The first crystal structures of iNKT cell TCRs bound to complexes of CD1d and microbe-derived glycolipids provide insight into the structural basis of iNKT cell microbial antigen recognition.
Abstract
Invariant natural killer T cells (iNKT cells) rapidly produce effector cytokines. In this study, we report the first crystal structures of the iNKT cell T cell receptor (TCR) bound to two natural, microbial glycolipids presented by CD1d. Binding of the TCR induced CDR3-α–dependent structural changes in the F′ roof of CD1d; these changes resemble those occurring in the absence of TCR engagement when the highly potent synthetic antigen α-galactosylceramide (α-GalCer) binds CD1d. Furthermore, in the Borrelia burgdorferi α–galactosyl diacylglycerol–CD1d complex, TCR binding caused a marked repositioning of the galactose sugar into an orientation that closely resembles α-GalCer. The TCR-dependent reorientation of the sugar, together with the induced CD1d fit, may explain the weaker potency of the microbial antigens compared with α-GalCer. We propose that the TCR of iNKT cells binds with a conserved footprint onto CD1d, regardless of the bound glycolipid antigen, and that for microbial antigens this unique binding mode requires TCR-initiated conformational changes.
Type I, or invariant natural killer T cells (iNKT cells) are an evolutionary conserved population of lymphocytes (Bendelac et al., 2007). Mouse iNKT cells express a semi-invariant TCR encoded by a Vα14-Jα18 rearrangement, whereas humans express an orthologous Vα24 segment also with an invariant rearrangement to Jα18 (Bendelac, 1995; Bendelac et al., 2007). iNKT cells are reactive to CD1d, a nonclassical MHC class I antigen–presenting molecule. Unlike MHC class I antigen–presenting molecules, which are encoded by highly polymorphic genes and which present peptides, CD1d is part of the nonpolymorphic CD1 gene family whose proteins are specialized for presenting various lipids (Brigl and Brenner, 2004). The antigens are bound to CD1 molecules with the lipid backbone inside a hydrophobic binding groove that is formed between two antiparallel helices that sit above an antiparallel β-sheet platform. The glycolipid head group, typically a carbohydrate, is presented at the CD1 surface for interaction with the TCR (Moody et al., 2005; Zajonc and Kronenberg, 2007; Godfrey et al., 2010).
iNKT cells respond well to glycosphingolipids (GSLs) presented by CD1d, including the model synthetic antigen α-galactosylceramide (α-GalCer) and microbial antigens such as α-galacturonosylceramide (GalA-GSL) from Sphingomonas spp. bacteria (Kawano et al., 1997; Kinjo et al., 2005). They also recognize an α-galactosyl diacylglycerolipid (α-GalDAG), called BbGL-2c, from Borrelia burgdorferi (Kinjo et al., 2006), which is the causative agent of Lyme disease. These glycolipids share a molecular pattern, in which a hexose sugar is linked to a lipid backbone, either ceramide or diacylglycerol, via an α-glycosidic bond. The CD1d antigen–binding groove has two large pockets, A′ and F′. Although GSLs bind in a conserved orientation in the CD1d-binding groove, with the fatty acid chain always inserted into the A′ pocket and the sphingosine chain accommodated within the F′ pocket, binding of diacylglycerol-based ligands is more flexible. We recently showed that that the A′ pocket favors binding of a C18:1 monounsaturated fatty acid of borrelial α-GalDAG. Depending on the position of the glycerol to which this fatty acid is attached, either sn-1 or sn-2, the glycolipid can bind in opposite orientations in the CD1d groove, which can greatly affect antigenicity because of a drastically different presentation of the sn-3–linked sugar head group (Wang et al., 2010).
Although crystal structures of human CD1a, -b, and -d, as well as mouse CD1d (mCD1d) in complex with different glycolipids exist (Moody et al., 2005; Zajonc and Kronenberg, 2007, 2009; Zajonc and Wilson, 2007), α-GalCer is the only antigen that has been crystallized bound to both mouse and human CD1d, as well as in complex with the iNKT cell TCR (Koch et al., 2005; Zajonc et al., 2005a; Borg et al., 2007; Pellicci et al., 2009). The crystal structures of the mouse and human TCR ternary complexes revealed an evolutionarily conserved binding mode, where the TCR adopts a unique, parallel orientation above the CD1d molecule, unlike the more commonly found diagonal footprint for peptide MHC-restricted TCRs (Rudolph et al., 2006). Although the TCR-α chain is situated directly above the opening to the CD1d-binding groove, perfectly positioned to interact with the galactose and polar regions of α-GalCer, the TCR-β chain is offset to the C-terminal end of the α1 helix of CD1d, above the closed end of F′ pocket of CD1d. As a result, the complementarity-determining regions (CDRs) of the TCR-α chain primarily interact with the glycolipid (CDR1-α and CDR3-α), whereas the TCR-β chain forms contact through CDR2-β with CD1d residues. CDR3-β does not interact with the antigen but, instead, modulates the TCR affinity (Mallevaey et al., 2009). The TCR of Vα14 iNKT undergoes no conformational changes in the CDR loops upon antigen binding, unlike classical peptide MHC-restricted TCRs, which show a high degree of plasticity in the receptor–ligand interface (Pellicci et al., 2009). Instead, the iNKT cell TCR binds in a lock and key mode and is therefore considered to display innate-like properties.
iNKT cells are important for the clearance of microbes that have antigens for the invariant TCR (Kinjo et al., 2005, 2006; Mattner et al., 2005; Tupin et al., 2008), although the affinity of these natural antigens for the invariant TCR is reduced ∼50–500-fold compared with the binding to α-GalCer complexes with mCD1d (Wang et al., 2010). The structural basis of microbial antigen recognition and the reasons for the decreased TCR affinity for these antigens remain unknown. In this study, we address these issues by determining the crystal structures of two of the known microbial antigens, Sphingomonas GalA-GSL and B. burgdorferi BbGL-2c, in ternary complexes with mCD1d and the semi-invariant TCR. Our data demonstrate a conserved binding mode with significant conformational changes induced by the TCR that explains the weaker TCR affinity of the natural antigens.
RESULTS
Overall structures
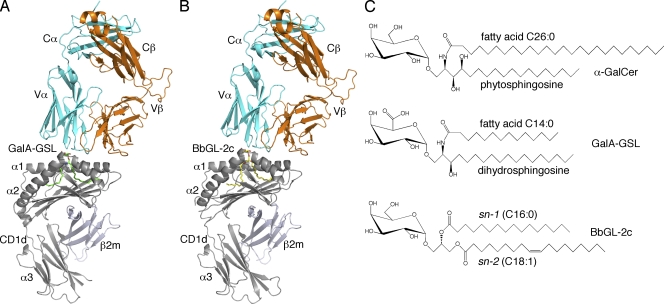
To address the structural basis for microbial glycolipid antigen recognition by mouse iNKT TCRs, we have determined the crystal structures of ternary complexes with Sphingomonas GSL antigen, mCD1d–GalA-GSL–TCR, and B. burgdorferi diacylglycerol antigen, mCD1d–BbGL-2c–TCR, at a resolution of 2.74 Å and 2.8 Å, respectively (Fig. 1 and Table I). Both complex structures are highly similar overall to the mCD1d–α-GalCer–TCR complex structure previously described (Pellicci et al., 2009): the iNKT cell TCR docks parallel to the CD1d-binding cleft, with the α chain above the F′ pocket (Fig. 1). Only the TCR α chain contacts the glycolipid (discussed in detail in TCR–lipid interaction and the TCR footprint), whereas the β chain forms conserved contacts with CD1 residues, mainly through CDR2-β. In addition, the unique CDR3-β of the TCR used in this study also forms contacts with CD1d (Table S1). The electron densities corresponding to both glycolipid ligands, especially for the polar moieties, are surprisingly very well defined, in contrast to previous structures determined in the absence of bound TCR (Fig. 2 and Fig. S1; Wu et al., 2006; Wang et al., 2010). In both structures, the electron density fully engulfs the galactose or galacturonic head groups, as well as the lipid backbones. As reported previously for GSL antigens (Zajonc and Wilson, 2007), the fatty acid and sphinganine tails of GalA-GSL are inserted into the A′ and F′ pockets, respectively. The palmitic acid spacer lipid, previously found in the A′ pocket of the mCD1d–GalA-GSL complex (Wu et al., 2006), was not observed here. Instead, the shorter fatty acid (C14:0) is inserted straight into the A′ pocket in a clockwise rather than the previously reported counterclockwise orientation (Wu et al., 2006). In that binding orientation, the fatty acid does not form the typical kink as is observed for the BbGL-2c ligand (Fig. 2). The reason for the different binding modality of the fatty acid in the A′ pocket of mCD1d is currently unknown, but it does not appear to alter the overall position of the lipid backbone inside the mCD1d groove. In the ternary complex with BbGL-2c, the sn-1–linked oleic acid was bound inside the A′ pocket and sn-2–linked palmitic acid in the F′ pocket, similar to the mCD1d–BbGL-2c complex in the absence of the TCR (Wang et al., 2010). Overall, the better-defined and unbiased electron density for both ligands after TCR binding suggests that upon TCR engagement, the polar moieties of both ligands are locked in a rigid binding orientation, whereas each ligand bound to CD1d only is slightly more flexible, resulting in less well-defined electron density (Wu et al., 2006; Wang et al., 2010).
Figure 1.
Recognition of microbial glycolipids by the iNKT cell TCR. (A) Structure of the TCR bound to mCD1d–GalA-GSL. (B) Structure of the TCR bound to mCD1d–BbGL-2c. (A and B) GalA-GSL is shown in green; BbGL-2c is shown in yellow; mCD1d heavy chain and β2m are shown in gray; TCR α chain is shown in cyan; TCR β chain is shown in orange. (C) Chemical structures of α-galactosyl–containing sphingolipids (α-GalCer and GalA-GSL) and diacylglycerolipid BbGL-2c.
Table I.
Data collection and refinement statistics
| Data collection and refinement statistics | mCD1d–BbGL-2c–TCR | mCD1d–GalA-GSL–TCR |
| Data collection | ||
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a, b, c (Å) | 78.00, 188.37, 149.79 | 78.93, 191.24, 151.21 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 40.0–2.80 [2.90–2.80] | 50.0–2.74 [2.84–2.74] |
| Number of reflections | 27,545 | 29,275 |
| Rmerge (%) | 11.7 [64.1] | 10.4 [46.4] |
| Multiplicity | 7.3 [7.4] | 5.5 [5.5] |
| Average I/σI | 19.7 [3.0] | 15.3 [2.7] |
| Completeness (%) | 99.9 [99.9] | 95.9 [97.5] |
| Refinement | ||
| Number of atoms | 6,599 | 6,606 |
| Protein | 6,413 | 6,371 |
| Ligand | 53 | 48 |
| Carbohydrate | 80 | 80 |
| Waters | 53 | 107 |
| R/Rfree | 0.203/0.259 | 0.198/0.253 |
| Ramachandran plot | ||
| Favored (%) | 96.6 | 96.7 |
| Allowed (%) | 99.8 | 100.0 |
| RMSDs | ||
| Bonds (Å) | 0.008 | 0.012 |
| Angles (°) | 1.13 | 1.43 |
| B-factors | ||
| Protein (Å2) | 48.0 | 41.4 |
| Ligand (Å2) | 63.7 | 26.3 |
| Carbohydrate (Å2) | 68.5 | 56.1 |
| Waters (Å2) | 40.8 | 33.7 |
Numbers in brackets refer to the highest resolution shell.
Figure 2.
Electron density maps for the microbial glycolipids. (A–D) Representation of the final 2Fo-Fc maps drawn around glycolipids GalA-GSL (A and C) and BbGL-2c (B and D) from the ternary mCD1d–glycolipid–TCR complexes. The maps are contoured at 1 σ and shown as a blue mesh in side view (A and B) and looking down into the binding groove from the top (C and D). Hydrophobic mCD1d residues interacting with the lipid backbone and charged residues contacting the polar moieties of the glycolipid sugar are depicted. The α2 helix is removed for clarity in A and B. Relative positions of TCR CDR1-α, CDR3-α, and CDR3-β are depicted for general orientation in A and B.
Comparison of the complexes with and without the TCR
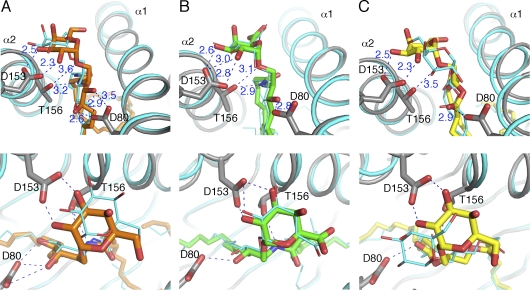
Previously, the structure of the mCD1d–PBS25 complex, which is an analogue of α-GalCer with an eight-carbon instead of C26:0 fatty acid chain, has been determined (Zajonc et al., 2005a). Likewise, the structure of the mCD1d–α-GalCer–TCR ternary complex, with the GSL antigen having the full C26:0 fatty acid chain, has also been determined (Pellicci et al., 2009). We compared three pairs of mCD1d complex structures to evaluate the position of the different glycolipid antigens, α-GalCer, GalA-GSL, and BbGL-2c, binding to mCD1d before and after interacting with the TCR (Fig. 3). Upon TCR binding to α-GalCer, the head group of α-GalCer is shifted by 1 Å toward the α1 helix, as reported previously (Pellicci et al., 2009). Interestingly, GalA-GSL has very minimal conformational change upon TCR binding. However, BbGL-2c shows the most significant change upon interacting with the TCR. The galactose moiety tilts toward the α2 helix by ∼30° (angle of the glycosidic bond), and furthermore, rotates 60° in a clockwise orientation when viewed from above. This causes the 6′ OH group to point toward the α1 helix instead of the α2 helix. Consequently, the 2′ and 3′ galactose hydroxyl groups are moved closer to the α2 helix to form H bonds with Asp153 of mCD1d, as has been observed for α-GalCer and for GalA-GSL, even without bound TCR. In contrast, the 2′ OH loses an H bond to Arg79 because it moves away from the α1 helix. Overall, BbGL-2c forms less extensive H bonds than either α-GalCer or GalA-GSL and has the biggest repositioning of the galactose group upon TCR binding, which involves the breaking of two H bonds, together with the loss of several van der Waals interactions (involving in particular residues Phe70, Tyr73, and Ser76 of mCD1d) while forming four new polar interactions.
Figure 3.
Comparison of α-GalCer, GalA-GSL, and BbGL-2c binding to mCD1d before and after TCR binding. (A–C) H-bond interactions between glycolipids and mCD1d in a front (top) and top view (bottom). mCD1d–glycolipid structures before TCR binding are shown in cyan. In each of the three ternary complexes, mCD1d is labeled in gray, and glycolipids are shown in sticks and different colors. α-GalCer is shown in orange; GalA-GSL is shown in green; BbGL-2c is shown in yellow. H bonds are illustrated as blue dashed lines with distances labeled in angstroms.
TCR–lipid interaction and the TCR footprint
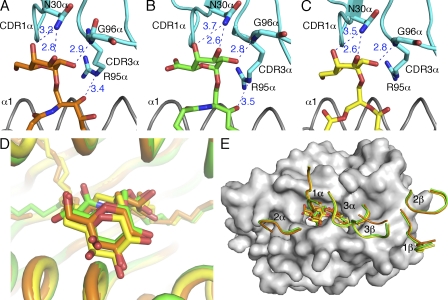
To compare TCR recognition of the different antigens, the interactions between TCR and the three ligands were analyzed (Fig. 4 and Table S1). In each of the three complexes, the 2′ hydroxyl of the galactose forms an H bond with Gly96-α of CDR3-α, whereas the 3′ and 4′ hydroxyl groups H bond to Asn30-α of CDR1-α. However, note that the H bond between the 4′ OH of GalA-GSL and N30-α is 3.7 Å, which slightly exceeds the maximal distance (3.5 Å) for typical H bonds. TCR residue Arg95-α (CDR3-α) H bonds with the 3′ hydroxyl of the sphingosine chain in α-GalCer and GalA-GSL ternary structures, whereas in the BbGL-2c complex, this moiety is not present. As a result, although both α-GalCer and GalA-GSL form four H bonds with the TCR, BbGL-2c forms only three and, as such, therefore may interact less avidly with the TCR.
Figure 4.
Interactions of the TCR with mCD1d-presented glycolipids. (A–C) H-bond interactions between the mCD1d-bound glycolipids and the TCR are depicted as blue dashed lines with distances labeled in angstroms. The CDR1-α and CDR3-α regions are shown in cyan. α-GalCer is shown in orange; GalA-GSL is shown in green; BbGL-2c is shown in yellow. The side chains of residue N30-α, G96-α, and R95-α that make contact with the glycolipids are shown. (D) Comparison of the glycolipids in the ternary complex structures after superimposition. Glycolipids are shown in sticks. (E) Comparison of footprints of the TCR on the surface of mCD1d. CDR loops are labeled and colored identically to the glycolipids. Note that the H bond between the 4′ OH of GalA-GSL and N30-α is 3.7 Å, which slightly exceeds the maximal distance (3.5 Å) for typical H bonds.
The presentation modes of the three ligands in the ternary complexes could be compared by superimposing the three structures. Even though the three ligands have different conformations when bound to mCD1d, they are adjusted into almost identical positions above the mCD1d-binding groove when the TCR is bound (Fig. 4 D; maximum root mean square deviation [RMSD] of 1.1 Å between all carbohydrate moieties). Comparing the three lipids, the conformation of GalA-GSL shows very little change when interacting with the TCR. These data suggest that this lipid is displayed in an optimized position in the binding groove of mCD1d, although the electron density in the TCR uncomplexed structure was the least defined. Therefore, GalA-GSL binding to mCD1d may be the most flexible, or alternatively, the ligand occupancy was not close to 100%, suggesting that there was still some endogenous ligand bound to mCD1d, which would affect the interpretation of the electron density map (Wu et al., 2006). The overall orientations of the TCR’s six CDR loops on the surface of mCD1d were compared when viewed from the side of the TCR (Fig. 4 E). All of the three CDR-α loops and CDR1-β and CDR2-β loops adopt almost identical conformations (RMSDs <0.5 Å in pairwise comparisons of the Cα atoms). However, the CDR3-β loop in the α-GalCer structure is partially disordered and not aligned with the two microbial lipid structures. This conformation difference is not due to the difference of TCR recognition footprint but a result of different CDR3-β usage. The TCR in the complex of α-GalCer has a different CDR3-β sequence (CASGDAGGNYAEQFF), which results in a 3-aa-longer CDR3-β loop, compared with the Vα14Vβ8.2 TCR from the 2C12 hybridoma used in this study (CASGDEGYTQYF). Not surprisingly, our TCR has a higher affinity toward mCD1d–α-GalCer (KD = 11 nM instead of KD = 29–69 nM; Pellicci et al., 2009; Wang et al., 2010), as the CDR3-β loops form additional contacts, including one salt bridge with CD1d (Table S1). Overall, the converged glycolipid conformations and TCR footprint reveal a highly conserved TCR recognition pattern, even for antigens that initially bind in a different orientation to mCD1d. In conclusion, the TCR has the capacity to reorient antigens to a preferred position for a conserved interaction, rather than exhibiting a plasticity in ligand recognition, which would have resulted in slightly different CDR loop conformations and possibly a variable binding footprint.
A preformed F′ roof may modulate TCR affinity
Previously, we have reported the equilibrium binding kinetics of the Vα14Vβ8.2 TCR used in this study for mCD1d loaded with α-GalCer, GalA-GSL, or BbGL-2c (Wang et al., 2010). The TCR shows the highest binding affinity for mCD1d–α-GalCer complexes (KD = 11 nM; Ka = 1.3 × 105 [M−1s−1]; Kd = 1.45 × 10−3 [s−1]) and the lowest affinity for mCD1d–BbGL-2c complexes (KD = 6.2 µM; Ka = 0.216 × 105 [M−1s−1]; Kd = 0.165 [s−1]). GalA-GSL has an intermediate binding to the invariant TCR, with a KD of 0.69 µM (Ka = 1.43 × 105 [M−1s−1]; Kd = 0.094 [s−1]; Wang et al., 2010). Both α-GalCer and GalA-GSL have similar fast TCR association rates, likely because of the relatively little reorientation of the glycoepitope of these antigens by the TCR (Fig. 3, A and B). In contrast, binding of the TCR to mCD1d–BbGl-2c is much slower, as the TCR has to reorient the BbGl-2c head group to obtain the conserved interaction (Figs. 3 C and 4 E). Interestingly, the dissociation rate is much faster for both GalA-GSL and BbGL-2c, even though all three ligands bind in a highly similar orientation in the TCR ternary complex (Fig. 4 D). We have further calculated the activation energies of TCR binding to the various mCD1d–glycolipid complexes from the kinetic equilibrium binding data (Table S2). We calculated the variation of the free energy standard (ΔG°) from the KD and converted Ka to an activation energy standard (Eα°), similar as reported previously (Wu et al., 2002; Sidobre et al., 2004). The comparison, calculated as ΔEα° normalized by ΔΔG° and expressed as phi (Φ) values, allowed us to investigate the effect of the different glycolipids moieties on the initial association phase. A Φ value of 0 indicates that the contact is not formed in the transition state nor participates in the initial association, whereas contacts for which values are higher are involved in the initial interaction. As GalA-GSL has a Φ value close to 0, the GalA-GSL modifications appear to have no effect on the initial TCR association. Given the peripheral position of the modification at the 6′ of the GalA-GSL sugar, this finding rules out a role for the 4-OH group of the α-GalCer ceramide backbone (missing in GalA-GSL) in promoting the initial phase of complex formation,. For BbGL-2c, the calculated Φ value of 0.28 suggests instead that the BbGL-2c ligand is partially involved in the transition intermediate. As observed for other ligands (Sidobre et al., 2004), the polar head is likely to be contacted first by the TCR in the initial association phase, and it is therefore possible that, in the case of BbGL-2c, it directly influences the Ka because of the need of the TCR to rearrange its orientation for proper binding.
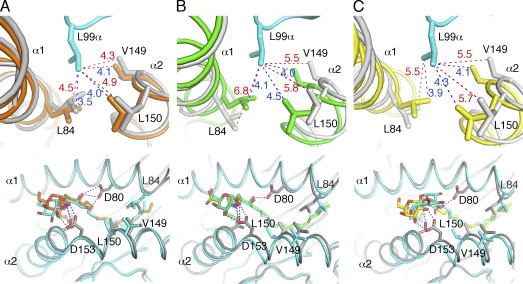
To address why the TCR dissociates faster from mCD1d when either GalA-GSL or BbGl-2c is bound, we compared the molecular surfaces of each pair of mCD1d–glycolipid and mCD1d–glycolipid–TCR ternary structures to reveal differences in the mCD1d–glycolipid complex before and after TCR binding (Fig. 5). As previously reported, for the mCD1d–PBS25 complex, mCD1d forms a roof over the F′ pocket (F′ roof), which buries the tail of the sphingosine chain underneath (Zajonc et al., 2005a). Interestingly, the roof is not present when either GalA-GSL or BbGL-2c binds to mCD1d (Fig. 5, top; Zajonc et al., 2005a; Wang et al., 2010), suggesting that α-GalCer, or in this case the short acyl chain variant PBS25, is exclusively able to induce structural changes above the F′ pocket. However, surprisingly, the F′ roof also forms above the two microbial ligands upon TCR binding (Fig. 5, bottom). Detailed structural comparison of the mCD1d structure before and after TCR binding identified major side chain as well as main chain movement of mCD1d residues L84, V149, and L150. This movement is likely induced upon insertion of a key TCR residue, L99 of CDR3-α, into the mCD1d-binding groove, which acts as a hydrophobic finger to attract those residues through nonpolar van der Waals interactions (Fig. 6). The importance of L99-α has previously been revealed by a site mutagenesis study (Scott-Browne et al., 2007), with an L99A substitution resulting in complete loss of T cell activation by α-GalCer. In each of the ternary complex structures, L99-α was positioned right above the roof and directly made contact with the roof-forming residues through nonpolar van der Waals interactions. The side chains of L84, V149, and L150 were in comparable distances (between 4 and 4.5 Å) to L99-α before and after TCR docked into the mCD1d–α-GalCer complex. However, for GalA-GSL and BbGL-2c, the distances between the side chains of L99-α to those of L84, V149, and L150 were >5.5 Å and shortened to around 4.0 Å only after TCR binding. Therefore, the roof formed by these three residues over the F′ pocket in mCD1d appears to be a structural feature that is required for optimal interaction with the TCR. α-GalCer, the only glycolipid that induces the formation of the roof upon mCD1d binding in the absence of the TCR, readily endows mCD1d with an optimized TCR recognition surface and thus has the highest binding avidity toward the TCR, in part because of the slow TCR dissociation. However, the microbial antigens GalA-GSL and BbGL-2c lack the preformed hydrophobic roof over the F′ pocket when bound inside the mCD1d groove, which is energetically less favorable for the recognition by the TCR, and therefore, they are relatively weaker antigens. The hydrophobic finger L99 of the TCR has to maintain the F′ roof, likely by paying an energetic penalty that leads to faster TCR dissociation.
Figure 5.
Comparison of the mCD1d antigen–binding grooves before and after TCR binding. (A–C) Binding groove portals of mCD1d with bound glycolipids α-GalCer (A), GalA-GSL (B), and BbGL-2c (C) before (top) and after (bottom) TCR binding. mCD1d is shown as a gray surface. Lipid antigens are shown as sticks, with α-GalCer in orange, GalA-GSL in green, and BbGL-2c in yellow.
Figure 6.
Comparison of the induced fit on the α1 and α2 helices caused by TCR binding. (A–C) CD1d-glycolipid structures (gray) are superimposed with ternary complexes after TCR binding. Interactions of TCR residue Leu99-α with the side chains of Leu84, Val149, and Leu150 of mCD1d (top). TCR is shown in cyan; mCD1d complexed with TCR is shown in orange when presenting α-GalCer (A), in green for GalA-GSL (B), and in yellow for BbGL-2c (C). The bottom panels show TCR view of the CD1d–glycolipid complexes (cyan) superimposed onto the CD1d–glycolipid structures after TCR binding (CD1d is shown in gray, and glycolipids colored as in the top panels). Van der Waals interactions are depicted as dashed lines and are labeled with distances in angstroms before (red) and after (blue) TCR binding.
The different abilities to preform the roof likely arise from the structural differences of the three glycolipids. GalA-GSL contains a sphinganine chain with a 3-OH group, whereas short-chain and full-length α-GalCer have a phytosphingosine tail with both 3-OH and 4-OH groups. When α-GalCer is bound, the three H bonds formed by both the 3-OH and 4-OH groups with Asp80 in the mCD1d, together with the extensive H bond formed by α-GalCer with the α2 helix, likely pull the backbone segments of α1 and α2 helices over the F’ pocket closer toward each other. This backbone change is accompanied by the repositioning of the side chains of L84, V149, and L150, causing formation of the roof (Zajonc et al., 2005a). The preset favorable conformation on the mCD1d surface does not change when the TCR binds, and there are no further gross changes in the distance between the backbones of α1 and α2 helices delineating the F′ pocket. GalA-GSL, which lacks the 4-OH group, forms only one H bond with 3-OH to Asp80 in the α1 helix, which appears not strong enough to induce the mCD1d conformational change. TCR residue L99-α inserts into the F pocket and pulls the side chains of L84, V149, and L150 closer, narrowing the backbones of α1 and α2 helices over the F′ pocket to form the F′ roof (Fig. 6).
For BbGL-2c, the 2′ hydroxyl group of the galactose forms one H bond with Asp80 and another H bond with Arg79. However, there is only one weak H bond formed between the oxygen atom of the sn-2–linked fatty acid and Thr156 on the α2 helix. Thus, the weak interaction between BbGL-2c and mCD1d is not able to cause induced formation of the roof. Additionally, the head group of BbGL-2c is not positioned similar to α-GalCer or GalA-GSL, thereby losing two crucial H bonds with Asp153. The interaction of the TCR with mCD1d presenting BbGL-2c will induce dramatic rotation of the galactose. As a result, the unfavorable conformation of BbGL-2c in the mCD1d-binding groove results in the weakest binding affinity to the TCR among the three glycolipid antigens.
DISCUSSION
We have proposed that the semi-invariant TCR of iNKT cells is selected for the recognition of glycolipid antigens from pathogenic microbes, which most likely have a diacylglycerol rather than a ceramide lipid, as ceramides are confined to relatively nonpathogenic Sphingomonas spp. (Kinjo et al., 2005; Mattner et al., 2005; Sriram et al., 2005). However, the first antigen discovered, which is the most potent and also the one most widely used in various experimental studies and clinical trials, is the ceramide-containing antigen α-GalCer (Kawano et al., 1997). Despite information on α-GalCer recognition, the structural basis for the TCR-mediated recognition of natural microbial antigens, including those with a diacylglycerol lipid, has remained unknown. Furthermore, it has been uncertain why complexes of these microbial antigens with mCD1d have a KD for the TCR that is ∼50–500-fold reduced compared with α-GalCer (Wang et al., 2010). In this study, we have addressed these important issues by determining the three dimensional structures of two natural microbial antigens in ternary complexes with mCD1d and the TCR of iNKT cells.
Although the glycolipid antigens from Sphingomonas spp. and B. burgdorferi have structural similarity to the model antigen α-GalCer, at least in terms of having a hexose sugar in α-linkage to a lipid with two aliphatic chains, their binding orientation to mCD1d was previously shown to be different, most notably for the borrelial α-GalDAG antigen BbGL-2c (Wang et al., 2010). Therefore, it is surprising that, upon TCR binding to the mCD1d–glycolipid complexes, most TCR contacts are conserved among all three structures. Furthermore, comparison of the crystal structures of those glycolipid antigens bound to mCD1d before and after TCR binding enabled us to gain insight into the mechanism of TCR recognition, including the remarkable capability of the TCR to reorient suboptimally bound glycolipids to obtain the conserved orientation of the sugar and the TCR footprint on mCD1d. Unexpectedly, for the diacylglycerol-based glycolipid BbGL-2c, the TCR apparently is able to induce the breaking of existing H bonds between the glycolipid and mCD1d while enabling the formation of new H bonds after a repositioning of the glycolipid in the mCD1d antigen–binding groove. This reflects the increased flexibility of binding of glycerol-based antigens within the groove of mCD1d. Consistent with this flexibility, we have previously observed two distinct binding modes for closely related, synthetic α-GalDAG antigens that were based on the structure of material purified from B. burgdorferi (Wang et al., 2010). These two antigens can bind with their glycolipid backbones oriented 180° rotated from one another inside the mCD1d-binding groove. The orientation depends on the fatty acids linked to the sn-1 and sn-2 positions of the glycerol (Wang et al., 2010), with the preference of the mCD1d A′ pocket for monounsaturated C18:1 fatty acids over fully saturated or diunsaturated fatty acids. The different orientations lead to different presentations of the galactose head group, with a nonantigenic glycolipid placing it rotated ∼120° away from the ideal position. It is likely that the TCR has a limit in its capacity to reorient the galactose moiety of such an antigen to be able to interact and bind in a conserved footprint, rendering it incapable of stimulating iNKT cells.
In contrast, with diacylglycerol antigens, the ceramide backbones of GSLs are bound in one orientation, with the sphingosine in the F′ pocket and sugar exposed in a more favorable orientation for carbohydrate presentation and TCR binding to the CD1d glycolipid complex. Therefore, it is not surprising that the most potent iNKT cell antigens contain ceramide backbones rather than diacylglycerol moieties. This more optimal placement of GSL antigens is in agreement with the results from earlier surface plasmon resonance–based studies, which indicated little temperature dependence in the binding of the TCR to mCD1d–α-GalCer complexes (Sidobre et al., 2002; Cantu et al., 2003). This suggested that there is little accommodation during the interaction and is consistent with a lock and key mechanism for TCR binding when α-GalCer is the antigen. Furthermore, when antigens with different sugars but the same GSL lipid were compared, calculation of the differences in the activation energy suggested that the TCR recognizes the exposed sugar during the initial phase of the interaction (Sidobre et al., 2004). This agrees not only with the structural data indicating that the sugar is placed in the near optimal position for TCR interaction for both α-GalCer and GalA-GSL but not for BbGL-2c, but also with the kinetic data indicating a similar Ka for the two GSL antigens but a much slower one for BbGL-2c.
A second, marked conformational change upon TCR binding observed in our study is the closing of the roof over the F′ pocket. This is likely mediated for recognition of the two natural microbial antigens by Leu99 in the CDR3 region of the TCR α chain. α-GalCer binding to mCD1d uniquely caused this change in the absence of the TCR. The F′ roof formation has not been observed for other structurally similar ligands, such as OCH or the α-GalCer phenyl analogues (Schiefner et al., 2009; Sullivan et al., 2010). However, in the case of OCH, a spacer molecule was described in the F′ pocket that likely prevents the roof formation. Examination of the electron density maps for the phenyl analogues revealed not interpreted electron density in the vicinity of the F′ roof that could possibly be attributed to a solvent molecule that interferes with the roof formation, likely DMSO, which has also been observed in a similar position in the mCD1d–BbGL-2f structure (Wang et al., 2010). Interestingly, structural changes above the F′ pocket have previously been suggested to be involved in modulating TCR affinities for a panel of α-GalCer analogues (McCarthy et al., 2007). Because TCR interaction with both the mCD1d–GalA-GSL and mCD1d–BbGL-2c complexes has a faster Kd, we propose that although the position of the sugar has a greater influence on the Ka, the ability to form the F′ roof independently of the TCR has a greater influence on TCR dissociation. This model, derived from our structural experiments, accounts for the intermediate KD of the TCR for mCD1d–GalA-GSL complexes, the weaker KD for mCD1d–BbGL-2c, and the observed kinetic properties of TCR binding to the different mCD1d–glycolipid antigen complexes.
In addition to the conserved footprint onto mCD1d, the TCR also has a few conserved amino acid hot spots where it contacts portions of the glycolipid antigen, namely the 2′-, 3′-, and 4′-OH groups of the galactose or galacturonic acid. Although all of these OH groups of the sugar participate in the TCR interaction, analysis of synthetic GSL compounds with individual positions altered indicates that the 2′ position is most important (Wu et al., 2005; Raju et al., 2009). These hot spots are also maintained by the TCR in at least one other nonglycosidic ligand, which was suggested to be a structural mimetic of an incomplete galactose ring (Silk et al., 2008). As a result the iNKT cell epitope is not strictly limited to α-anomeric galactose and structurally related sugars, but can also contain other polar moieties that maintain those conserved contacts upon TCR engagement.
The conserved TCR footprint has first been suggested based on site-directed mutagenesis experiments of the individual TCR CDR residues, which affected structural related glycolipids to a similar degree (Scott-Browne et al., 2007). However, this study did not include the borrelial α-GalDAG ligand, which binds differently to mCD1d compared with α-GalCer. A subsequent study revealed differences in the contributions of different CDR-α and -β region amino acids in the activation of T cell hybridomas when α-GalCer and α-GalDAG were compared (Florence et al., 2009). This was proposed to be caused by a different contribution of those TCR residues to antigen binding, suggesting a different orientation of the sugar of α-GalDAG when bound to mCD1d or induced conformation of mCD1d. However, in light of the structures presented in this study, we propose that those residues are likely important instead for facilitating the reorientation of the galactose moiety of α-GalDAG to give the conserved orientation. It appears that the TCR–CD1d contact interface has been optimized during the course of evolution and that those conserved hot spot interactions outweigh the steric hindrances caused by suboptimally bound ligands to the extent possible.
However, it has not formerly been demonstrated that this conserved footprint is also true for a complex β-anomeric carbohydrate–containing antigen, the self-antigen isoglobotrihexosyl ceramide (iGb3). However, the unique binding mode of the TCR of iNKT cells, being off-set to the C-terminal end of the α1 helix combined with a slight vertical tilt, opens an escape hatch between CDR1-α and the A′ roof of CD1d, through which longer antigens can escape. As the structure of iGb3 bound to mCD1d illustrates, those ligands would clash with the TCR, assuming the conserved footprint (Zajonc et al., 2008). However, previous modeling attempts (Zajonc et al., 2008), as well as recent data on α-anomeric forms of iGb3 (Yin et al., 2009), suggest that β-linked glycolipids will be, to the extent possible, flattened by the TCR to mimic the α-anomeric glycolipid pattern, while possibly conserving those TCR hot spots on the ligand, and further by exposing the elongated head group through the escape hatch. However, how specificity for the terminal galactose of iGb3 can then be retained is currently unknown, but it could entail specific binding to either CD1d for a more stable conformation or interaction with framework residues of the TCR, as no CDR is close enough for direct interaction. Likewise, the mechanism for TCR recognition by some cultured human iNKT cells for lyso-phosphatidylcholine presented by human CD1d (Fox et al., 2009) remains to be determined, and this could involve an entirely different contact point and/or footprint on CD1d.
In summary, we have demonstrated an unanticipated degree of structural changes induced by the TCR in complexes of two microbial glycolipids bound to mCD1d. These changes involve the sugar moiety of the antigen itself and mCD1d amino acids in the roof over the F′ pocket. Because these changes are preformed when α-GalCer, the most potent antigen, binds to mCD1d, they could form the basis for rational drug design of more potent antigens for activating human iNKT cells.
MATERIALS AND METHODS
Protein expression and purification.
Fully glycosylated mCD1d protein and Vα14/Vβ8.2 TCR were expressed and purified as described previously (Zajonc et al., 2005b; Wang et al., 2010). In brief, mCD1d was secreted from 5 liters of baculovirus-infected SF9 (Spodoptera frugiperda 9) insect cells (2 × 106 cells/ml) in shaking flasks (145 rpm) at 28°C after 3 d of infection at a multiplicity of infection of approximately three. SF9 cells were spun down (1,000 g for 10 min at 4°C), and the media were further concentrated to 0.4 liter and exchanged against PBS buffer using tangential flow-through filtration (Millipore). The protein was purified from the concentrated media by Ni2+ affinity chromatography (using His-Bind Fractogel; EMD) in 50 mM Tris-HCl, pH 8.0, buffer followed by anion-exchange chromatography using MonoQ HR 10/10 (GE Healthcare) in 10 mM Tris-HCl buffer at pH 8.0 with a linear gradient of 0–250 mM NaCl.
For TCR refolding, inclusion bodies of the α and β chains were dissolved in 50 mM Tris-HCl, 5 mM EDTA, 2 mM DTT, and 6 M guanidine-HCl, pH 7.0, and stored at −80°C. 32 mg of α chain and 48 mg of β chain were thawed, mixed, pulsed with 1 mM DTT, and then added drop wise to 1 liter of refolding buffer (50 mM Tris-HCl, 0.4 M l-Arg, 5 M urea, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, and 0.2 mM PMSF, pH 8.0, at room temperature) under constant stirring at 4°C. After 16 h, the same amount of α and β chains was added again and continued to stir for an additional 8 h. The refolding mix was then dialyzed overnight against 18 liters of 10 mM Tris-HCL, 0.1 M urea, pH 8.0, and then again for 8 h against the same fresh buffer. Finally the refolding mix was dialyzed against 18 liters of 10 mM Tris-HCl, pH 8.0, for 24 h. The refolded TCR was either centrifuged for 30,000 g for 10 min or directly filtered through 0.22-µm filter. DEAE Sepharose beads (GE Healthcare; 5 ml settled resin) were added to the dialyzed refolding mix and stirred for 2–4 h at 4°C. DEAE beads were collected with a 40–60-µm Buchner funnel, washed with 10 mM Tris-HCl, pH 8.0, and transferred to an Econo column (Bio-Rad Laboratories). The refolded TCR was eluted with 100 mM NaCl in 10 mM Tris-HCl, pH 8.0, diluted fourfold with 10 mM Tris-HCl, pH 8.0, and injected into a MonoQ 5/50 GL (GE Healthcare). The TCR was eluted using a linear NaCl gradient (0–300 mM NaCl). TCR-containing fractions were pooled, concentrated, and further purified by size-exclusion chromatography using a Superdex S200 10/300 GL (GE Healthcare).
Glycolipid loading and ternary complex formation.
The synthetic GalA-GSL was dissolved at 3 mg/ml in DMSO. The B. burgdorferi diacylglycerolipid (BbGL-2c) was dissolved in 0.5% Tween and 0.9% NaCl at a concentration of 0.22 mg/ml. mCD1d was incubated overnight with 3–6 M excess of GalA-GSL and BbGL-2c in the presence of 0.05% Tween 20 and 100 mM Tris-HCl, pH 7.0, respectively. GalA-GSL–loaded CD1d protein was then incubated with equimolar amounts of TCR for 30 min at room temperature, and the ternary mCD1d–GalA-GSL–TCR complex was isolated by size-exclusion chromatography using a Superdex S200 10/300 GL. BbGL-2c–loaded CD1d was purified by size-exclusion chromatography first and then incubated with the same amount of TCR for 30 min without further purification. Both the complexes were concentrated to 4 mg/ml for crystallization.
Crystallization and structure determination.
Crystals of both mCD1d–glycolipid–TCR complexes were grown at 22.3°C by sitting drop vapor diffusion while mixing 0.5 µl of protein with 0.5 µl of precipitate (18% polyethylene glycol 3350 and 0.2 M ammonium citrate dibasic for GalA-GSL; 20% polyethylene glycol 3350 and 0.1 M citrate, pH 5, for BbGL-2c). Crystals were flash-cooled at 100 K in mother liquor containing 20% glycerol. Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory beamline 7.1 and processed with the HKL2000 software suite (Otwinowski and Minor, 1997). The mCD1d–GalA-GSL–TCR crystal belongs to space group C2221 with cell parameters a = 79.0 Å, b = 191.2 Å, and c = 151.2 Å. The mCD1d–BbGL-2c–TCR crystal also belongs to space group C2221 with cell parameters a = 78.0 Å, b = 188.4 Å, and c = 149.8.2 Å.
The asymmetric unit contains one mCD1d–glycolipid–TCR molecule with an estimated solvent content of 56.9% for GalA-GSL and 55.3% for BbGl-2c complex. The structures were determined by molecular replacement using MOLREP as part of the CCP4 suite (Collaborative Computational Project Number 4, 1994; Vagin and Teplyakov, 1997) using the protein coordinates from the mCD1d-iGB3 structure (Protein Data Bank ID 2Q7Y; Zajonc et al., 2008), followed by the variable domain of the mouse Vα14Vβ8.2 TCR (Pellicci et al., 2009) and, finally, the constant domain (from PDB ID 3HE6) as the search model. The molecular replacement solution with the complete TCR revealed that the constant domain was slightly tilted in both of our structures. After the molecular replacement solution was obtained containing both mCD1d and TCR, the model was rebuilt into σA-weighted 2Fo-Fc and Fo-Fc difference electron density maps using the program COOT (Emsley and Cowtan, 2004). The final refinement steps were performed using the TLS (translation, libration, and screw axis) procedure in REFMAC (Winn et al., 2001) with five anisotropic domains (α1-α2 domain of CD1d, including carbohydrates and glycolipid, α3 domain, β2M, variable domain, and constant domain of TCR). The mCD1d–GalA-GSL–TCR structure was refined to 2.74 Å with a final Rcryst and Rfree of 19.8% and 25.3%, respectively. The mCD1d–BbGL-2c–TCR structure was refined to 2.8 Å with a final Rcryst and Rfree of 20.3% and 25.9%, respectively. The quality of the model was examined with the program Molprobity (Lovell et al., 2003).
Protein Data Bank accession numbers.
Coordinates and structure factors for the mCD1d–BbGL-2c–Vα14 TCR and mCD1d–GalA-GSL–TCR ternary complexes have been deposited in the Protein Data Bank under accession codes 3O9W and 3O8X, respectively.
Online supplemental material.
Fig. S1 depicts omit maps for the glycolipid ligands, as well as electron density for the CDR residues of the TCR. Table S1 shows a list of contacts formed between CD1d, glycolipid, and TCR. Table S2 contains activation energies that were calculated from previously published surface plasmon resonance data (Wang et al., 2010). Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101335/DC1.
Acknowledgments
The authors would like to thank Stanford Synchrotron Radiation Laboratory staff of beamline 7.1 for assistance with remote data collection.
This work was funded by National Institutes of Health grants RO1 AI074952 (to D.M. Zajonc) and RO1 AI45053 and R37 AI71922 (to M. Kronenberg) and Foundation for Research Science and Technology grant C08X0808 (to G.F. Painter). D.M. Zajonc is a recipient of a CRI Investigator award from the Cancer Research Institute.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- α-GalCer
- α-galactosylceramide
- α-GalDAG
- α-galactosyl diacylglycerolipid
- CDR
- complementarity-determining region
- iGb3
- isoglobotrihexosyl ceramide
- GalA-GSL
- α-galacturonosylceramide
- GSL
- glycosphingolipid
- mCD1d
- mouse CD1d
- RMSD
- root mean square deviation
References
- Bendelac A. 1995. Mouse NK1+ T cells. Curr. Opin. Immunol. 7:367–374 10.1016/0952-7915(95)80112-X [DOI] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- Borg N.A., Wun K.S., Kjer-Nielsen L., Wilce M.C., Pellicci D.G., Koh R., Besra G.S., Bharadwaj M., Godfrey D.I., McCluskey J., Rossjohn J. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- Brigl M., Brenner M.B. 2004. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 22:817–890 10.1146/annurev.immunol.22.012703.104608 [DOI] [PubMed] [Google Scholar]
- Cantu C., III, Benlagha K., Savage P.B., Bendelac A., Teyton L. 2003. The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J. Immunol. 170:4673–4682 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. 1994. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Florence W.C., Xia C., Gordy L.E., Chen W., Zhang Y., Scott-Browne J., Kinjo Y., Yu K.O., Keshipeddy S., Pellicci D.G., et al. 2009. Adaptability of the semi-invariant natural killer T-cell receptor towards structurally diverse CD1d-restricted ligands. EMBO J. 28:3781 10.1038/emboj.2009.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox L.M., Cox D.G., Lockridge J.L., Wang X., Chen X., Scharf L., Trott D.L., Ndonye R.M., Veerapen N., Besra G.S., et al. 2009. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 7:e1000228 10.1371/journal.pbio.1000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Pellicci D.G., Patel O., Kjer-Nielsen L., McCluskey J., Rossjohn J. 2010. Antigen recognition by CD1d-restricted NKT T cell receptors. Semin. Immunol. 22:61–67 10.1016/j.smim.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E., et al. 1997. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 278:1626–1629 10.1126/science.278.5343.1626 [DOI] [PubMed] [Google Scholar]
- Kinjo Y., Wu D., Kim G., Xing G.W., Poles M.A., Ho D.D., Tsuji M., Kawahara K., Wong C.H., Kronenberg M. 2005. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 434:520–525 10.1038/nature03407 [DOI] [PubMed] [Google Scholar]
- Kinjo Y., Tupin E., Wu D., Fujio M., Garcia-Navarro R., Benhnia M.R., Zajonc D.M., Ben-Menachem G., Ainge G.D., Painter G.F., et al. 2006. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat. Immunol. 7:978–986 10.1038/ni1380 [DOI] [PubMed] [Google Scholar]
- Koch M., Stronge V.S., Shepherd D., Gadola S.D., Mathew B., Ritter G., Fersht A.R., Besra G.S., Schmidt R.R., Jones E.Y., Cerundolo V. 2005. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 6:819–826 10.1038/ni1225 [DOI] [PubMed] [Google Scholar]
- Lovell S.C., Davis I.W., Arendall W.B., III, de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. 2003. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 50:437–450 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Mallevaey T., Scott-Browne J.P., Matsuda J.L., Young M.H., Pellicci D.G., Patel O., Thakur M., Kjer-Nielsen L., Richardson S.K., Cerundolo V., et al. 2009. T cell receptor CDR2 β and CDR3 β loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 31:60–71 10.1016/j.immuni.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J., Debord K.L., Ismail N., Goff R.D., Cantu C., III, Zhou D., Saint-Mezard P., Wang V., Gao Y., Yin N., et al. 2005. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 434:525–529 10.1038/nature03408 [DOI] [PubMed] [Google Scholar]
- McCarthy C., Shepherd D., Fleire S., Stronge V.S., Koch M., Illarionov P.A., Bossi G., Salio M., Denkberg G., Reddington F., et al. 2007. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J. Exp. Med. 204:1131–1144 10.1084/jem.20062342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody D.B., Zajonc D.M., Wilson I.A. 2005. Anatomy of CD1-lipid antigen complexes. Nat. Rev. Immunol. 5:387–399 10.1038/nri1605 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pellicci D.G., Patel O., Kjer-Nielsen L., Pang S.S., Sullivan L.C., Kyparissoudis K., Brooks A.G., Reid H.H., Gras S., Lucet I.S., et al. 2009. Differential recognition of CD1d-α-galactosyl ceramide by the V β 8.2 and V β 7 semi-invariant NKT T cell receptors. Immunity. 31:47–59 10.1016/j.immuni.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Castillo B.F., Richardson S.K., Thakur M., Severins R., Kronenberg M., Howell A.R. 2009. Synthesis and evaluation of 3″- and 4″-deoxy and -fluoro analogs of the immunostimulatory glycolipid, KRN7000. Bioorg. Med. Chem. Lett. 19:4122–4125 10.1016/j.bmcl.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.G., Stanfield R.L., Wilson I.A. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Schiefner A., Fujio M., Wu D., Wong C.H., Wilson I.A. 2009. Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands. J. Mol. Biol. 394:71–82 10.1016/j.jmb.2009.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne J.P., Matsuda J.L., Mallevaey T., White J., Borg N.A., McCluskey J., Rossjohn J., Kappler J., Marrack P., Gapin L. 2007. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8:1105–1113 10.1038/ni1510 [DOI] [PubMed] [Google Scholar]
- Sidobre S., Naidenko O.V., Sim B.C., Gascoigne N.R., Garcia K.C., Kronenberg M. 2002. The V α 14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J. Immunol. 169:1340–1348 [DOI] [PubMed] [Google Scholar]
- Sidobre S., Hammond K.J., Bénazet-Sidobre L., Maltsev S.D., Richardson S.K., Ndonye R.M., Howell A.R., Sakai T., Besra G.S., Porcelli S.A., Kronenberg M. 2004. The T cell antigen receptor expressed by Valpha14i NKT cells has a unique mode of glycosphingolipid antigen recognition. Proc. Natl. Acad. Sci. USA. 101:12254–12259 10.1073/pnas.0404632101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk J.D., Salio M., Reddy B.G., Shepherd D., Gileadi U., Brown J., Masri S.H., Polzella P., Ritter G., Besra G.S., et al. 2008. Cutting edge: nonglycosidic CD1d lipid ligands activate human and murine invariant NKT cells. J. Immunol. 180:6452–6456 [DOI] [PubMed] [Google Scholar]
- Sriram V., Du W., Gervay-Hague J., Brutkiewicz R.R. 2005. Cell wall glycosphingolipids of Sphingomonas paucimobilis are CD1d-specific ligands for NKT cells. Eur. J. Immunol. 35:1692–1701 10.1002/eji.200526157 [DOI] [PubMed] [Google Scholar]
- Sullivan B.A., Nagarajan N.A., Wingender G., Wang J., Scott I., Tsuji M., Franck R.W., Porcelli S.A., Zajonc D.M., Kronenberg M. 2010. Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J. Immunol. 184:141–153 10.4049/jimmunol.0902880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupin E., Benhnia M.R., Kinjo Y., Patsey R., Lena C.J., Haller M.C., Caimano M.J., Imamura M., Wong C.H., Crotty S., et al. 2008. NKT cells prevent chronic joint inflammation after infection with Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA. 105:19863–19868 10.1073/pnas.0810519105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30:1022–1025 10.1107/S0021889897006766 [DOI] [Google Scholar]
- Wang J., Li Y., Kinjo Y., Mac T.T., Gibson D., Painter G.F., Kronenberg M., Zajonc D.M. 2010. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc. Natl. Acad. Sci. USA. 107:1535–1540 10.1073/pnas.0909479107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Isupov M.N., Murshudov G.N. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57:122–133 10.1107/S0907444900014736 [DOI] [PubMed] [Google Scholar]
- Wu D., Xing G.W., Poles M.A., Horowitz A., Kinjo Y., Sullivan B., Bodmer-Narkevitch V., Plettenburg O., Kronenberg M., Tsuji M., et al. 2005. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc. Natl. Acad. Sci. USA. 102:1351–1356 10.1073/pnas.0408696102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Zajonc D.M., Fujio M., Sullivan B.A., Kinjo Y., Kronenberg M., Wilson I.A., Wong C.H. 2006. Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc. Natl. Acad. Sci. USA. 103:3972–3977 10.1073/pnas.0600285103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.C., Tuot D.S., Lyons D.S., Garcia K.C., Davis M.M. 2002. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 418:552–556 10.1038/nature00920 [DOI] [PubMed] [Google Scholar]
- Yin N., Long X., Goff R.D., Zhou D., Cantu C., III, Mattner J., Mezard P.S., Teyton L., Bendelac A., Savage P.B. 2009. Alpha anomers of iGb3 and Gb3 stimulate cytokine production by natural killer T cells. ACS Chem. Biol. 4:199–208 10.1021/cb800277n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc D.M., Kronenberg M. 2007. CD1 mediated T cell recognition of glycolipids. Curr. Opin. Struct. Biol. 17:521–529 10.1016/j.sbi.2007.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc D.M., Kronenberg M. 2009. Carbohydrate specificity of the recognition of diverse glycolipids by natural killer T cells. Immunol. Rev. 230:188–200 10.1111/j.1600-065X.2009.00802.x [DOI] [PubMed] [Google Scholar]
- Zajonc D.M., Wilson I.A. 2007. Architecture of CD1 proteins. Curr. Top. Microbiol. Immunol. 314:27–50 10.1007/978-3-540-69511-0_2 [DOI] [PubMed] [Google Scholar]
- Zajonc D.M., Cantu C., III, Mattner J., Zhou D., Savage P.B., Bendelac A., Wilson I.A., Teyton L. 2005a. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat. Immunol. 6:810–818 10.1038/ni1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc D.M., Maricic I., Wu D., Halder R., Roy K., Wong C.H., Kumar V., Wilson I.A. 2005b. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 202:1517–1526 10.1084/jem.20051625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc D.M., Savage P.B., Bendelac A., Wilson I.A., Teyton L. 2008. Crystal structures of mouse CD1d-iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J. Mol. Biol. 377:1104–1116 10.1016/j.jmb.2008.01.061 [DOI] [PMC free article] [PubMed] [Google Scholar]