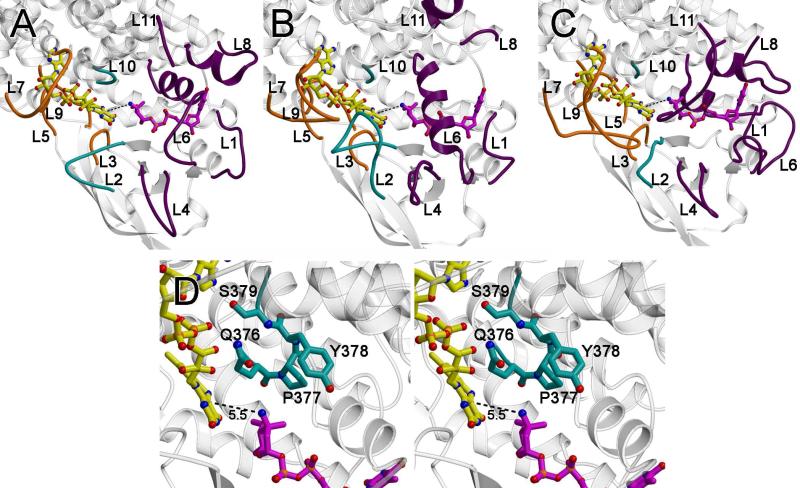
Figure 7.
Active site loops. The loops that form the active site are as follows: the loops between α3 and α4 (loop L1; residues 106 – 109), β1 and β2 (loop L2; residues 134-142), β3 and β4 (loop L3, residues 158-164), β4 and β5 (loop L4; residues 178-180), β6 and β7 (loop L5; residues 201-213), β9 and α6 (loop L6; residues 248-253), α6’ and α7’ (where the ’ indicates this is from an adjacent monomer; loop L7; residues 272-280), α7 and α8 (loop L8; residues 310-317), α8’ and α9’ (loop L9, residues 351-353), α9 and α10, (loop L10; residues 375-379) and at the C-terminus (loop L11; residues 390-412). (A) A ribbon diagram of ORF36 shown with modeled flavin (yellow carbons) and TDP-l-evernosamine (magenta carbons) as sticks. Active site loops L3, L5, L7, and L9, are predicted to interact with flavin and are colored orange, active site loops L1, L4, L6, L8, and L11 are predicted to interact with substrate and are colored purple, and active site loops L2 and L10 are predicted to interact with both substrate and cofactor and are colored teal. (B) A ribbon diagram of human short branched-chain acyl Co-A dehydrogenase (PDB entry 2JIF) (35), highlighting loops L1 to L11 colored are colored as in (A). (C) A ribbon diagram of A. baumannii 4-hydroxyphenylacetate monooxygenase (PDB entry 2JBT) (24) highlighting loops L1 to L11 colored as in (A). (D) Stereoview of the ORF36 loop L10 containing a tandem cis-peptide. Loop L10 is shown in sticks with teal carbons. The Q376-P377 and P377-Y378 bonds both adopt a cis conformation. Modeled flavin and substrate are displayed as in (A).