAD and TDP-l-evernosamine were modeled into the active site using a combination of manual and automated methods (Figures 7A and Figures S2 and S3 of the Supporting Information). Information from an examination of the co-structures of flavin-containing monooxygenases in complex with their substrates was used to identify ideal cofactor-substrate distances and geometry (24, 50-52).
Comparison of ORF36 to acyl-CoA dehydrogenses and flavin-containing monooxygenases allowed us to identify pre-formed pockets in the active site of ORF36 that likely bind the isoalloxazine ring of the flavin and the TDP of TDP-l-evernosamine. Residues near the predicted isoalloxazine binding pocket are highly conserved in nitrososynthases and are similar to flavin-containing monooxygenases and acyl-CoA dehydrogenases (Figures S4-6, Supporting Information), particularly at loops L3, L5 and L9. In contrast, the predicted binding location for the adenine nucleotide of the flavin is in a region of the active site where the loops are splayed open. In both acyl-CoA dehydrogenases and flavin-containing monooxygenases, loop L7 (Figure 7A) forms specific contacts between flavin and protein, is shorter in enzymes that preferentially utilize FAD and longer in enzymes that preferentially use FMN, and can undergo modest conformational adjustment following flavin binding to optimize contacts with the nucleotide. Despite the in vitro ability of ORF36 to use either FMN or FAD as a cofactor, loop L7 resembles loops from acyl-CoA dehydrogenase superfamily members that use FAD. In this structure, it adopts a conformation that suggests an adjustment of the main chain will accompany flavin binding.
Active site
Residues contributing to the presumed active site emanate predominantly from the N-terminal helical domain and the β-sheet domain, but include residues from the C-terminal domain of an adjacent monomer (Figure S2, Supporting Information) (21). The active site is built along the length of the α4 helix, which forms the base of the active site cleft. Side chains from α4, β1, α6 and α8 likely contribute to flavin binding, while side chains from the α4, α6, α7, and α8 helices could contribute to substrate binding (Figure S2B). Importantly, the loops between the secondary structural elements surrounding the active site (loops L1-L11; Figure 7A-D, Figure S3 of the Supporting Information) likely play an important role in substrate binding and catalysis. In striking contrast to the main scaffold of the protein, loops L1, L2, L4, L5, L6, L7, L8 and L11 substantially differ in structure amongst related enzymes (Figure 7A,B,C, Figure S3 and Supplementary Movie 1 of the Supporting Information), suggesting that they may be important for conferring substrate and cofactor specificity.
In both ORF36 and the nitrososynthase KijD3, the active site measures 30 Å wide by 20 Å deep and appears to be solvent exposed as compared to acyl-CoA dehydrogenases and flavin-containing monooxygenases (Figure 8). The unusually expansive active site portico physically originates from the conformations of active site loops L1, L2, L4, L6, L7, and L11 (Figure 7A), which appear splayed in ORF36 and KijD3 as compared to the acyl-CoA dehydrogenases and flavin-containing monooxygenases (Figure 7 A,B,C). With the exception of L7, these loops are located in the region of the active site known to bind substrate in the acyl-CoA dehydrogenase superfamily, suggesting that significant main chain adjustments could accompany substrate binding. Of particular note is loop L10 in ORF36, which is located on the floor of the active site cleft. This tight turn contains tandem cis-peptide bonds between Gln 376 and Pro 377 and between Pro377 and Tyr 378 (Figures 7D and S7 of the Supporting Information). While it is somewhat speculative to assign a cis-peptide bond at the resolution of this structure, modeling the loop with trans-peptide bonds or with only a single cis-proline resulted in disallowed Ramachandran angles for Tyr 378 and poor agreement of the main chain with the electron density. Corroborating our interpretation, the sequences of KijD3 and ORF36 are identical in loop L10 (Figure S5, Supporting Information), and the structure of KijD3 also contains a tandem cis-peptide bond in this location (11).
In both the ORF36 and KijD3, the temperature factors of the residues of the loops surrounding the active site are significantly elevated as compared to the temperature factors in the remainder of the protein (Figure 8). Crystallographic temperature factors, also known as atomic displacement parameters, can reflect the thermal motion of atoms and hint at regions of increased relative mobility in a given structure.
DISCUSSION
Substrate selectivity of ORF36
Data presented in this work verify that TDP-l-epi-vancosamine 10 is a substrate for the nitrososynthase ORF36. This was an expected outcome, as the A-ring des-4-O-Me congener of everninomicin is a naturally occurring variant in M. carbonacea var. africana (5). That the biosynthetic progenitors of TDP-l-epi-vancosamine 10 were not fully oxidized upon incubation with ORF36 suggests that related enzymes in kijanimicin and rubradirin biosynthesis act not on a common precursor, but on substrates at least closely related, if not identical, to the amino congeners of the final nitrosugars in these molecules. While other possibilities are conceivable, the apparent abortive oxidation of the ketoamine substrate analog 9 also serendipitously aided in the potential deconvolution of a major shunt reaction product, the C-4 ketone 12, in which the 4-hydroxy position of TDP-l-epi-vancosamine is oxidized, perhaps via dehydrogenation or hydroperoxylation of the C-4 hydroxyl to the ketone oxidation state. While we have yet to be able to generate the authentic substrate TDP-l-evernosamine, we expect that reactions of this substrate will reveal that blocking of the C-4 hydroxyl group at its methyl ether will protect this sugar from spurious oxidation. The potential C-4 oxidation reaction is unprecedented and is interesting in the context of acyl-CoA dehydrogenase homologs found associated with other secondary metabolic gene clusters in actinomycetes that apparently do not contain amino sugar biosynthetic genes, expanding the potential scope of this enzyme to other oxidation reactions.
Structural comparison of ORF36 and KijD3
ORF36 has 65% identity to KijD3, the nitrososynthase homolog from the kijanimicin biosynthetic pathway. As expected, their overall structures are similar, with an RMS deviation of 0.76 Å between the Cα atoms of ORF36 and KijD3 tetramers. The greatest differences in both sequence and structure between these related enzymes are located within the regions that map to the active site cleft, and may be related to differences in function and substrate binding. For example, while residues lining the predicted isoalloxazine and TDP binding sites are almost completely conserved between ORF36 and KijD3 (Figure S8 of the Supporting Information), the interface of those two binding sites, where the nitrosugar is presumed to bind, has higher sequence variation and may reflect differences in preferred substrate. The main structural differences between ORF36 and KijD3 occur in active site loops L2, L4 and L6 located within the central β-sheet domain, and L7 and L8 of the C-terminal domain. These regions also display increased crystallographic temperature factors in both structures, which may reflect flexibility within these active site loops.
The active site
Examination of the ORF36 and KijD3 (11) chemical reactions and active sites identify common features that suggest large conformational adjustments could accompany productive cofactor and substrate binding (Figures 7 and 8). Both nitrososynthase active sites are wide and highly solvent-accessible, surrounded by loops that adopt splayed conformations with elevated temperature factors as compared to the remainder of the protein. In addition, both enzymes contain a tandem cis-peptide bond within active site loop L10 (Figure 7D, Supporting Information Figure S7), which could undergo isomerization to initiate a conformational change. Finally, the chemistry catalyzed by both these nitrososynthases presumably requires exclusion of solvent from the active site; in the acyl-coA dehydrogenases and class D flavin-containing monooxygenases this is achieved by the conformations of loops L2, L4, L6, L7, L8 and L11 (Figures 7A-C and 8). Taken together, these observations suggest that it is reasonable, while still somewhat speculative, to expect a rearrangement of the active site loops following cofactor and substrate binding.
Many enzymes undergo conformational adjustments upon substrate binding that influence catalysis (53, 54), suggesting that the open active site architecture could be physiologically relevant in both ORF36 and KijD3 (11). One possible role for the open loop positions may be to allow entry of large ligands to the active site. Another prospect is that the active site loops in these structures may adopt a non-productive conformation in order to prevent unwanted side reactions or futile cofactor cycling in the absence of substrate. These intriguing possibilities clearly can only be addressed with further experimentation.
Proposed minimal mechanism of ORF36
Results from our biochemical investigations of ORF36 indicate that during the reaction sequence, the enzyme first oxidizes TDP-l-epi-vancosamine 10 to the diffusible hydroxylamine intermediate 13, which is then further oxidized to the nitroso sugar 14. Moreover, we have demonstrated that the oxygen atom transfer reaction and the successive oxidative dehydrogenation result in single incorporation of oxygen from 18O2. Correspondingly, the general mechanism we envision for this transformation (Figure 9), and for oxidation of the biologically relevant substrate TDP-l-evernosamine, centers on an oxygen atom transfer reaction in which flavin-C4a-hydroperoxide serves as the electrophile in a nucleophilic substitution reaction (23, 55).
Figure 9.
Proposed monooxygenase mechanism of nitrososynthase. Pathway A denotes the first oxidation of substrate by ORF36, and pathway B shows the second oxidation step of the reaction cycle from substrate TDP-l-epi-vancosamine to the final nitrososugar product.
In overview, ORF36 likely first binds reduced flavin possibly provided by an unidentified external reductase (several candidate reductases exist in the genome of M. carbonacea var. africana, GenBank accession number ACES00000000.1 or NCBI Reference Sequence NZ_ACES00000000.1) and promotes reaction with molecular oxygen to form a flavin-C4a-hydroperoxide, a well-known process in flavin monooxygenases (56, 57). Substrate then binds and undergoes oxidation, after which the hydroxylamine 13 and oxidized flavin are released to allow recycling of the cofactor, as evidenced by the accumulation of the hydroxylamino sugar 13 in reactions and everninomicin D analogs. The second oxidative dehydrogenation sequence may result from an iterative monooxygenase reaction followed by dehydration or alternatively, via a dehydrogenase mechanism akin to that catalyzed by the homologous acyl-CoA dehydrogenases. Based on the indirect observation that hydroxylamine 13 does not react with ORF36 and oxidized flavin (generated by limiting NADPH in the reaction)(8), we favor a monooxygenase mechanism in which a second round of oxidation generates nitroso sugar 14 (Figure 9B) perhaps via a dihydroxylamino or aminohydroperoxide intermediate (not shown). This mechanism is supported by the observed incorporation of oxygen from 18O2, but not H 182O (data not shown), into the final product (Figure 4). The consistent stoichiometry of 18O2 incorporation may reflect the exquisite ability of the acyl-CoA dehydrogenase fold to control O2 access to the active site.
Summary and Conclusions
This study has characterized the conversion of the TDP-l-epi-vancosamine to the corresponding nitrososugar by the nitrososynthase ORF36, which we propose undergoes conformational adjustments upon substrate binding prior to catalyzing monooxygenation of substrates specific to the everninomicin biosynthetic pathway. Here, we conclusively show a monooxygenase mechanism and identify the architectural constraints of the enzyme housing this reaction. To the best of our knowledge, these results comprise the first detailed biochemical characterization of a functionally demonstrated nitrososynthase enzyme. These insights are particularly relevant given the known species targeting effects of the N-oxidation state at C-3 in the evernitrose ring (5) in this late stage III antibacterial clinical candidate. Nitrososynthase articulates a new class of enzyme revealing increasingly diverse flavin-dependent monooxygenase reactivity in enzymes with an acyl-CoA dehydrogenase fold. The results presented here have implications for the growing number of enzymes in secondary metabolic gene clusters that have been automatically annotated as acyl-CoA dehydrogenase-like enzymes, but may instead be performing oxidation reactions with little resemblance to the dehydrogenase paradigm. Future studies of everninomicin biosynthesis will lead to a better understanding of the effects of variability of C-4 O-methylation and C-5 stereochemistry on enzyme oxidation cycles and the protein structural determinants controlling selectivity and catalysis by this new family of enzymes. The delineation of this minimal mechanism, based on biochemical and structural studies, forms the basis for future more detailed mechanistic studies.
Supplementary Material
ACKNOWLEDGEMENT
We thank Prof. Michael Burkart, University of California, San Diego and Prof. Hung-wen Liu, University of Texas, Austin for generously providing for providing Eva A-E and RfbB expression constructs, respectively. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). Use of the Stanford Synchrotron Radiation Lightsource supported by Stanford University and the U.S. Department of Energy. We thank Gary Cecchini, Bill McIntire, and Carmello Rizzo for critical reading.
Footnotes
This work was supported by Office of Naval Research grant N000140610144 to BOB and NIH grant GM077189 to TMI.
The atomic coordinates and structure factors for ORF36 (code 3MXL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
SUPPORTING INFORMATION AVAILABLE One movie, eight figures and one table are available free of charge at http://pubs.acs.org.
References
- 1.Ganguly AK. Ziracin, a novel oligosaccharide antibiotic. J. Antibiot. 2000;53:1038–1044. doi: 10.7164/antibiotics.53.1038. [DOI] [PubMed] [Google Scholar]
- 2.Jones RN, Hare RS, Sabatelli FJ. In vitro Gram-positive antimicrobial activity of evernimicin (SCH 27899), a novel oligosaccharide, compared with other antimicrobials: a multicentre international trial. J. Antimicrob. Chemother. 2001;47:15–25. doi: 10.1093/jac/47.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Chu M, Mierzwa R, Jenkins J, Chan TM, Das P, Pramanik B, Patel M, Gullo V. Isolation and characterization of novel oligosaccharides related to Ziracin. J. Nat. Prod. 2002;65:1588–1593. doi: 10.1021/np020093t. [DOI] [PubMed] [Google Scholar]
- 4.McNicholas PM, Najarian DJ, Mann PA, Hesk D, Hare RS, Shaw KJ, Black TA. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents. Chemother. 2000;44:1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waitz JA, Horan AC. Micromonospora carbonacea var. africana. Schering Corporation; U.S.: 1988. [Google Scholar]
- 6.Ganguly AK, Girijavallabhan VM, Miller GH, Sarre OZ. Chemical modification of everninomicins. J. Antibiot. 1982;35:561–570. doi: 10.7164/antibiotics.35.561. [DOI] [PubMed] [Google Scholar]
- 7.Poulet FM, Veneziale R, Vancutsem PM, Losco P, Treinen K, Morrissey RE. Ziracin-induced congenital urogenital malformations in female rats. Toxicol. Pathol. 2005;33:320–328. doi: 10.1080/01926230590925061. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y, Al-Mestarihi A, Grimes CL, Kahne D, Bachmann BO. A Unifying Nitrososynthase Involved in Nitrosugar Biosynthesis. J. Am. Chem. Soc. 2008;130:15756–15757. doi: 10.1021/ja8051415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CG, Lamichhane J, Song KI, Nguyen VD, Kim DH, Jeong TS, Kang SH, Kim KW, Maharjan J, Hong YS, Kang JS, Yoo JC, Lee JJ, Oh TJ, Liou K, Sohng JK. Biosynthesis of rubradirin as an ansamycin antibiotic from Streptomyces achromogenes var. rubradiris NRRL3061. Arch. Microbiol. 2008;189:463–473. doi: 10.1007/s00203-007-0337-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, White-Phillip JA, Melancon CE, Kwon HJ, Yu WL, Liu HW. Elucidation of the kijanimicin gene cluster: Insights into the biosynthesis of spirotetronate antibiotics and nitrosugars. J. Am. Chem. Soc. 2007;129:14670–14683. doi: 10.1021/ja0744854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruender NA, Thoden JB, Holden HM. X-ray structure of kijd3, a key enzyme involved in the biosynthesis of D-kijanose. Biochemistry. 2010;49:3517–3524. doi: 10.1021/bi100318v. [DOI] [PubMed] [Google Scholar]
- 12.Parry RJ, Li W. Purification and characterization of isobutylamine N-hydroxylase from the valanimycin producer Streptomyces viridifaciens MG456-hF10. Arch. Biochem. Biophys. 1997;339:47–54. doi: 10.1006/abbi.1996.9857. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Zhao H. Mechanistic studies on the conversion of arylamines into arylnitro compounds by aminopyrrolnitrin oxygenase: identification of intermediates and kinetic studies. Angew. Chem., Int. Ed. Engl. 2006;45:622–625. doi: 10.1002/anie.200502903. [DOI] [PubMed] [Google Scholar]
- 14.Simurdiak M, Lee J, Zhao H. A new class of arylamine oxygenases: evidence that p-aminobenzoate N-oxygenase (AurF) is a di-iron enzyme and further mechanistic studies. Chembiochem. 2006;7:1169–1172. doi: 10.1002/cbic.200600136. [DOI] [PubMed] [Google Scholar]
- 15.Korboukh VK, Li N, Barr EW, Bollinger JM, Jr., Krebs C. A long-lived, substrate-hydroxylating peroxodiiron(III/III) intermediate in the amine oxygenase, AurF, from Streptomyces thioluteus. J. Am. Chem. Soc. 2009;131:13608–13609. doi: 10.1021/ja9064969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem. Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 17.Johnson HD, Thorson JS. Characterization of CalE10, the N-oxidase involved in calicheamicin hydroxyaminosugar formation. J. Am. Chem. Soc. 2008;130:17662–17663. doi: 10.1021/ja807557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips IR, Shephard EA. Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol. Sci. 2008;29:294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Meneely KM, Barr EW, Bollinger JM, Jr., Lamb AL. Kinetic mechanism of ornithine hydroxylase (PvdA) from Pseudomonas aeruginosa: substrate triggering of O2 addition but not flavin reduction. Biochemistry. 2009;48:4371–4376. doi: 10.1021/bi900442z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Berkel WJ, Kamerbeek NM, Fraaije MW. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 21.Thorpe C, Kim JJ. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 1995;9:718–725. doi: 10.1096/fasebj.9.9.7601336. [DOI] [PubMed] [Google Scholar]
- 22.Ghisla S, Massey V. Mechanisms of flavoprotein-catalyzed reactions. Eur. J. Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 23.Palfey BA, McDonald CA. Control of catalysis in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 2010;493:26–36. doi: 10.1016/j.abb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 24.Alfieri A, Fersini F, Ruangchan N, Prongjit M, Chaiyen P, Mattevi A. Structure of the monooxygenase component of a two-component flavoprotein monooxygenase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1177–1182. doi: 10.1073/pnas.0608381104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SH, Hisano T, Takeda K, Iwasaki W, Ebihara A, Miki K. Crystal structure of the oxygenase component (HpaB) of the 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8. J. Biol. Chem. 2007;282:33107–33117. doi: 10.1074/jbc.M703440200. [DOI] [PubMed] [Google Scholar]
- 26.Webb BN, Ballinger JW, Kim E, Belchik SM, Lam KS, Youn B, Nissen MS, Xun L, Kang C. Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Biol. Chem. 2010;285:2014–2027. doi: 10.1074/jbc.M109.056135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dresen C, Lin LY, D'Angelo I, Tocheva EI, Strynadka N, Eltis LD. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 2010 doi: 10.1074/jbc.M109.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballou DP, Entsch B, Cole LJ. Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem. Biophys. Res. Commun. 2005;338:590–598. doi: 10.1016/j.bbrc.2005.09.081. [DOI] [PubMed] [Google Scholar]
- 29.Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. Kinetic mechanisms of the oxygenase from a two-component enzyme, p-hydroxyphenylacetate 3-hydroxylase from Acinetobacter baumannii. J. Biol. Chem. 2006;281:17044–17053. doi: 10.1074/jbc.M512385200. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Thomas MG, Hubbard BK, Losey HC, Walsh CT, Burkart MD. Deoxysugars in glycopeptide antibiotics: enzymatic synthesis of TDP-L-epivancosamine in chloroeremomycin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11942–11947. doi: 10.1073/pnas.210395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi H, Liu YN, Liu HW. A two-stage one-pot enzymatic synthesis of TDP-L-mycarose from thymidine and glucose-1-phosphate. J. Am. Chem. Soc. 2006;128:1432–1433. doi: 10.1021/ja0562144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romana LK, Santiago FS, Reeves PR. High level expression and purification of dthymidine diphospho-D-glucose 4,6-dehydratase (rfbB) from Salmonella serovar typhimurium LT2. Biochem. Biophys. Res. Commun. 1991;174:846–852. doi: 10.1016/0006-291x(91)91495-x. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pike ACW, Hozjan V, Smee C, Niesen FH, Kavanagh KL, Umeano C, Turnbull AP, Von Delft F, Weigelt J, Edwards A, Arrowsmith CH, Sundstrom M, Oppermann U. Crystal structure of human short-branched chain acyl-coA dehydrogenase. To be published.
- 36.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 39.Kraulis PJ. MOLSCRIPT - a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 40.Merritt EA, Murphy ME. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 41.Kleywegt GJ. Crystallographic refinement of ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2007;63:94–100. doi: 10.1107/S0907444906022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 44.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press; New York: 1965. pp. 97–166. [Google Scholar]
- 45.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 46.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Timmons SC, Thorson JS. Increasing carbohydrate diversity via amine oxidation: aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr. Opin. Chem. Biol. 2008;12:297–305. doi: 10.1016/j.cbpa.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chow YL, Chen SC, Mojelsky T. Nitrosamine photoaddition to norbornene and the mechanism of nitrosoalkane cleavage. J. Chem. Soc., Chem. Commun. 1973:827–828. [Google Scholar]
- 49.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics (Oxford, England) 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatti DL, Entsch B, Ballou DP, Ludwig ML. pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis. Biochemistry. 1996;35:567–578. doi: 10.1021/bi951344i. [DOI] [PubMed] [Google Scholar]
- 51.Lindqvist Y, Koskiniemi H, Jansson A, Sandalova T, Schnell R, Liu Z, Mantsala P, Niemi J, Schneider G. Structural basis for substrate recognition and specificity in aklavinone-11-hydroxylase from rhodomycin biosynthesis. J. Mol. Biol. 2009;393:966–977. doi: 10.1016/j.jmb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 52.McCulloch KM, Mukherjee T, Begley TP, Ealick SE. Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications. Biochemistry. 2009;48:4139–4149. doi: 10.1021/bi900149f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson KA. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J. Biol. Chem. 2008;283:26297–26301. doi: 10.1074/jbc.R800034200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weikl TR, von Deuster C. Selected-fit versus induced-fit protein binding: kinetic differences and mutational analysis. Proteins. 2009;75:104–110. doi: 10.1002/prot.22223. [DOI] [PubMed] [Google Scholar]
- 55.Massey V. Activation of Molecular-Oxygen by Flavins and Flavoproteins. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 56.Bruice TC. Oxygen-flavin chemistry. Isr. J. Chem. 1984;24:54–61. [Google Scholar]
- 57.Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Djordjevic S, Pace CP, Stankovich MT, Kim JJ. Three-dimensional structure of butyryl-coA dehydrogenase from Megasphaera elsdenii. Biochemistry. 1995;34:2163–2171. doi: 10.1021/bi00007a009. [DOI] [PubMed] [Google Scholar]
- 59.Kim JJ, Wang M, Paschke R. Crystal structures of medium-chain acyl-coA dehydrogenase from pig liver mitochondria with and without substrate. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satoh A, Nakajima Y, Miyahara I, Hirotsu K, Tanaka T, Nishina Y, Shiga K, Tamaoki H, Setoyama C, Miura R. Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA. J. Biochem. 2003;134:297–304. doi: 10.1093/jb/mvg143. [DOI] [PubMed] [Google Scholar]
- 61.Battaile KP, Molin-Case J, Paschke R, Wang M, Bennett D, Vockley J, Kim JJ. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases. J. Biol. Chem. 2002;277:12200–12207. doi: 10.1074/jbc.M111296200. [DOI] [PubMed] [Google Scholar]
- 62.Chang C, Skarina T, Kagan O, Savchenko A, Edwards AM, Joachimiak A. Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1. To be published.
- 63.Tan K, Skarina T, Kagen O, Savchenko A, Edwards A, Joachimiak A. The crystal structure of a putative hydroxylase from Rhodococcus sp. RHA1. To be published.
- 64.Nagpal A, Valley MP, Fitzpatrick PF, Orville AM. Crystal structures of nitroalkane oxidase: insights into the reaction mechanism from a covalent complex of the flavoenzyme trapped during turnover. Biochemistry. 2006;45:1138–1150. doi: 10.1021/bi051966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.