Abstract
Pore-forming toxins, many of which are pathogenic to humans, are highly dynamic proteins that adopt a different conformation in aqueous solution than in the lipid environment of the host membrane. Consequently, their crystal structures obtained in aqueous environment do not reflect the active conformation in the membrane, making it difficult to deduce the molecular determinants responsible for pore formation. To obtain structural information directly in the membrane, we introduce a fluorescence technique to probe the native topology of pore-forming toxins in planar lipid bilayers and follow their movement during pore formation. Using a Förster resonance energy transfer (FRET) approach between site-directedly labeled proteins and an absorbing compound (dipicrylamine) in the membrane, we simultaneously recorded the electrical current and fluorescence emission in horizontal planar lipid bilayers formed in plastic chips. With this system, we mapped the topology of the pore-forming domain of Cry1Aa, a biological pesticide from Bacillus thuringiensis, by determining the location of the loops between its seven α helices. We found that the majority of the toxins initially traverse from the cis to the trans leaflet of the membrane. Comparing the topologies of Cry1Aa in the active and inactive state in order to identify the pore-forming mechanism, we established that only the α3–α4 hairpin translocates through the membrane from the trans to the cis leaflet, whereas all other positions remained constant. As toxins are highly dynamic proteins, populations that differ in conformation might be present simultaneously. To test the presence of different populations, we designed double-FRET experiments, where a single donor interacts with two acceptors with very different kinetics (dipicrylamine and oxonol). Due to the nonlinear response of FRET and the dynamic change of the acceptor distribution, we can deduce the distribution of the acceptors in the membrane from the time course of the donor fluorescence. We found that Cry1Aa is present on both membrane leaflets.
INTRODUCTION
The method of choice in determining the three-dimensional structure of a membrane protein is solving the high-resolution crystal structure. However, crystallization is time consuming and crystal structures are difficult to obtain. Moreover, in the case of more flexible proteins, such as pore-forming toxins, crystal structures often do not reflect the structure in the native membrane. Many pore-forming toxins adopt in solution a different conformation than in their target membrane, for they undergo major conformational changes that are often related to the pore-forming mechanism. This class of proteins includes the toxins of the pathogens Clostridium botulinum (Botulinum neurotoxins), Clostridium tetani (Tetanus toxin), and Bacillus anthracis (Anthrax toxin), which are all potentially lethal to humans, but also the toxins of Bacillus thuringiensis, one of which (Cry1Aa) we studied here.
Cry1Aa is widely used as a biological pesticide in forestry due to its high specificity for insects belonging to the family of lepidoptera (Schnepf et al., 1998), whose larvae damage forests and crop. Its protoxin is taken up by the larvae through ingestion and is activated by proteases in the digestive tract (Ogiwara et al., 1992; Knowles, 1994). It unfolds its cytolytic activity in the midgut of the larvae. Then, the toxins lead, after binding to a specific receptor and pore formation in the membrane, to cell destruction and finally death (Peyronnet et al., 2001).
The crystal structure of Cry1Aa has been solved in 1995 (Grochulski et al., 1995), followed by the crystal structures of several other toxins of B. thuringiensis (Li et al., 1991; Boonserm et al., 2005; Akiba et al., 2008; Cohen et al., 2008; Liu et al., 2008). Cry1Aa shows a typical three-domain structure (Fig. 1 B). Domains II and III are responsible for receptor binding (Aronson et al., 1995; Lee et al., 1995; Schnepf et al., 1998), and Domain I, consisting of seven α helices α1–α7, is responsible for pore formation (Schwartz et al., 1997). Pores formed by Cry1Aa have been well characterized by electrophysiology. Schwartz and Laprade (2000) demonstrated that the pores show well-defined conductivity and selectivity for cations. The permeability, as shown by Peyronnet et al. (2001), is dependent also on the size of the cation. At higher concentrations, however, the conductivity increases, suggesting larger pore diameter (Peyronnet et al., 2001). The pore-forming entity for the well-defined pores has been suggested to be tetrameric based on atomic force microscopy (Vie et al., 2001). Finally, the helix α4 has been shown by means of mutagenesis and electrophysiology to be the pore-lining helix (Vachon et al., 2004). The presence of the receptors favors annealing of the toxin to the membrane but is not necessary for pore formation (Peyronnet et al., 2001).
Figure 1.
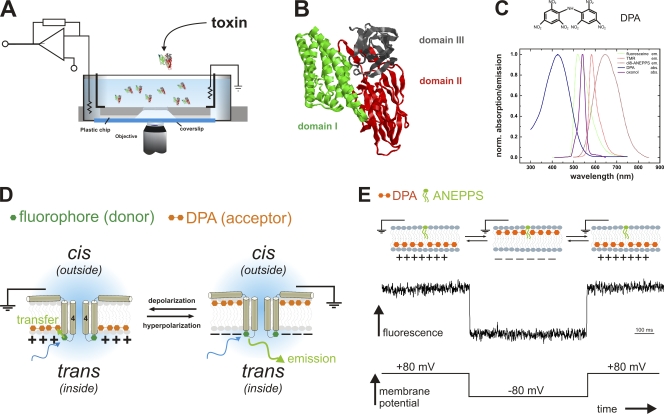
Fluorescence topology assay. (A) Horizontal planar lipid bilayer configuration for optical access: The bilayer is formed in the aperture (Ø = 80–200 µm) of a small plastic chip (gray). The chip with access channels on the bottom is placed on a glass coverslip, facilitating the bilayer to be imaged with a high NA objective. Electrical currents are recorded with a patch-clamp amplifier. The configuration is mounted on the stage of an inverted microscope. (B) Structure of Cry1Aa (Grochulski et al., 1995). The three domains are shown in green (I), red (II), and gray (III). (C) Structure of dipicrylamine (DPA, top), absorption spectrum of DPA (blue), and emission spectra of tetramethylrhodamine (TMR, red), fluorescein (green) and di-8-ANEPPS (brown). DPA is a negatively charged amphiphatic compound, which absorbs in the visible spectral range. It overlaps with TMR as well as with fluorescein emission wavelengths with an R0 of 35 and 45 Å, respectively. (D) “Umbrella” model and DPA assay. According to the umbrella model, the helices 4 and 5 translocate through the membrane and form the ion-conducting pore. With the DPA assay, we can detect the location of a labeled residue (green hexagon) in the toxin by depolarizing and hyperpolarizing the membrane, and thereby moving the DPA (double orange hexagon) from one leaflet to the other. It only comes to energy transfer (and thus reduced donor fluorescence) if the DPA and the fluorophore are located on the same side of the membrane (see text for details). (E) Control experiment with a membrane-fixed dye. Di-8-ANEPPS inserts into the membrane between the headgroups. Due to its long hydrophobic tail, it cannot translocate to the inner leaflet. In response to a hyperpolarizing voltage pulse (bottom), the DPA moves to the outer leaflet close to the fluorophores resulting in energy transfer and reduced fluorescence (center). Upon returning the voltage, the signal recovers. Please note that the fluorophore is opposite to D.
Based on the above results, the “umbrella” model has been proposed. According to this model, the seven α helices of domain I anneal to the outer leaflet of the membrane and form tetramers (Gazit et al., 1998), and in order to achieve pore formation, the α4–α5 hairpins translocate through the bilayer to form an ion-conducting pore in the center (Fig. 1 D). Recent findings that helices other than α4 also insert into the membrane, however, suggest that the refolding of Cry1Aa may be more complex than has been previously recognized (Tomimoto et al., 2006; Nair and Dean, 2008). Yet, attempts at uncovering the molecular mechanism of pore formation of Cry1Aa have been hampered by the fact that the refolding occurs after binding to the membrane, and, thus, cannot be presently imaged using crystallography, which provides the structure in solution, i.e., before entering the membrane.
Here, we present a novel technique using Förster resonance energy transfer (FRET) between site-directedly labeled toxins and the membrane, which allows probing of the topology of Cry1Aa in a planar lipid bilayer, where they are assumed to adopt their native conformation. We used a horizontal bilayer chamber design with which we can image a planar lipid bilayer with a high-numerical aperture objective (Fig. 1 A). We have simultaneous electrical and optical access to the membrane so that we can control the membrane potential and measure electric currents as well as record the fluorescence changes. With this technique, we have been able to observe the topology of Cry1Aa after its insertion into the membrane, and to successfully examine and elucidate its pore-forming mechanism.
MATERIALS AND METHODS
Mutagenesis, purification, and labeling of Cry1Aa
Cry1Aa constructs in plasmid vectors pMP39 and pBA1 were described previously (Masson et al., 1990; Bah et al., 2004). Cysteine mutations were introduced using Quikchange (Strategene) and amplified using Qiagen Miniprep Kits following standard protocols. Constructs were sequenced in order to verify mutations. Most mutants were transformed into B. thuringiensis except S39C, which was grown in Escherichia coli strain DH5. Bacteria were grown 3 d in YT at 37°C containing 10 µg/ml tetracycline or 100 µg/ml ampicilline for DH5 and B. thuringiensis, respectively. Protoxins were isolated as described by Masson et al. (1990). Protoxins were digested with 0.125 mg/ml trypsin (Invitrogen) in 50 mM NaCl, 50 mM NaCO3, adjusted to pH 10.5 for 90 min at 37°C. Activated toxins were centrifuged for 90 min at 200,000 g to remove lipids and other insoluble material. Toxins were then purified by fast protein liquid chromatography using a Mono-Q anion exchange column. Toxin was eluted with a 50–500 mM NaCl gradient in carbonate buffer (pH 10.5). Precipitates formed by dialyzing purified toxins against distilled water were collected and stored at 4°C. Purity was verified by SDS gel electrophoresis.
Toxins were labeled for 1 h with a 10-fold excess of TMR- or fluorescein-maleimide (Invitrogen). Unbound dye was removed by incubation with biobeads (62 mg/ml) for two successive rounds of 90 min each, or by buffer exchange using concentrators (25 kD, Amicon, Millipore). The labeling ratio was determined by comparing the protein concentration (Bradford assay, Thermo Fisher Scientific) with the fluorophore concentration (absorption). For fluoresceine labeling, W219C had low accessibility and showed a labeling ratio after removal of excess label of 0.32. The other mutants had a labeling ratio of 0.9 ± 0.3. Cry1Aa-WT had low unspecific labeling (labeling ratio 0.06 ± 0.03) that was negligible with respect to the specific labeling. Spectra were measured in a SPEX fluorolog spectrometer. Fluorophores were dissolved in octanol in order to mimic lipid environment.
Fluorescence setup for planar lipid bilayer
The fluorescence setup is based on a Carl Zeiss, Inc. Axiovert 200 inverted microscope with a 60x water objective (NA = 0.9). For TMR excitation, a 30-mW laser (532 nm, World Star Technology) was extended to fill out the back pupil of the objectives and focused onto the back-focal plane. For blue excitation, a mercury lamp (Carl Zeiss, Inc.) was used. Excitation light was filtered with a 470/40 and 545/25 excitation filter, and emission by a 504/12 and 605/70 for fluorescein and TMR, respectively. Emission was collected with a Photomax 200 (Dagan Corp.) fluorescence amplifier with an avalanche photodiode as a detector. For the general experiments, emission was focused onto the detector. For confocal measurements, the focusing lens was removed, and the detector placed in the image plane and aligned in xyz direction to the excitation spot. For the confocal scan, the motorized z drive of a Carl Zeiss, Inc. Observer microscope was used to adjust the z position in steps of 0.1 µm, and the fluorescence intensity was recorded for each level. The “point spread function” for our configuration was determined by scanning a single fluorescent bead (1 µm).
Bilayers were formed in a horizontal orientation in polymer chips (150 µm thickness) with a geometry similar to the one described previously (Pantoja et al., 2001). They had a small conical chamber on the bottom with a 150–250-µm aperture to the top. The plastic chip was placed above a coverslip, giving optical access for the objective of an inverted microscope (Fig. 1 A). The horizontal bilayer is formed inside of the top round aperture. For electrical control, the electrodes of a patch-clamp amplifier are connected to the cis and trans chambers via agar bridges. Small access channels allowed fluid exchange and electrical access to the bottom chamber. The chips were glued to standard 35-mm plastic Petri dishes. As trans chamber, a 50-mm Petri dish with a concentric hole was used, at the bottom of which a glass coverslip was glued. Current was recorded using an Axopatch-1D (Axon Instruments) and was stored on a PC using GPatchM (University of California at Los Angeles).
FRET measurements with DPA
Lipids were prepared from chloroform stock solution by evaporation of the solvent and resuspension of the lipids in decane (Sigma-Aldrich). Bilayers were formed by painting the lipids over the hole with a small glass rod. Only synthetic phospholipids and purified cholesterol (Avanti Polar Lipids) in the composition POPE:POPC:cholesterol 7:2:1 (wt:wt:wt) were used. Bilayer formation was monitored by capacitance measurements. Recording solutions were cis 150 mM N-methyl-d-glucamine(NMG)-Cl, 10 mM Hepes, 1 mM CaCl2, and trans 150 mM KCl, 10 mM Hepes, 1 mM CaCl2; solutions were adjusted with HCl or KOH to respective pH. Dipicrylamine (DPA) (10 µM) was added to the solution before bilayer formation. Toxins were added to the cis chamber from 2 mg/ml stock solution to a final concentration of 30–40 nM. After 8–10-min incubation time, nonannealed toxin was vigorously washed away (four to five times). The relative fluorescence change was determined from the difference of the steady-state fluorescence values before the pulse and at the end of the pulses normalized to fluorescence F before application of the pulse. Pore-forming ability of all labeled mutants was verified by electrophysiology in planar lipid bilayer.
We verified that not a “leak” of the oxonol fluorescence into donor emission was responsible for the signals observed in double-FRET experiments. We determined the emission intensity of oxonol through either direct excitation or energy transfer from the toxin at blue excitation used for the double-FRET experiments. The ratio of oxonol to fluorescein emission was 1–10 dependent on the toxin concentration. The fluorescein emission filter will let pass maximally 0.006% of the oxonol emission. The contribution of oxonol to total fluorescence during the experiments was thus <0.06%, which would be twofold the maximal dF/F that could be generated by the oxonol in the fluorescein signal. It is thus negligible with respect to the signals.
Energy transfer efficiency for two acceptors
For double FRET experiments, the toxins were labeled with fluorescein-maleimide, which transfers energy both to oxonol and DPA (Fig. 1 C). The situation is different than in the case of a single donor and acceptor, as both oxonol and DPA act as acceptors, while the toxin-bound fluorophore is the donor. We, therefore, have to consider two energy transfers from the toxin to two different acceptors. The energy transfer efficiency ET is given by
| (1) |
where τD is the fluorescence lifetime of the donor in the absence of acceptors and the kT are the rate constants to the respective acceptors i. These rate constants are a function of the R0i of the donor–acceptor pair and the mean distance as a function of time ri(t) between the donor and the respective acceptor i.
| (2) |
The mean distance ri(t) between the donor and the acceptor depends on the number of acceptors in the respective leaflet (see Appendix). With Eq. 2, the energy transfer efficiency ET can be written as
| (3) |
Online supplemental material
The online supplemental material shows the different time constants for DPA and oxonol. Both compounds were added simultaneously to the bilayer, and FRET efficiency was monitored via oxonol emission. The fluorescence shows two positive transients occuring while oxonol and DPA are on opposite sides of the bilayer. The on- and offset of the transient represent the DPA and oxonol time constants, respectively. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.200910347/DC1.
RESULTS
Experimental design
Fig. 1 D illustrates the principle of the method. It is a FRET assay between the fluorescently labeled toxin and an acceptor located in the membrane. To site directedly label the toxin, a cysteine was introduced in the loops between the helices of domain I. An organic thiol-reactive fluorophore (tetramethylrhodamine-maleimide or fluorescein-maleimide), which acted as a donor in the FRET system, was covalently bound to the cysteine. DPA, an amphiphatic negatively charged compound that partitions into the membrane, served as acceptor. Due to their net negative charge, DPA molecules are distributed between the outer and the inner membrane leaflet according to the membrane potential (Fernandez et al., 1983; Chanda et al., 2005a,b), where they are located between the headgroups on either side but are not found in the hydrophobic core. Throughout the manuscript, we use the convention that the potential is measured at the inside (trans) with respect to ground on outside (cis; Fig. 1 D) in analogy to the cell model. At saturating negative potentials, the DPA is, therefore, located at the headgroups of the outer leaflet (cis) and, at positive potentials, at the headgroups of the inner leaflet (trans).
Because the absorption of DPA overlaps with the emission spectra of both fluorescein and TMR (Fig. 1 C), FRET occurs between the fluorophore attached to the toxin and DPA located in the membrane (R0 = 45 Å and 35 Å, respectively). DPA itself is an absorber and not fluorescent. FRET efficiency is, thus, monitored by donor emission intensity. The position of the fluorophore, and thereby also that of the mutated residue in the protein, relative to the membrane normal can be detected by the direction of the fluorescence change upon reversal of the membrane potential (Chanda et al., 2005a,b). A change in the membrane potential moves the DPA either closer to or further from the fluorophore (Fig. 1 D). Let us assume that the labeled position is located on the internal leaflet (Fig. 1 D, green hexagon). Applying a positive membrane potential (left) attracts the negatively charged DPA to the internal leaflet. The distance between the fluorophore and the DPA is short (<R0), resulting in high energy transfer efficiency and, thus, low donor emission. If membrane polarity is reverted to negative potential, the DPA moves to the outer leaflet (Fig. 1 D, right). Then, the distance between the donor (toxin) and acceptor (DPA) is at least the thickness of the bilayer (>50 Å) and therefore larger than the R0, which means low energy transfer efficiency and high donor emission. Consequently, we will observe a pulse to higher fluorescence in the donor emission signal during a negative test pulse from +100 to −80 mV and back to +100 mV. For reasons of symmetry, a fluorophore located in the external leaflet will result in lower donor fluorescence in response to a negative voltage pulse.
To verify that our technique correctly reports the position of a fluorophore in the bilayer, we tested the fluorescence response of DPA and a static fluorophore, of which we know the location. We added the static fluorescent probe di-8-ANEPPS (Invitrogen) that intercalates into the cis leaflet of the bilayer between the headgroups (Ries et al., 2004), and added to the solution DPA (10 µM), which subsequently enters the membrane. Fig. 1 E shows the voltage dependence of the signal. (Note that the fluorophore is now located on the opposite side compared with Fig. 1 D.) At positive potential (inside, trans), the DPA occupied the inner leaflet far from the ANEPPS on the outer leaflet (>50 Å), leading to very little FRET and high donor (ANEPPS) fluorescence (Fig. 1 E). During a negative pulse to −80 mV, the DPA moved to the outer leaflet and came close to ANEPPS. Energy transfer efficiency was high and fluorescence decreased (Fig. 1 E). Upon return to +80 mV, the fluorescence recovered (DPA returned to the inner leaflet). The ANEPPS experiment shows that we can determine whether a fluorophore is located in the inner or outer leaflet by the change in fluorescence upon voltage inversion; a fluorophore located in the outer leaflet provokes a negative fluorescence change when pulsing from +80 to −80 mV (Fig. 1 E), while a fluorophore located in the inner leaflet will generate a positive fluorescence change (Fig. 1 D). It is important to note that these signals, per se, are caused not by the movement of the toxin/fluorophore itself but merely by the movement of our probe (DPA). Moving the probe has the great advantage that we can determine not only the movement of any labeled residue but also the location of any static labeled position with respect to the membrane surface.
On the other hand, it also means that careful attention has to be paid to nonspecific labeling since any fluorophore in the membrane will produce a signal. We, therefore, started by determining whether any signal originated from unspecific labeling. As wild-type Cry1Aa (Cry1Aa-WT) does not contain any native cysteines, any labeling of Cry1Aa-WT toxin would be unspecific. For each preparation, we, therefore, also labeled Cry1Aa-WT parallel to the cysteine mutants in order to ascertain that the ratio between specific and unspecific labeling was sufficiently high. Excess dye was removed (see Materials and methods) and the efficiency of labeling was determined (EOL, number of fluorophores per toxin protein). EOL of Cry1Aa-WT was <0.06 ± 0.03 and the EOL of the cysteine mutants ≈ 1 (0.9 ± 0.3). This meant that although it was impossible to remove all fluorophores from the Cry1Aa-WT sample, only a very small amount of nonspecifically bound fluorophores remained in the sample. Nevertheless, since our method is sensitive enough to detect the signal originating from all static fluorophores, we had to ensure that the nonspecific signal does not alter our results. We found a normalized fluorescence change of labeled Cry1Aa-WT incorporated into a horizontal bilayer of 0.5 ± 0.3% (Fig. 2 C). Since we used only samples for experiments where the EOL of the cysteine mutants was at least 8–10-fold higher than WT, the dF/F of 0.5% originating from the nonspecific signal contributed <1/10 to 1/8 to the final signal corresponding to a normalized fluorescence change of 0.05–0.06%. This value is significantly lower than the values detected in the cysteine mutants (see below), showing that nonspecific labeling had no significant influence on the specific signal.
Figure 2.
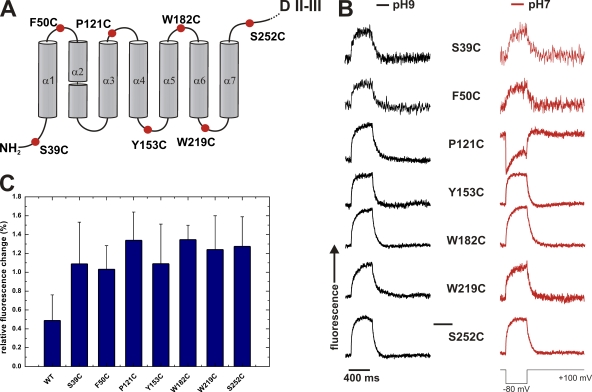
FRET signals of Cry1Aa mutants. (A) Position of cysteine mutants of domain I. Mutants in every loop were investigated except the α2–α3 loop, as mutants in this loop did not express. (B) Fluorescence traces (TMR) of the mutants in A. Fluorescence traces were taken for a hyperpolarizing pulse (+100 mV/−80 mV/+100 mV) at pH 9.0 (black traces) and after exchange to pH 7.0 (red traces). (C) Relative fluorescence changes (dF/F; mean ± SD) of the cysteine mutants in A.
Topology of domain I of Cry1Aa in planar lipid bilayer
The pore-forming domain I consists of seven α helices (α1–α7) connected by short loops of four to six amino acids (Grochulski et al., 1995). α2–α7 have the typical length for transmembrane helices (22–32 amino acids; α2 is broken in the center and α1 is only 12 amino acids long). Assuming that the helices do not significantly bend in the native conformation, one can construct the topology of Cry1Aa once the vertical positions of the short loops in the membrane are known.
To identify the location of all loops of domain I, we created a mutant for each loop by introducing a single cysteine to the respective loop (Fig. 2 A). Cysteine mutants for all loops, except for the α2–α3 loop, expressed functionally. The conductance of the pores in 150 mM KCl was between 266 ± 50 pS and 444 ± 79 pS. In asymmetric 150 mM NMG/150 mM K, the reversal potential was −26.6 mV, indicating pK/pNMG = 2.8. These electrophysiological properties of the pores are in accordance with those reported previously (Peyronnet et al., 2000, 2001). The structure and pore-forming mechanism of the toxin remained thus largely unaltered by the mutation or the subsequent labeling with the fluorophore.
To identify the positions of the toxin that change conformation during pore formation, ideally, we would follow the evolution of the signals with time. However, the toxins all arrive at the membrane at different times and their translocation is, thus, not synchronized. Taking a cue from a previous study showing the pH dependence of the velocity of pore formation, we attempted to better synchronize the translocation of toxins. Vachon et al. (2004) showed that although no significant effect of pH on pore-forming activity of Cry1Aa-WT has been observed in insect brush border membranes (BBMs) in the steady state, the rate of pore formation of both Cry1Aa-WT and mutants is generally accelerated in BBM at neutral pH in comparison to basic pH. Vachon’s finding implies that toxins can be kept in a state of low activity at basic pH during the annealing to the membrane before pore formation is “allowed” at neutral pH. We, therefore, incubated the toxin at pH 9.0 for 10–15 min, thus minimizing the number of toxins translocating during the incubation period. We then washed excess toxin, took a first measurement, and subsequently varied the proton concentration to pH 7.0. Although pore formation was observed also at pH 9.0 (unpublished data), the rate of pore formation was increased at pH 7.0, and thus any difference between basic and neutral pH is likely related to conformational changes linked to pore formation. We do not know the origin of the pH dependence, but the toxins thus treated show their typical pore-forming behavior, indicating that they still assume their native functional conformation at pH 7.0.
Following this procedure for the loop mutants, we incubated planar lipid bilayer with the tetramethylrhodamine-maleimide–labeled mutant toxins (30–40 nM) and measured current and fluorescence intensity. We applied a negative test pulse (+100 mV/−80 mV/+100 mV) both in the absence and presence of DPA. A negative, hyperpolarizing pulse provokes, as outlined above, a decrease in fluorescence for the duration of the pulse if the labeled position is located in the outer (cis) leaflet, and, conversely, an increase in fluorescence if the labeled position is located in the inner (trans) leaflet. In absence of DPA, no voltage-dependent fluorescence change was observed.
In the presence of DPA, the fluorescence signal at pH 9.0 for all labeled positions in the different loops of domain I initially increased during a depolarizing pulse, indicating that they are mostly located on the inner leaflet (trans) with respect to the midline of the bilayer (Fig. 2 B). The average normalized fluorescence change (dF/F) was 1.2 ± 0.2% with little variation among the mutants (Fig. 2 C). These results suggested that the toxins do not simply initiate their pore-forming action from the outer surface of the bilayer but rather that the entire domain I unfolded once it reached the membrane and inserted into the membrane, crossing the midline of the bilayer such that the helices of domain I became immersed in the inner leaflet.
A more detailed consideration of the fluorescence signals indicate that the fluorescence does not stay constant after the rapid translocation of DPA but that still some variations are observed (e.g., Fig. 2 B, P121C). This might be a hint that the toxin changes its conformation in the bilayer dependent on the voltage. This was to be expected if the bilayers are crossing the membrane suggesting a high mobility.
Loop α3–α4 translocates through the membrane for pore formation
The fact that pore formation is accelerated at pH 7.0 in comparison to pH 9.0 leads to the conclusion that differences in the topology at both pH values should be correlated with pore formation. When we altered the proton concentration from pH 9.0 to pH 7.0, most loop positions remained on the intracellular side with the exception of S39C (N terminus) and P121C (α3–α4 loop). The signal of S39C changed in 50% of all cases to the outer leaflet. S39 is located in the N terminus of helix α1 and may therefore be more flexible. The signal of P121C reversed its direction in 58% of the cases (Fig. 2 B, n = 24), indicating that the majority of the α3–α4 loops was located in the outer leaflet. The majority of them must therefore have moved through the membrane from the inner to the outer leaflet. The transition of P121C is a clear indication that the helices α3 and α4 are involved in pore formation. In addition, we never observed pore formation before the signal of P121C was reversed, suggesting that translocation of the α3–α4 hairpin is a prerequisite for pore formation.
Nevertheless, occasionally pore formation was observed already at pH 9.0 because the toxins, in spite of being kept in a state of low activity at pH 9.0, were not fully inactive. Although pore formation was never observed without prior movement of the α3–α4 hairpin, we still wanted to ensure that the difference in fluorescence at both pHs is truly due to the pore-forming process. We, therefore, sought a mutation that kept the channel in a fully inactive state at pH 9.0. The R131E mutant, inserting a charge reversal in the helix α4, had been shown in a volumetric assay to be biologically active only at neutral pH 7.0 and completely inactive in basic conditions (pH 9.0) (Vachon et al., 2004). As this result was obtained in BBM in the presence of receptors, it remained to be verified whether the biological activity is directly correlated with pore formation. We, thus, tested the pore-forming activity of Cry1Aa-R131E in planar lipid bilayer, and found that no pore formation of Cry1Aa-R131E occurred at pH 9.0, whereas in 45% of all experiments, pore formation was observed after switching to pH 7.0 (Cry1Aa-WT: 30%). Note that the percentages are underestimated, as tests were stopped when bilayers broke and were counted as negative if no pore formation was yet observed, although breakage might have been induced by too strong Cry1Aa pore formation. Since activity of mutants is defined by its ability to form pores, the results demonstrated that the R131E mutant remained in a nonactive conformation at pH 9.0 and became active at pH 7.0. Thus with R131E, we now have a mutant that we can switch between an inactive and an active state.
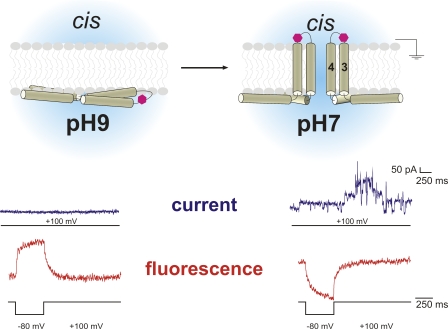
To follow the movement of the α3–α4 loop, which we found to be important for pore formation, we added a cysteine in the α3–α4 loop at position T122 in the R131E mutant (R131E-T122C). When T122C-R131E was exposed to the pH protocol, we found that at pH 9.0 (inactive), the fluorescence signal in the presence of DPA indicated that the α3–α4 loop was confined to the inner leaflet in all cases. After changing to pH 7.0, in 7 of 10 experiments, the signal switched from the internal to the external leaflet (Fig. 3, bottom). Please note that in the three experiments during which the signal remained the same, no pore formation was observed also. The switch of the signal when changing to the active conformation confirms the data we obtained from position P121 and demonstrates that the α3–α4 loop moves through the membrane for pore formation (Fig. 3, top). Since the loop is very short, the C-terminal α3 and N-terminal α4 also have to follow this movement, suggesting translocation of the α3–α4 hairpin during pore formation. This agrees with the umbrella model in so far as the α3–α4 loop is external and the α4–α5 loop internal. Helix α4 forms the pore-lining helix as demonstrated earlier (Vachon et al., 2004). Nevertheless, we found the other helices to be internal in this study. The translocation process is, thus, initiated from the inside (Fig. 3, top). The α3–α4 helices and partly α1 were also the only ones we found to be transmembrane, indicating that they are responsible for pore formation. As we observed transition of the hairpin before pore formation, the translocation step is necessary but not sufficient for pore formation.
Figure 3.
Translocation of the α3–α4 hairpin. Results for the T122C-R131E mutant. The fluorophore translocates through the membrane upon activation by pH change. The fluorescence signal (red) reverses as a response to a negative pulse (+100 mV/−80 mV/+100 mV), and pore formation is observed after pH change. Pore formation (blue, 100 mV) is observed only at pH 7.0. Considering that the other positions are located at the intracellular leaflet, we suggest a model for pore formation as depicted here (top). The entire domain I translocates to the internal leaflet. Subsequently, helices α3 and α4 reach through the membrane and form the ion-conducting pore from the internal side. The magenta hexagon depicts the fluorophore attached to the α3–α4 loop. Two toxins are shown on the right as it is thought that toxins oligomerize for pore formation. Please note that the pulse protocols for the current and fluorescence traces are not identical. Currents are shown only from +100 mV.
Background fluorescence is not responsible for low dF/F
At a concentration of 10 µM DPA, we expect ∼16,600 DPA molecules per µm2 (Chanda et al., 2005b). From this number, we can calculate the mean area per DPA molecule and, thereby, the mean distance between donor and acceptor (see Appendix). The maximal fluorescence change due to FRET between DPA and TMR (R0 = 35 Å) for a 50-Å movement of DPA through the membrane was calculated to be dF/F = 18%, assuming that the donors are located close to the headgroups of one leaflet of the membrane (Lakowicz, 1999). In our experiments above, however, our mean relative fluorescence change was only 1.2 ± 0.2%.
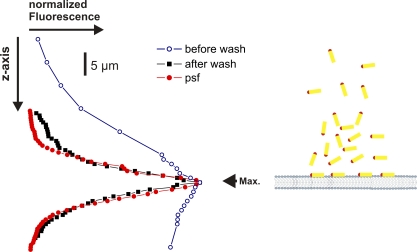
One possible explanation for the relatively low relative fluorescence change dF/F is high background fluorescence from unspecific labeling or from unbound toxins in the cis chamber. Unspecific labeling was above excluded as a source of background fluorescence and, thus, cannot cause a low dF/F. To test, then, whether additional background fluorescence arises from unbound toxins in the cis chamber, we made a confocal z-scan of the fluorescence across the bilayer. We formed a diffraction-limited spot with a green laser (532 nm) and used the sensitive surface of the photodiode as pinhole detector (Ø ≈ 100 µm). We then measured fluorescence originating only from the diffraction-limited spot every 0.1 µm in z direction. Fig. 4 shows the fluorescence distribution before and after vigorous washing. After washing, the fluorescence intensity agrees well with the point spread function (Fig. 4, red) determined with a fluorescent bead, indicating that the fluorescence originated mainly from the bilayer and that background fluorescence from the cis chamber was negligible. The diminished dF/F thus has a different origin than background fluorescence.
Figure 4.
Confocal z-scan of labeled toxin in the bilayer. Confocal z-scans through a bilayer after incubation with labeled toxin. The fluorescence was concentrated to the bilayer after rigorous washing of the toxin as described in the Materials and methods. In red, the “point spread function” (psf) is shown. It was determined with a 1-µm fluorescent bead.
Double FRET measurements
The value of dF/F must, then, be intrinsically relatively low, which, in turn, suggests that our assumptions above are not applicable to Cry1Aa. Our assumptions, to repeat, were that the fluorophores were located between the headgroups of the lipids furthest away from the DPA and that they were located only in one leaflet of the membrane. If either one of the assumptions is not true, the intrinsic dF/F decreases. First, due to the distance dependence of FRET, a large fluorescence change is expected only if a large distance change occurs. If, however, the fluorophore is located closer to the centerline rather than between the headgroups of the lipids, the distance between DPA and fluorophore does not change as much as predicted (Fig. 5 A, center). In the extreme case that the fluorophore is located directly on the centerline, there will be no distance change at all in the steady state.
Figure 5.
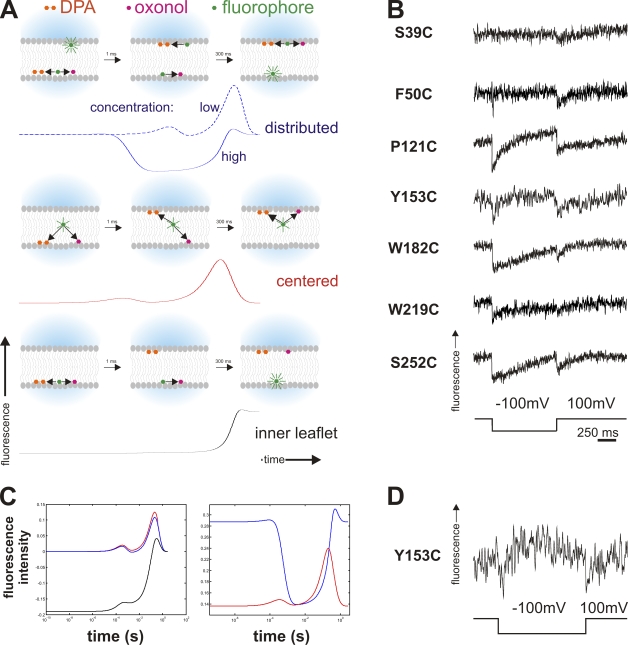
Double FRET experiments. (A) Timeline of double FRET in response to a hyperpolarizing pulse. Depicted are the principle states (as cartoons) and respective simulated fluorescence double FRET signals below for high acceptor concentration for the instances where the fluorophores are distributed between the two leaflets (top), located close to the center (center), or located in just one leaflet (bottom). Black arrows indicate energy transfer between toxin (fluorophore) and acceptor (oxonol and DPA) for close or medium distance. While in the centered case, the fluorescence will show two transients (on a logarithmic scale) during the movement of the acceptors, distributed toxins will lead to a transient fluorescence decrease while fluorophores on both sides are transferring energy to oxonol or DPA. A fluorophore located on only one side will lead to a monotonous increase as the acceptors move away. For distributed donors, the fluorescence response for low acceptor concentrations is shown as a dashed line for comparison (see text for details). (B) Results of the double FRET experiments for the cysteine mutants (Fig. 2 A). The traces show a double-negative transient as a response to a depolarizing pulse. (C) Simulated fluorescence response (semi-logarithmic scale) of toxins located close to the center (red), distributed between both leaflets (blue), and located on only one side (black) for a low (6,400 acceptors/µm2, left) and high (22,300 acceptors/µm2, right) concentration in response to a hyperpolarizing pulse. The negative transient in the fluorescence response for distributed toxins occurs only at high concentrations. (D) Trace of double FRET of Y153C where negative and positive transients are visible. This occurs at intermediate acceptor concentrations.
Second, we may have more than one population of toxins. Toxins are very dynamic and behave differently than integral membrane proteins expressed in the plasma membrane. Consequently, not all toxins need to have the same conformation or to be located at the same position in the membrane. This is particularly likely if the toxin, as we found above, binds to the membrane and relocates to the opposite leaflet. A certain percentage of the toxins will remain on the outer leaflet. In this case, the acceptors move away from one donor population but at the same time closer to the opposite one (Fig. 5 A, top). Consequently, the superposed fluorescence change of both will be dependent on their distribution between both leaflets. If there were exactly the same amount of toxin on both sides, no fluorescence change would be observed in the steady state.
With the steady-state measurements we performed above, it was not possible to explore whether one of the two cases mentioned is responsible for the diminished dF/F or even to distinguish between them. To study the distribution and location of the toxins in more detail, we devised “double FRET” experiments using a single donor (fluorescein) attached to the toxins with two different acceptor molecules with very different translocation time constants, DPA and oxonol (DiSBAC2, Anaspec). DPA and oxonol transit through the membrane with a time constant of 0.5 and 300 ms, respectively (see Supplemental material). The different time constants of DPA and oxonol produce two effects during the translocation leading to a dynamic fluorescence change (see Appendix for details): (1) a transient “dilution” of the oxonol during its slow transition occurs that temporarily increases the donor acceptor distance; and (2) a transient situation arises where DPA is on one and oxonol on the other side. Due to these two effects, different fluorescence signals will be evoked by differently distributed donors. The donors can be distributed between both leaflets (Fig. 5 A, top), centered along the midline of the membrane (Fig. 5 A, center), or located on only one surface of the membrane (Fig. 5 A, bottom).
In case of “distributed” donors, the system is symmetric so that the initial and final fluorescence will be identical. However, during the movement of oxonol, the concentration in both layers is decreased leading to a transient increase in distance to all donors and, thereby, to a transient increase in the donor fluorescence, as shown in the simulated fluorescence traces (Fig. 5 C, left, blue trace). During DPA translocation, the same effect occurs, but it is too rapid to resolve (note that the graph is in semi-logarithmic scale).
In addition, during the interval when DPA is in one leaflet and oxonol on the other, donors on both sides of the bilayer (both populations) are close to an acceptor (Fig. 5 A, top), leading (transiently) to higher transfer efficiency and lower fluorescence (Fig. 5 C, right, blue trace). Although both the positive and negative transients occur at all acceptor concentrations, their relative amplitude differs with the concentration. At lower concentrations, the transient increase is observed, and at high concentrations mainly the transient decrease (see Appendix for details).
For “centered” donors, initial and final donor fluorescence are also equal (Fig. 5 A, centered), and the transient increase due to the “dilution” of the oxonol is observed (Fig. 5 C, left, red trace). In contrast, the fluorescence is not decreased in the interval when DPA and oxonol occupy opposite leaflets. For a fluorophore at the centerline, the distance between donor and acceptor on either leaflet is the same (Fig. 5 C, right, red trace). Finally, a donor located only on one surface of the bilayer still has a small transient due to the dilution of oxonol, but it is insignificant in relation to the large fluorescence change due to the DPA and oxonol translocations (Fig. 5 C, left, black trace).
Thus, investigating the dynamic fluorescence response in double FRET experiments allows us to distinguish between the cases of donors only at one surface of the membrane (Fig. 5 A, bottom), “distributed” donors on both surfaces (Fig. 5 A, top), or “centered” donors located nearer to the centerline of the membrane (Fig. 5 A, center). If we observe a positive transient at low acceptor concentrations, we can be sure that the low dF/F is caused by either distributed toxins or because they are close to the centerline. If we observe a negative transient at higher concentrations, we can be certain that those toxins are distributed on both sides of the membrane.
Distribution of the toxin in the membrane
We tested the double FRET experiments with all our loop mutants and observed for all except S39C (N terminus) a clear transient decrease in donor fluorescence (Fig. 5 B). The transient decrease unambiguously indicated that the toxin is distributed between both leaflets. In P121C, Y153C, and W182C, we occasionally also observed positive transients, which was likely due to a lower acceptor concentration in those bilayers (bleaching), or a negative and positive simultaneously (intermediate concentrations, Fig. 5 D). As mentioned above, positive transients can occur at both centered and distributed toxins. It is not surprising to find the toxin distributed between both sides if it is added to the cis side and has to translocate to the trans leaflet.
The two N-terminal positions F50C and S39C were difficult to recognize. The signal of F50C was not as clear as the other positions. For S39C, we were not able to obtain clear signals in either direction. Instead, we observed slow conformational changes that indicated that the N terminus seems highly movable in the membrane, which is in accordance with our findings in the single FRET measurements. S39C was, apart from the α3–α4 loop, the only position changing its position.
We observed one peculiarity with the position W219C. When polarizing the membrane from positive potentials, the expected negative transient was observed, however, when returning to positive potential, the fluorescence only showed a very slow signal (Fig. 5 B). When inversing the polarities, the slow signal occurs first, followed by the expected fast transient. This signal is explained if the W219 position is moving more toward the midline during the negative membrane potential. Upon return of the DPA, only very small signals will be observed (close to midline), followed by a superposition of the return of the W219 to its original position (closer to the DPA, negative) and the transition of the oxonol (further from the toxin, positive).
In summary, the transients that we observed in the majority of cases indicated that the reduced relative fluorescence change was caused not by additional background fluorescence but rather by the donor (labeled position of the toxin) being distributed between both leaflets. We, thus, have two populations of toxins, one which already translocated and another which remained on the cis side.
DISCUSSION
The FRET measurements in a horizontal planar lipid bilayer geometry presented in this study facilitate rapid determination of the topology of purified toxins while simultaneously monitoring their function via electrophysiological access to the bilayer. We investigated topology and pore formation mechanism of the Cry1Aa toxin of B. thuringiensis and found that domain I of Cry1Aa of B. thuringiensis inserts into the membrane and accumulates in its inner (trans) leaflet. From the data of α1–α2 and DI–DII loops, we know that a fraction of the toxin remains in the external leaflet, which is consistent with the toxin moving through refolding from the outer to the inner leaflet.
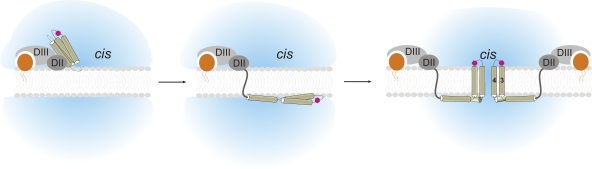
Comparing “inactive” (pH 9) and active (pH 7) conformation, the α3–α4 loop and variably also the N terminus were the only ones that switched their position and were found in the outer leaflet. Because at the same time the α4–α5 loop remained on the internal leaflet, the helix α4 had to be transmembrane and would be the pore-lining helix as demonstrated earlier (Vachon et al., 2004). We cannot, with certainty, decide whether α3 is transmembrane since the α2–α3 loop mutants did not express functionally. However, either α2 or α3 also has to be transmembrane because the α1–α2 loop was localized in the internal leaflet. As the α3–α4 loop is initially located on the internal leaflet, it is more likely that only one hairpin (α3–α4) traverses the membrane in order to form the pores. Moreover, transmembrane structures are a prerequisite for ion conduction, and both α3 and α4 have the correct length for transmembrane helices (22–32 amino acids). The α5–α6 loop was also on the internal side of the membrane but showed some mobility in the double FRET experiments. Mobility of α5 had been shown to be crucial for the pore-forming activity of Cry1Aa (Schwartz et al., 1997). We found that α5–7 are all located in the inner leaflet. The linker between domains I and II, on the other hand, was very flexible, which may be caused by the missing receptor binding of domains II and III. These results suggest that domain I of Cry1Aa traverses by refolding through the membrane to the inner leaflet and forms the pore from the inside by sliding the helices α3 and α4 through the membrane (Fig. 6).
Figure 6.
The proposed mechanism of pore formation. The Cry1Aa toxin first binds to the membrane-bound receptor with domains II and III. In the subsequent step, the α-helical domain I unfolds and transits to the internal (trans) leaflet. For pore formation to occur, the α3–α4 hairpin inserts into the membrane and reaches to the other side. Several toxins oligomerize in order to form the water-filled pore.
This view is reinforced by the functional data. We never observed pore formation when the α3–α4 loop was not located on the external leaflet, although they were located initially in the inner leaflet. We found that the translocation step takes place before observation of pores. We can, thus, conclude that the initial step is translocation of the α3–α4 hairpin. The final step might be oligomerization as the pores are thought to be formed by oligomers; however, from our data we cannot yet decide during which step the toxins oligomerize. Future FRET experiments between toxin monomers may give an answer here. The strict correlation between translocation of the α3–α4 hairpin from the inside to the outside and the subsequent pore formation confirm that α3–α4 translocation is the initial pore-forming mechanism.
It was unexpected to find such a large domain (28 kD) translocating through the membrane. This is a modification of the umbrella model, according to which the helices α4–α5 traverse the membrane from the outside. Also in the umbrella model, the α3–α4 loop is located on the external and the α4–α5 loop on the internal side of the membrane. The difference to our data is the location of the other helices of domain I (i.e., helices not directly involved in pore formation). However, the presence of helices α1–α2 and α5–α7 on the internal leaflet would explain why in atomic force microscopy measurements of Cry1Aa, the observed structures were too small to accommodate the extra helices (Vie et al., 2001; Laflamme et al., 2008).
Cry1Aa had also been proposed to be inserted into the membrane previously (Tomimoto et al., 2006; Nair and Dean, 2008). Nair and Dean (2008) recently used fluorescently labeled cysteine mutants of Cry1Aa and Cry1Ab to monitor whether the fluorophores remained in aqueous solution or entered into the membrane by quenching with external iodide. They found, as us, that the positions that they tested were not exposed to external solution anymore. While their results agree with our findings, they could not further identify the location of the residues with their quenching assay in vesicles. Our method is not restricted to external accessibility, but may be used for any position of the protein within the membrane.
Similarly, Tomimoto et al. (2006) suggested insertion of domain I of Cry1Aa based on digestion patterns (“buried dragon” model). According to Tomimoto’s model, α1 lies on the external surface whereas α2–α7 all are anti-parallel transmembrane helices. Domains II and III are external. The model is based on three digestible sites in the α1–α2 loop, the α3–α4 loop, and domain II. We agree with the α3–α4 loop being accessible. The α1–α2 loop might also in part be accessible for enzymes, considering that we showed that the toxin, and in particular the position F50 in the α1–α2 loop, distributes between both leaflets, so that a population on the external leaflet exists.
While the pore-forming mechanism was already identified using the single FRET assay, the double FRET technique added information about the distribution of the proteins in the membrane. This information is not necessary for well-defined integral membrane proteins, such as ion channels whose topology remains, in general, static, apart from small conformational changes. Toxins, in contrast, are highly dynamic proteins in the membrane, and thus their topology needs to be investigated in more detail. The double FRET technique provides a possibility to obtain this information. The double FRET experiments allowed us to distinguish clearly between background fluorescence and signals coming from the membrane. We were also able to determine the distribution within the membrane, information that was not available with previous methods. While lipids fluorescently labeled at different position in the acyl chain have been used in previous studies, they provide only an averaged static signal lacking the information about the distribution. We found the toxin to be very mobile within the membrane, which may be related to its pore-forming activity. Domains II and III bind in the native environment to the receptor on the external side of the host membrane. Consequently, either domain II or III has to be transmembrane. We tested a few residues on domains II and III (M283, S373, N462, A606; unpublished data), but they also were located in the inner leaflet. However, the bilayer system is receptor free, possibly allowing domains II and III a higher mobility than in the native system.
For proteins expressed in mammalian cells, the dynamic FRET method between protein and membrane can be used in patch-clamped cells (Blunck et al., 2004; Chanda et al., 2005b) or voltage-clamped oocytes (Chanda et al., 2005a). In a cell system, however, the labeling is more susceptible to background fluorescence due to surface cysteines and unspecific binding to the membrane surface. Both effects are minimized in the bilayer system. Background labeling is a major concern for all FRET measurements because nonspecifically attached fluorophores in close proximity to the donor or acceptor would lead to high transfer efficiency and thus to false results. In the bilayer environment, we can verify the specific labeling of the proteins before intercalation into the membrane. The membrane was formed from synthetic lipids only in order to ensure that no background fluorescence originates from the lipids. Black lipid membranes, however, contain a residual amount of solvent (here decane), which slightly increases membrane thickness.
The strategy we chose here closes the gap between the more indirect biochemical approaches and the structure determination by x-ray crystallography or electron microscopy. While it does not provide Å-resolution, it provides structural information in the native conformation. Fluorescence spectroscopy of horizontal planar lipid bilayers gives us electrical control of the membrane simultaneously with optical access. This way, we can observe the conformational changes of the membrane proteins and correlate them with the function (electrical current). To our knowledge, this is the first report of monitoring conformational changes of membrane proteins in planar lipid bilayers under electrophysiological control using fluorescence spectroscopy.
The technique is not restricted to toxins alone, but may be applied to all membrane proteins that can be reconstituted in planar lipid bilayer. We also performed, for instance, fluorescence measurements on reconstituted ion channels in the horizontal planar lipid bilayers (unpublished data). The technique may be transferred even to mammalian cells, as toxins would be labeled before addition to the cells and background fluorescence would be kept to a minimum. We expect the technique to find a widespread application in the investigation of molecular mechanisms of membrane proteins.
APPENDIX A: DISCUSSION OF THE NONLINEAR EFFECTS IN DOUBLE FRET EXPERIMENTS
Transient decrease in acceptor concentration during translocation
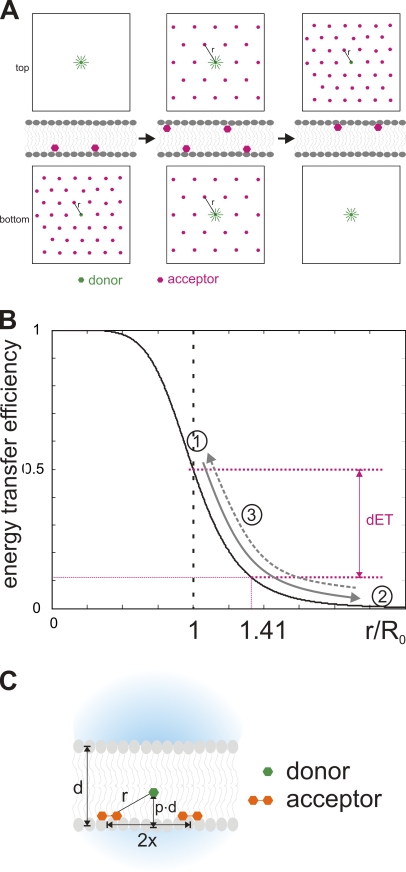
Two effects arise in double FRET experiments with two types of acceptors in the membrane. The first occurs during the transition of either of the two acceptors, and the second is a steady-state effect occurring after the fast translocation of DPA but before the slow one of oxonol. Let us consider first the effect caused by the “dilution” effect of the acceptors during its translocation. We will discuss only one single acceptor type with two donors in opposite leaflets of the membrane upon a hyperpolarizing voltage step. For reasons of symmetry, reversed potentials result in identical signals. Initially, all acceptors are on the internal leaflet (inside is positive). The acceptors have thus a long distance to the donors on the outer leaflet and these are fully fluorescent. On the inner leaflet, the donors have a mean distance r from an acceptor, which is inversely proportional to the square root of the acceptor concentration (see Appendix B). Fig. 7 B shows the dependence of the ET efficiency on the distance. Let us assume that, at the initial concentration, r = R0. Then, 50% of the fluorescence is transferred to the acceptor.
Figure 7.
Transient concentration decrease during translocation. (A) Schematic of the dynamics of acceptor distribution during the translocation process. At positive potential (inside), the negatively charged acceptors are all in the inner leaflet (left). Once the potential reverses to negative, they exponentially relocate to the outer leaflet. At some time during the translocation process, the concentration on both sides is equal (center). Now the mean distance to the acceptor r(t) is 1.41 times longer than initially (inner leaflet). In the new steady state (right), all acceptors are in the outer leaflet, and the situation is inverted to the initial one. (B) Relation between energy transfer efficiency and distance between donor and acceptor. The distance is a function of the concentration of the acceptors in the membrane. Numbers indicate distances (and ET) at the initial state for the inner (1) and outer (2) leaflet, as well as at equal concentrations for both leaflets (3), assuming the concentration is such that the initial value of r(t) = R0. The arrows indicate how the distance and ET change during a voltage pulse (dynamic fluorescence response) for the inner (solid) and outer (dashed) leaflet. dET indicates the difference in energy transfer for the donors in the inner leaflet between initial (1) and equal state (3). A positive transient is generated as a result of the change of ET between (1) and (3) being larger than between (2) and (3). (C) Variables used in formulas: d, thickness of the bilayer; r, distance between donor and acceptor; 2x, mean distance between two acceptors in one leaflet; p, fraction of the bilayer at which the fluorophore is located.
When the potential is reversed, the acceptors leave the inner leaflet and translocate to the outer leaflet with a certain transition probability. Hence, the concentration in the inner leaflet decreases exponentially at the same rate as the concentration in the outer leaflet increases. When half the acceptors have moved, the concentration, and with it the mean distance r, is equal in both leaflets (Fig. 7 A, center). Now donors on the inner and the outer leaflet will also have the same ET efficiency and the same fluorescence intensity (Fig. 7 B, 3). As the concentration is halved on the internal leaflet, the distance is √2 = 1.41 times longer. In our example (r = R0), r increased to 1.41R0 and the energy transfer decreased from 50 to 11.2%. Thus, instead of the outer donors emitting nearly 100% and the inner ones emitting 50%, all donors emit 89.8%. (In fact, if the thickness of the bilayer is ≈R0, it follows from Pythagoras’ rule that r ≈ 1.41R0 and ET ≈ 11%. Thus there is no change yet in the fluorescence of the outer donors.) The overall fluorescence intensity is therefore increased. The underlying reason is that the slope is steeper between R0 and 1.41R0 than at longer distances (Fig. 7 B). However, this is only a transient situation as donors continue to translocate to the outer leaflet. Once all donors are in the outer leaflet, the situation is inverted to the initial one (Fig. 7 A, right). The donors on the outer leaflet now have a mean distance r to the acceptors while the donors on the inner leaflet are relatively far away (fully fluorescent). Thus, due to the nonlinearity in the distance dependence of the ET efficiency, the temporary concentration and distance change during the redistribution of the acceptors lead transiently to higher fluorescence. This effect is significant only if the distance r is close to R0. At higher and lower concentrations/distances, the transient becomes smaller (results of simulations on the concentration dependence are shown in Appendix B).
As we have two acceptors, this positive fluorescence transient occurs twice per reversal of the potential, once when the DPA translocates and a second time when oxonol translocates (Fig. 5 C, left). The DPA transient, however, is too fast to be evident in our system (please note that the time scale is logarithmic). For this reason, it was not detectable in the simple FRET experiments. For a pulse from positive to negative and back to positive potentials, we still obtain two transients for the oxonol, one at the beginning and another at the end of the pulse. For this effect to occur, it makes no difference whether we have two donors equally distributed between the two leaflets or one donor located close to the midline of the bilayer (Fig. 5 C, blue and red traces). The decisive factor is that the system is symmetric and, thus, the averaged distances to acceptors in the initial and final state are (approximately) equivalent. If, in contrast, one donor is located in just one of the leaflets, a steady increase or decrease of donor fluorescence occurs (Fig. 5 C, black trace).
Acceptor distribution leads to transient fluorescence decrease during potential reversal
In the presence of two acceptor types, there develops a second effect originating from the coordination between the two acceptors (Fig. 5 A), which allows us to distinguish between a centered fluorophore or two distributed ones. Let us first assume two donors on opposite leaflets (Fig. 5 A, top). The location of the two types of acceptors on the internal leaflet leads to energy transfer from the fluorophores to both oxonol and DPA. Upon applying a hyperpolarizing pulse, first DPA translocates with a time constant of 0.5 ms, creating a transient situation where the oxonol is located on the inner and DPA on the outer leaflet. Thus, fluorophores (donors) on the inner leaflet will transfer their energy to oxonol and the ones on the outer to DPA with an efficiency dependent on the acceptor concentration. While donors in the inner leaflet will have increased their fluorescence, donors on the outer leaflet will have decreased theirs. The effect on the overall fluorescence is always a decrease but the amount varies with acceptor concentrations.
Let us assume that the concentration is such that r = 0.5R0 for both acceptor types. Then according to Eq. 3, the inner leaflet donors would transfer more than 99% of their energy to the acceptor and less than 1% of the total fluorescence intensity would remain. Under the assumption that R0 equals approximately the bilayer thickness, r would equal 1.41R0 for the outer donors. Hence their fluorescence intensity would be at 67%. After DPA has translocated, donors in both the inner and outer leaflet would fluoresce with 1.5% of their maximal fluorescence intensity. The overall fluorescence intensity would be very low (3% compared with 67%). Once the oxonol has followed to the outer leaflet, the intensity would return to its initial value (Fig. 5 C, right, blue trace). The actual differences in fluorescence intensity are, in fact, slightly lower than calculated above because DPA and oxonol both follow exponential decays. The idealized situation, depicted in our consideration, where all DPA is on one side and all oxonol on the other, never occurs in that clarity. (The calculations for exact simulations are given in Appendix B.)
If a donor is located in the center of the bilayer (Fig. 5 A, center), the distances to the acceptors in the outer or inner leaflet are equal, and thus, no decrease in fluorescence intensity occurs. Here, only the transient increases due to the temporary concentration decreases as described above are observed. Likewise, no negative transient is observed when one fluorophore is located in either the outer or the inner leaflet. During translocation, the acceptors move steadily further away (or closer toward) the fluorophore and a steady increase (decrease) in fluorescence is observed (Fig. 5 A, bottom). Thus, if we observe a negative transient in the double FRET experiments, the donors are distributed between the outer and inner leaflets of the membrane.
Both effects the transient increase as well as the transient decrease occur at all concentrations, but their amplitudes shift from one to the other dependent on the range of concentrations. The two effects have their origin in the nonlinearity of the ET–distance relationship and are optimized at the respective saturation ranges at short and long distance. While at low concentrations, mainly the transient increases are observed, at high concentrations the transient decrease predominates (Fig. 5 C and Fig. 8, E and F).
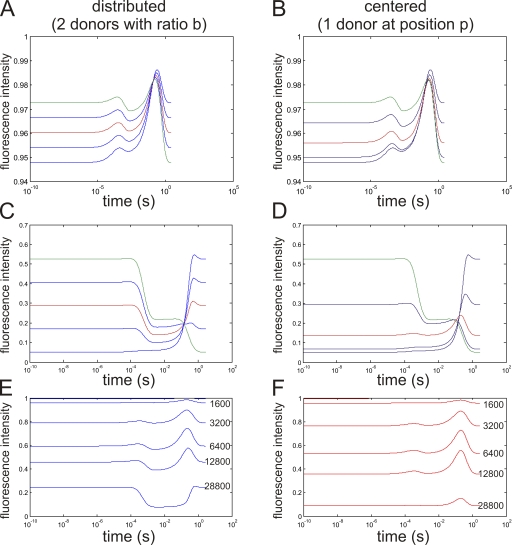
Figure 8.
Simulated double FRET responses. Simulated fluorescence traces (semi-logarithmic scale) for donors distributed between both leaflets (A, C, and E) and donors located at a fraction p of the membrane (B, D, and F). (A and B) Concentration of 1,600 acceptors/µm2 with a distribution between both leaflets of b/(1 − b) or located at a position p in the bilayer. The red curve indicates b = 0.5 (equal distribution) or p = 0.5 (center of the bilayer), respectively. (C and D) Same as A and B, but with a concentration of 22,300 acceptors/µm2. (E and F) b or p = 0.5, with increasing concentrations x acceptors/µm2 (x as indicated).
APPENDIX B: SIMULATIONS OF DONOR FLUORESCENCE IN DOUBLE FRET EXPERIMENTS
Upon inversing the polarity of the membrane from positive to negative potentials, the acceptors translocate from the inner to the outer leaflet following an exponential decay so that the number of acceptors on the inner leaflet for oxonol (Ox(t)i) and DPA (DPA(t)i) is given by
| (A1) |
| (A2) |
where τOx/DPA is the time constant of translocation and Ox(0)/DPA(0) the total number of the respective receptor in the bilayer. The number of acceptors on the outer leaflet is given by
| (A1a) |
| (A2a) |
The mean distance 2x(t) between two acceptors in a leaflet is given by the acceptor concentration:
| (A3) |
with A representing the surface of the bilayer and N the number of acceptors of a specific type in the bilayer leaflet. N(t) will have to be replaced with Ox(t)i, Ox(t)o, DPA(t)i, or DPA(t)o.
Thus, the mean distance between the donor and the nearest acceptor parallel to the membrane is x(t). Acceptors other than the nearest one will not have any significant influence on the FRET efficiency since the rate constant (see below) would be <1.6% of the nearest one.
Thus, we will obtain for each donor four distances to oxonol and DPA on the inner and outer leaflet. If the donor is located at a fraction p of the thickness d of the membrane, the distances r(t) can be calculated using simple geometric considerations (Fig. 7 C):
| (A4) |
and
| (A5) |
dependent on which side the acceptor is located (i: inner; o: outer leaflet). xi and xo are calculated from Eq. A3 with Ni(t) given by Eqs. A1 and A2 for oxonol and DPA, respectively. No(t) is accordingly given by Eqs. A1a and A2a.
The four rate constants of energy transfer kT(t) for each r(t) are given by
| (A6) |
with τD being the fluorescence lifetime of the donor in the absence of the acceptors. The energy transfer efficiency ET is then given by
| (A7) |
With these formulas, the development over time of the fluorescence for donors located at different positions in the membrane can be calculated. (1) A donor located close to the centerline of the membrane (p ≈ 0.5) would have a very small difference whether the acceptors are located outside or inside (p ≈ 1 – p). (2) A donor located inside (p = 0) would always remain relatively far from an acceptor on the outside (r(t) > d) and vice versa for a donor located outside (p = 1). (3) If the donors are distributed on two positions, one inside (p = 0) and another outside (p = 1), the fluorescence intensity Fi/o for both donors are calculated separately and added to the overall fluorescence intensity Ftotal according to the distribution ratio b (number of donors inside/total number of donors): Ftotal = bFi + (1 − b)Fo.
We simulated the time course of fluorescence for fluorophores either located at a single location in the membrane or distributed on both leaflets with variable distribution between both leaflets. The acceptors were all located on the internal leaflet at start and translocated to the outer leaflet with time constants of 0.5 and 300 ms for DPA and oxonol, respectively. The R0 were 45 and 51 Å for DPA and oxonol, respectively. The bilayer was assumed to be 150 µm in diameter with a thickness of 50 Å.
The results are summarized in Fig. 8. In A, C, and E, the results for distributed donors are shown, and in B, D, and F, the results are shown for one donor located at different positions in the membrane. The case of one donor on the outside is included in A–D as a limiting value (b = 0, p = 1). It is marked in green. Red marks the “ideal” case (b = 0.5 and p = 0.5, respectively).
In Fig. 8 (A and B), the fluorescence intensity at a low acceptor concentration (1,600/µm2) is shown. The distribution between the two leaflets (Fig. 8 A) varied from b = 0 to 1 in steps of 0.25. The red trace shows equal distribution between both leaflets. In Fig. 8 B, the fluorescence responses for donors positioned at a fraction p within the bilayer are shown. The fluorescence trace in red here represents the donor located in the center of the bilayer (p = 0.5). In both cases, the trace shows two distinct transient fluorescence increases. The x axis is displayed logarithmic, which means that the first transient occurs extremely fast, while the second, more pronounced one, is slow. The cause for the positive transients is the temporary decrease in concentration of the acceptors. While the concentration in the inner leaflet decreases, it increases on the outer leaflet. During the translocation, the closest acceptor is therefore relatively far from the donors (see Fig. 7). Thus, if a clear transient is observed during a voltage step, the donors, and hence the labeled position of the toxin, have to be either located in the center of the bilayer or distributed between both leaflets (Fig. 5 B).
In Fig. 8 (C and D), the same cases as above are shown for higher concentrations (22,300 acceptors/µm2). For a donor in the center of the bilayer (Fig. 8 D), the shape of the fluorescence intensity curve has not changed qualitatively. For the distributed donors (Fig. 8 C), in contrast, the positive transients have diminished in relation to the total fluorescence change. Instead, a negative transient, most prominent in the red trace, developed. This negative transient occurs during the temporary situation after DPA, but before oxonol, has translocated. In this situation, every donor is close to an acceptor so that overall fluorescence is decreased. If we can observe a negative transient in our experiment, the respective labeled position of the toxin is distributed between both leaflets of the membrane (Fig. 5 D).
In Fig. 8 (E and F), the dependence of the fluorescence responses on the total acceptor concentration is shown. We always kept both acceptors at the same concentration in order to mimic the experimental conditions most accurately. Fig. 8 F shows that there is an optimal concentration of ∼6,400 acceptors/µm2, where the positive transients are highest. At higher concentrations, the negative transient in Fig. 8 E develops while the positive transients decrease.
Acknowledgments
We would like to thank Mireille Marsolais for technical assistance and Luke Masson for providing the Cry1Aa-pMP39, −pBA1, and some of the cysteine mutants. We thank Drs. Raynald Lapraole and Jean-Louis Schwartz for helpful discussions.
This project was funded by the Natural Sciences and Engineering Research Council (327201DG), Canada Research Chairs (202965), and the Canadian Foundation for Innovation (202965) to R. Blunck and by GÉPROM (Fonds de la recherche en santé Québec). N. Groulx holds a student fellowship by the Fonds québécois de la recherche sur la nature et les technologies.
Footnotes
Abbreviations used in this paper:
- BBM
- brush border membranes
- DPA
- dipicrylamine
- EOL
- efficiency of labeling
- ET
- energy transfer
- FRET
- Förster resonance energy transfer
- NMG
- N-methyl-d-glucamine
References
- Akiba T., Abe Y., Kitada S., Kusaka Y., Ito A., Ichimatsu T., Katayama H., Akao T., Higuchi K., Mizuki E., et al. 2008. Crystal structure of the parasporin-2 bacillus thuringiensis toxin that recognizes cancer cells. J. Mol. Biol. 386:121–133 10.1016/j.jmb.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Aronson A.I., Wu D., Zhang C. 1995. Mutagenesis of specificity and toxicity regions of a Bacillus thuringiensis protoxin gene. J. Bacteriol. 177:4059–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A., Van F.K., Brousseau R., Masson L. 2004. The Bacillus thuringiensis Cry1Aa toxin: effects of trypsin and chymotrypsin site mutations on toxicity and stability. J. Invertebr. Pathol. 85:120–127 10.1016/j.jip.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Blunck R., Starace D.M., Correa A.M., Bezanilla F. 2004. Detecting rearrangements of shaker and NaChBac in real-time with fluorescence spectroscopy in patch-clamped mammalian cells. Biophys. J. 86:3966–3980 10.1529/biophysj.103.034512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonserm P., Davis P., Ellar D.J., Li J. 2005. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J. Mol. Biol. 348:363–382 10.1016/j.jmb.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Chanda B., Asamoah O.K., Blunck R., Roux B., Bezanilla F. 2005a. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 436:852–856 10.1038/nature03888 [DOI] [PubMed] [Google Scholar]
- Chanda B., Blunck R., Faria L.C., Schweizer F.E., Mody I., Bezanilla F. 2005b. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat. Neurosci. 8:1619–1626 10.1038/nn1558 [DOI] [PubMed] [Google Scholar]
- Cohen S., Dym O., Albeck S., Ben-Dov E., Cahan R., Firer M., Zaritsky A. 2008. High-resolution crystal structure of activated Cyt2Ba monomer from Bacillus thuringiensis subsp. israelensis. J. Mol. Biol. 380:820–827 10.1016/j.jmb.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Fernandez J.M., Taylor R.E., Bezanilla F. 1983. Induced capacitance in the squid giant axon. Lipophilic ion displacement currents. J. Gen. Physiol. 82:331–346 10.1085/jgp.82.3.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit E., La R.P., Sansom M.S., Shai Y. 1998. The structure and organization within the membrane of the helices composing the pore-forming domain of Bacillus thuringiensis delta-endotoxin are consistent with an “umbrella-like” structure of the pore. Proc. Natl. Acad. Sci. USA. 95:12289–12294 10.1073/pnas.95.21.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochulski P., Masson L., Borisova S., Pusztai-Carey M., Schwartz J.L., Brousseau R., Cygler M. 1995. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J. Mol. Biol. 254:447–464 10.1006/jmbi.1995.0630 [DOI] [PubMed] [Google Scholar]
- Knowles B.H. 1994. Mechanism of action of Bacillus thuringiensis insecticidal delta-endotoxins. Adv. in Insect Physiol. 24:275–308 10.1016/S0065-2806(08)60085-5 [DOI] [Google Scholar]
- Laflamme E., Badia A., Lafleur M., Schwartz J.L., Laprade R. 2008. Atomic force microscopy imaging of Bacillus thuringiensis Cry1 toxins interacting with insect midgut apical membranes. J. Membr. Biol. 222:127–139 10.1007/s00232-008-9106-8 [DOI] [PubMed] [Google Scholar]
- Lakowicz J.R. 1999. Principles of Fluorescence Spectroscopy. Second edition Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- Lee M.K., Young B.A., Dean D.H. 1995. Domain III exchanges of Bacillus thuringiensis CryIA toxins affect binding to different gypsy moth midgut receptors. Biochem. Biophys. Res. Commun. 216:306–312 10.1006/bbrc.1995.2625 [DOI] [PubMed] [Google Scholar]
- Li J.D., Carroll J., Ellar D.J. 1991. Crystal structure of insecticidal delta-endotoxin from Bacillus thuringiensis at 2.5 A resolution. Nature. 353:815–821 10.1038/353815a0 [DOI] [PubMed] [Google Scholar]
- Liu C.Q., Nuttall S.D., Tran H., Wilkins M., Streltsov V.A., Alderton M.R. 2008. Construction, crystal structure and application of a recombinant protein that lacks the collagen-like region of BclA from Bacillus anthracis spores. Biotechnol. Bioeng. 99:774–782 10.1002/bit.21637 [DOI] [PubMed] [Google Scholar]
- Masson L., Prefontaine G., Peloquin L., Lau P.C., Brousseau R. 1990. Comparative analysis of the individual protoxin components in P1 crystals of Bacillus thuringiensis subsp. kurstaki isolates NRD-12 and HD-1. Biochem. J. 269:507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair M.S., Dean D.H. 2008. All domains of Cry1A toxins insert into insect brush border membranes. J. Biol. Chem. 283:26324–26331 10.1074/jbc.M802895200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara K., Indrasith L.S., Asano S., Hori H. 1992. Processing of delta-endotoxin from Bacillus thuringiensis subsp. kurstaki HD-1 and HD-73 by gut juices of various insect larvae. J. Invertebr. Pathol. 60:121–126 10.1016/0022-2011(92)90084-H [DOI] [PubMed] [Google Scholar]
- Pantoja R., Sigg D., Blunck R., Bezanilla F., Heath J.R. 2001. Bilayer reconstitution of voltage-dependent ion channels using a microfabricated silicon chip. Biophys. J. 81:2389–2394 10.1016/S0006-3495(01)75885-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyronnet O., Vachon V., Schwartz J.L., Laprade R. 2000. Ion channel activity from the midgut brush-border membrane of gypsy moth (Lymantria dispar) larvae. J. Exp. Biol. 203:1835–1844 [DOI] [PubMed] [Google Scholar]
- Peyronnet O., Vachon V., Schwartz J.L., Laprade R. 2001. Ion channels induced in planar lipid bilayers by the Bacillus thuringiensis toxin Cry1Aa in the presence of gypsy moth (Lymantria dispar) brush border membrane. J. Membr. Biol. 184:45–54 10.1007/s00232-001-0071-8 [DOI] [PubMed] [Google Scholar]
- Ries R.S., Choi H., Blunck R., Bezanilla F., Heath J.R. 2004. Visualizing the substrate dependence on the structure and dynamics of black lipid membranes. J. Phys. Chem. B. 108:16040–16049 10.1021/jp048098h [DOI] [Google Scholar]
- Schnepf E., Crickmore N., Van R.J., Lereclus D., Baum J., Feitelson J., Zeigler D.R., Dean D.H. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J.L., Laprade R. 2000. Membrane permeabilization by Bacillus thuringiensis toxins: protein insertion and pore formation. Entomopathogenic Bacteria: From Laboratory to Field Application. Charles J.F., Delecluse A., Nielsen-Le Roux C., Kluwer Academic Publishers, New York: 199–218 [Google Scholar]
- Schwartz J.L., Lu Y.J., Sohnlein P., Brousseau R., Laprade R., Masson L., Adang M.J. 1997. Ion channels formed in planar lipid bilayers by Bacillus thuringiensis toxins in the presence of Manduca sexta midgut receptors. FEBS Lett. 412:270–276 10.1016/S0014-5793(97)00801-6 [DOI] [PubMed] [Google Scholar]
- Tomimoto K., Hayakawa T., Hori H. 2006. Pronase digestion of brush border membrane-bound Cry1Aa shows that almost the whole activated Cry1Aa molecule penetrates into the membrane. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 144:413–422 10.1016/j.cbpb.2006.04.013 [DOI] [PubMed] [Google Scholar]
- Vachon V., Prefontaine G., Rang C., Coux F., Juteau M., Schwartz J.L., Brousseau R., Frutos R., Laprade R., Masson L. 2004. Helix 4 mutants of the Bacillus thuringiensis insecticidal toxin Cry1Aa display altered pore-forming abilities. Appl. Environ. Microbiol. 70:6123–6130 10.1128/AEM.70.10.6123-6130.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vie V., Van M.N., Pomarede P., Dance C., Schwartz J.L., Laprade R., Frutos R., Rang C., Masson L., Heitz F., Le G.C. 2001. Lipid-induced pore formation of the Bacillus thuringiensis Cry1Aa insecticidal toxin. J. Membr. Biol. 180:195–203 10.1007/s002320010070 [DOI] [PubMed] [Google Scholar]