The actin cross-linking protein filamin A reduces migration, invasion, and metastasis of breast cancer cells.
Abstract
The actin cross-linking protein filamin A (FLNa) functions as a scaffolding protein and couples cell cytoskeleton to extracellular matrix and integrin receptor signaling. In this study, we report that FLNa suppresses invasion of breast cancer cells and regulates focal adhesion (FA) turnover. Two large progression tissue microarrays from breast cancer patients revealed a significant decrease of FLNa levels in tissues from invasive breast cancer compared with benign disease and in lymph node–positive compared with lymph node–negative breast cancer. In breast cancer cells and orthotopic mouse breast cancer models, down-regulation of FLNa stimulated cancer cell migration, invasion, and metastasis formation. Time-lapse microscopy and biochemical assays after FLNa silencing and rescue with wild-type or mutant protein resistant to calpain cleavage revealed that FLNa regulates FA disassembly at the leading edge of motile cells. Moreover, FLNa down-regulation enhanced calpain activity through the mitogen-activated protein kinase–extracellular signal-regulated kinase cascade and stimulated the cleavage of FA proteins. These results document a regulation of FA dynamics by FLNa in breast cancer cells.
The ability of cancer cells to invade surrounding normal tissues at primary sites requires a series of early events, including rapid reorganization of cell cytoskeleton, formation and extension of plasma membrane protrusions in response to chemotactic signals, stable focal adhesion (FA)–mediated cell attachment to extracellular matrix near the leading edge of the protrusions, and translocation of the cell body forward aided by release of FAs at the cell rear. FAs act as signaling centers in which multiple dynamic protein–protein interactions occur to regulate the assembly and disassembly of FA sites, which are essential for the control of cell movement and migration.
In contrast to FA assembly, which is primarily driven by the GTP-bound Rho GTPases (Ridley and Hall, 1992; Hall, 1998), the dynamic of disassembly of FA protein complexes is not fully understood. Earlier studies pinpoint a key role for cell cytoskeleton signaling both in FA formation and removal at FA sites (for review see Webb et al., 2002). In particular, actin-binding proteins such as filamins are crucial for cell adhesion to extracellular matrix and cell movement. They mediate cross-linking of cortical cytoplasmic actin into a dynamic three-dimensional structure and participate in the anchoring of several plasma membrane proteins, including integrins, to the cortical actin. These functions are essential for cell locomotion and migration in response to microenvironmental stimuli. Three highly conserved filamin isoforms exist in mammals: filamin A (FLNa), FLNb, and FLNc. These isoforms have a wide tissue expression, although FLNc is more restricted to skeletal and cardiac muscles in adults (Sheen et al., 2002; Feng and Walsh, 2004). Notably, FLNa is the dominant mammalian nonmuscle isoform of actin-binding proteins, which organizes actin filaments into orthogonal networks (Gorlin et al., 1990; Stossel et al., 2001; Nakamura et al., 2002).
The molecular function of FLNa in cell chemotaxis remains debated and seems to vary depending on FLNa expression levels and its interacting partners, particularly those that share overlapping binding sites on integrins such as talin. Of biological significance, FLNa missense mutations are associated with a wide spectrum of human disorders where loss of function mutations are regarded as a cause of impaired neural cell migration in response to microenvironmental cues, in addition to causing defects in the vascular system (Cunningham et al., 1992; Eksioglu et al., 1996; Fox et al., 1998; Sheen et al., 2002; Nagano et al., 2004; Feng et al., 2006). However, a role of FLNa in cell migration is contradicted by other studies showing that locomotion of cells derived from distinct FLNa knockout mice is not significantly different from WT cells (Feng et al., 2006; Hart et al., 2006). Moreover, overexpression of FLNa in FLNa-deficient M2 cells (Cunningham et al., 1992) or mouse cortical neurons (Sarkisian et al., 2006) resulted in inhibition rather than stimulation of cell migration. Strong binding of FLNa to integrin was reported to prevent cell migration (Pfaff et al., 1998; Calderwood et al., 2001). Another study using the HT-1080 human fibrosarcoma cells demonstrated that FLNa knockdown did not affect the speed but rather the initiation of migration (Baldassarre et al., 2009).
In this study, we provide evidence that low levels of FLNa correlate with human breast cancer progression. We report that FLNa down-regulation stimulates breast cancer cell migration and cell invasion in vitro and promotes metastasis formation in xenograft breast cancer mouse models. We demonstrate that FLNa regulates FA disassembly via a calpain-dependent mechanism.
RESULTS
Low expression levels of FLNa correlate with breast cancer progression
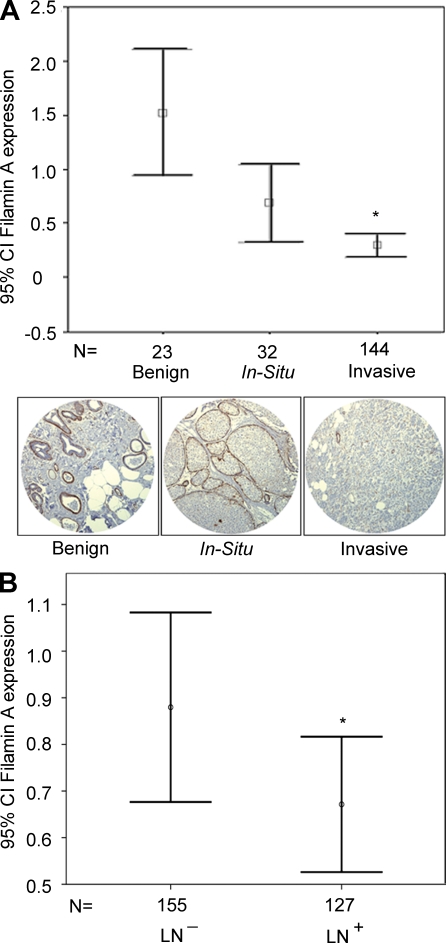
To examine the potential clinical relevance of FLNa to cancer progression, we investigated its expression using a human breast tissue microarray (TMA) from cancer patients with progressive breast disease: benign, in situ, and invasive breast carcinoma. Immunohistochemistry analysis using a monoclonal antibody that recognizes the N-terminal portion of FLNa revealed an inverse correlation between FLNa high expression levels and breast cancer progression (Fig. 1 A). The normalized mean epithelial expression of FLNa in invasive breast cancer (0.30 ± 0.625) was significantly lower than localized in situ breast cancer (0.69 ± 0.998) and benign tissue (1.52 ± 0.344; P < 0.005). In contrast to FLNa, immunohistochemical analysis using a polyclonal antibody against FLNb revealed a positive correlation between high expression levels of FLNb in epithelial cells and breast cancer progression (Fig. S1 A). The correlation between low expression levels of FLNa and invasiveness was also observed in an independent TMA of 258 lymph node–positive and –negative breast cancer patients’ samples. Expression levels of FLNa were significantly decreased in lymph node–positive breast cancer (n = 127) versus lymph node–negative cancer (n = 155; mean epithelial expression of FLNa was 0.89 ± 0.21 and 0.66 ± 0.14 for lymph node–negative and –positive breast cancer, respectively; P = 0.008; Fig. 1 B). A similar inverse correlation between FLNa expression levels and lymph node status was observed using an antibody that recognizes phospho-FLNa-Ser2152 (P = 0.004; Fig. S1 B).
Figure 1.
Low FLNa expression levels are correlated with breast cancer progression. (A) FLNa expression levels were examined by blinded pathology analysis of TMA (n = 222 samples) from breast cancer patients with progressive disease: benign, in situ carcinoma, and invasive carcinoma. Histogram represents normalized mean of epithelial expression of FLNa levels (confidence intervals [CI] of 95% ± SEM) using a monoclonal antibody that recognizes the FLNa N terminus. Images below the histogram correspond to representative immunostaining of breast cancer tissue cores from progressive breast cancers. Differences among the three groups were compared with one-way analysis of variance and the post-hoc Tukey honestly significant difference test for multiple comparisons (*, P < 0.005). (B) An independent TMA was assembled from a total of 258 lymph node (LN)–positive and –negative breast cancer patients’ samples. Histogram shown represents normalized mean of epithelial expression of FLNa (confidence intervals of 95% ± SEM) as determined by quantitative evaluation of immunostaining. A significant decrease of FLNa expression was observed in lymph node–positive (n = 127) versus lymph node–negative breast cancer (n = 155; *, P = 0.008).
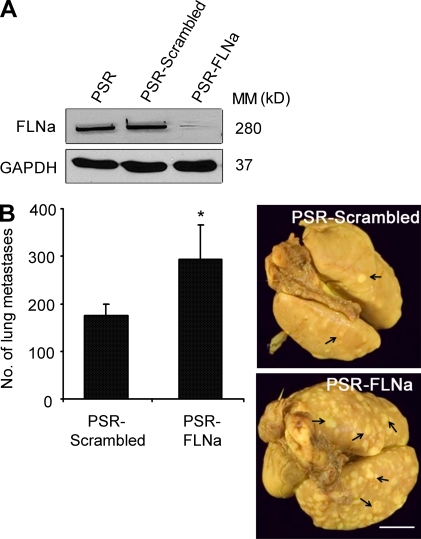
To further investigate the inverse correlation between low levels of FLNa expression and cancer invasiveness, we examined the impact of FLNa down-regulation on cancer progression using preclinical orthotopic breast cancer models. We compared the incidence of lung metastases after implantation of MDA-231-ErbB2 cells expressing either scrambled short hairpin RNA (shRNA; pSuper retro [PSR] scrambled) or FLNa shRNA (PSR-FLNa) into the mammary fat pad of SCID mice. A significant increase in the number of lung metastases was observed in the FLNa-silenced group compared with control mice (147 ± 17.68 vs. 293.33 ± 62.52 for PSR scrambled and PSR-FLNa, respectively; P < 0.005; Fig. 2). In a similar manner, silencing FLNa in the breast cancer cell line BT-20 enhanced the incidence of lung metastases after implantation into the mammary fat pad compared with control cells expressing scrambled shRNA (unpublished data).
Figure 2.
Inhibition of FLNa promotes cancer progression to metastases in a preclinical orthotopic breast cancer model. MDA-231-ErbB2 cells expressing empty retroviral particles (PSR) or particles containing control scrambled shRNA (PSR-scramble) or FLNa-specific shRNA (PSR-FLNa) were implanted into the mammary fat pad of 10 SCID mice per cell type. Primary tumors were excised when they reached a size of 0.6–0.8 cm3, and animals were kept under observation for a period of 3 mo to allow metastasis formation. Mice were then subjected to autopsy, and lung metastases were examined. (A) Representative Western blot analysis of total cell extracts from MDA-231-ErbB2 tumor cells isolated from lung metastatic nodules confirming efficient stability of FLNa knockdown after in vivo implantation. GAPDH was used as an internal control. MM, molecular mass. (B) Graphs show the mean number of lung metastases ± SEM (*, P < 0.005), and images show representative lungs with metastatic nodules. Arrows indicate surface lung metastatic nodules. Bar, 8 mm.
Inhibition of FLNa stimulates breast cancer cell motility and invasion
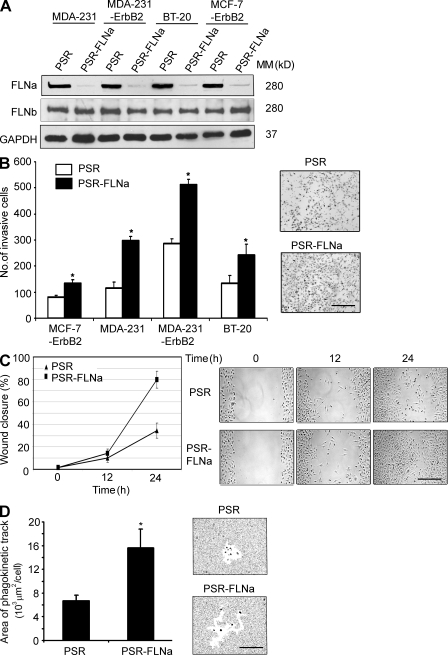
To investigate the function of FLNa in cancer progression, we used a panel of breast cancer cell lines with distinct invasive properties (MCF-7-ErbB2, MDA-231, MDA-231-ErbB2, and BT-20 cells) and their matched pairs in which FLNa was stably inhibited using an RNA interference approach. FLNa-specific shRNA expression resulted in >90% inhibition of endogenous FLNa protein compared with control cells (PSR; Fig. 3 A and Fig. S2). In contrast, no obvious compensation by FLNb expression was seen. In comparison with PSR control cells, the Boyden chamber assay revealed a significant stimulation of cell invasion after FLNa inhibition (P < 0.01 for all the cell types tested; Fig. 3 B).
Figure 3.
Inhibition of FLNa stimulates breast cancer cell migration and invasion. (A) Western blot showing FLNa and FLNb expression in MDA-231, MDA-231-ErbB2, MCF-7-ErbB2, and BT-20 cells stably engineered to express FLNa-shRNA (PSR-FLNa) and their matched control cells expressing PSR puro vector alone (PSR). GAPDH was used as an internal control. MM, molecular mass. (B) Invasion of MDA-231, MDA-231-ErbB2, MCF-7-ErbB2, and BT-20 cells expressing PSR (control) and FLNa-shRNA using the Boyden chamber assay as described in Materials and methods. Bar graph represents the mean number of invaded cells (*, P < 0.01), and representative images of invasive cells on the membrane are shown. Results are expressed as the mean ± SD of five independent experiments and five fields per condition. (C) Cell migration analyzed by the wound healing assay. MDA-231-ErbB2 cells were wounded and monitored over 24 h to determine the rate of migration into the scratched area. Bar graph represents the mean ± SD of five independent experiments, and representative images of the wound captured at different time points are shown. (D) Cell motility analyzed using the phagokinetic track assay as described in Materials and methods. Bar graphs shows a quantification (mean ± SD) of the cleared areas from four independent experiments with >100 tracks assessed per experiment (*, P < 0.01). Images on the right represent brightfield microscopic images of the cleared phagokinetic areas. Bars: (B and C) 150 µm; (D) 120 µm.
In a similar manner, cell motility using the wound healing assay and focusing on the MDA-231-ErbB2 cells revealed that ∼80% of the wound was closed within 24 h by FLNa-silenced cells compared with 34% only for control cells (Fig. 3 C). Consistent with these results, phagokinetic track assay indicated an increase in the motility of FLNa-silenced cells compared with control cells, as revealed by the free tracks generated by phagokinetic ingestion or attachment of the gold particles to the cell surface. Compared with PSR control cells, the mean area of migration increased by ∼2.5-fold in FLNa-silenced cells (Fig. 3 D).
Inhibition of FLNa accelerates FA disassembly
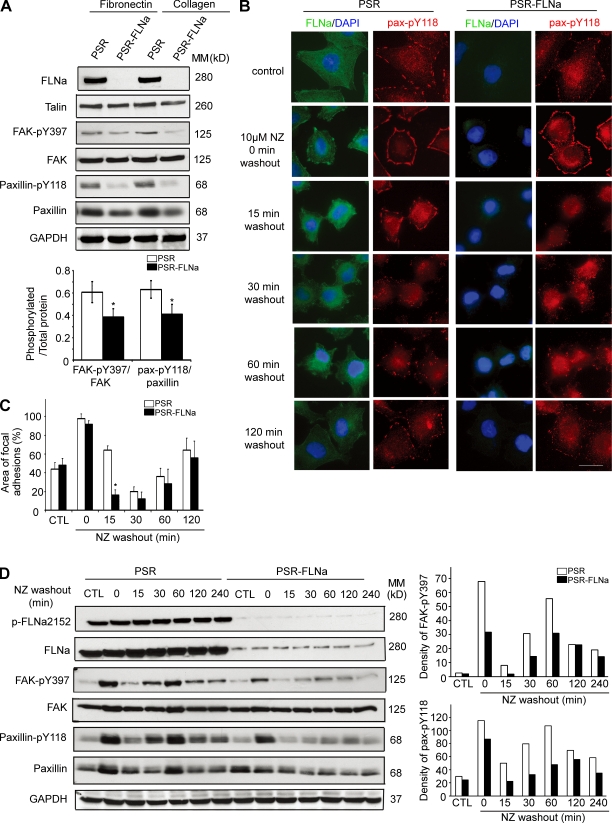
To understand the mechanism by which FLNa regulates cancer cell migration, we investigated FA turnover in matched control and FLNa shRNA–expressing BT-20 cells cultured on either fibronectin- or collagen-coated plates. Western blot analysis revealed a consistent decrease in FA kinase (FAK)–pY397 (the ratios of phosphorylated/total FAK were 0.617 ± 0.09 and 0.386 ± 0.073 for control and FLNa-silencing cells, respectively; P < 0.05) and paxillin-pY118 phosphorylation (the ratios of phosphorylated/total paxillin were 0.632 ± 0.079 and 0.407 ± 0.088 for control and FLNa-silenced cells, respectively; P < 0.05, n = 4) in FLNa shRNA–expressing cells compared with control cells (Fig. 4 A). Next we investigated FA turnover by first using the nocodazole-based assay. Treatment of cells with the microtubule-disrupting agent nocodazole activates RhoA GTPase, which then stimulates the formation of FAs. Nocodazole washout initiates a wave of microtubule regrowth and transient FA disassembly that is followed by FA recovery, a process which is FAK dependent (Ezratty et al., 2005; Yeo et al., 2006). Exposure of serum-starved control cells to nocodazole induced FA formation followed by FA disassembly and reassembly upon nocodazole washout (Fig. 4 B). FA reappearance after nocodazole washout was observed starting from 1 h and became more obvious at 2 h. In comparison with control cells, FLNa-silenced cells showed a rapid loss of paxillin-pY118 (more obvious at 15 min after nocodazole washout; Fig. 4 B). Similar results were obtained using an antibody against FAK-pY397 and vinculin (unpublished data). The areas of FAs formed under each condition were quantified in Fig. 4 C. FLNa silencing induced a strong reduction of FA area at 15 min after nocodazole washout (85% from 0-min level compared with 35% for control cells; P < 0.01).
Figure 4.
FLNa regulates FA disassembly. (A) BT-20 cells expressing empty retroviral particles (PSR) or FLNa-specific shRNA (PSR-FLNa) were seeded on fibronectin- or collagen-coated dishes and grown at 70% confluence. Total protein extracts were probed using antibodies against the indicated proteins. Bar graph shows quantification of ratio of phosphorylated/total proteins (mean ± SD) from four independent experiments (*, P < 0.05). (B) Serum-starved control (PSR) and FLNa-silenced (PSR-FLNa) BT-20 cells were incubated with 10 µM nocodazole (NZ) for 4 h. Control untreated cells and treated cells at the indicated times after nocodazole washout were fixed and immunoassayed with anti–paxillin-pY118 or anti-FLNa. Three independent experiments were performed. Bar, 20 µm. (C) Quantification of FA areas using Volocity software. Data are expressed as the area of FAs per DAPI-stained nucleus and were obtained from at least 20 cells per condition. Quantifications show mean ± SD from three independent experiments as in B (*, P < 0.01). (D) Representative Western blots of FAK and paxillin during FA disassembly. BT-20 cell lysates were collected at the indicated times as in B and immunoblotted with anti–FAK-pY397 or anti–paxillin-pY118. Bar graphs represent the densitometry result as percentage of density of PSR control at 0 min (which is 100%) from four independent Western blots. MM, molecular mass.
To confirm the impact of FLNa on FA disassembly, we monitored the level of paxillin-pY118 and FAK-pY397 of control and FLNa-silenced cells by Western blot analysis. Interestingly, the levels of both paxilin-pY118 and FAK-pY397 were decreased after nocodazole washout (Fig. 4 D), and this correlated with the loss of FA shown by immunofluorescence in Fig. 4 B. Densitometry quantification of FAK-pY397 and paxillin-pY118 is reported in Fig. 4 D (right). Similar results were observed in other breast cancer cell types, including MDA-231-ErbB2 cells (Fig. S3).
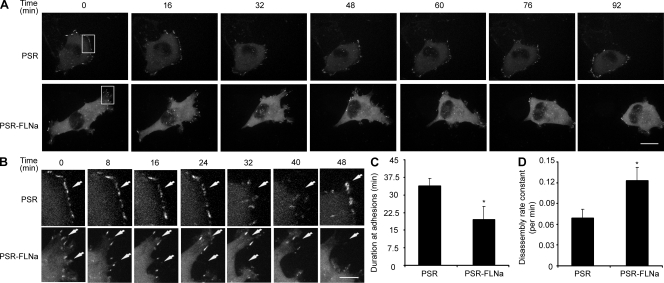
To further confirm the impact of FLNa on FA turnover, we investigated FA dynamics by time-lapse video spinning disk confocal microscopy using enhanced YFP (EYFP)–tagged paxillin (EYFP-paxillin) as an FA marker. In protrusive regions of FLNa-silenced cells, the intensity of paxillin adhesions decreased and eventually disappeared as new adhesions formed at the leading edge. FLNa-silenced cells showed a higher migratory rate and rapid disassembly of FAs compared with matched control cells, which have more stable FA structures (Fig. 5, A and B; and Videos 1 and 2). Further quantification of the duration of EYFP-paxillin intensity in forming or disassembling FAs was performed based on methodology reported by Franco et al. (2004b). A persistence of EYFP-paxillin in adhesion complexes of control cells was observed for an extended duration compared with FLNa-silenced cells (P < 0.01; Fig. 5 C). Moreover, the rate of EYFP-paxillin from adhesion sites (quantified by integrating the EYFP-paxillin fluorescent intensity in individual adhesions over time) was increased in FLNa-silenced cells compared with control cells (P < 0.01; Fig. 5 D). Similar results were obtained when cells were transfected with the FA marker EYFP-vinculin (unpublished data).
Figure 5.
Time-lapse quantification of FA dynamics after FLNa inhibition. (A) Control (PSR) and FLNa-silenced (PSR-FLNa) BT-20 cells transfected with EYFP-paxillin were plated on chamber slide, stimulated with 10 ng/ml EGF, and then analyzed by time-lapse spinning disk confocal microscopy. Representative frames are shown. (B) Enlargements of the boxed regions of A showing white arrows that indicate positions of paxillin-containing FAs. (C) Quantification of EYFP-paxillin persistence in FAs. Duration measurements were quantified by counting the amount of time lapsed between the first and last frames in which an individual adhesion was followed. (D) Rate constants of EYFP-paxillin disassembly at individual adhesions were calculated as described in Materials and methods. (C and D) Quantifications show the mean ± SD from three independent experiments (*, P < 0.01). Bars: (A) 15 µm; (B) 5 µm.
Regulation of FA turnover by FLNa is calpain dependent
The mechanism by which cytoskeletal proteins regulate FA dynamics is not fully understood. Microtubule disruption has been shown to induce Rho activation and the formation of FAs and stress fibers (Danowski, 1989; Bershadsky et al., 1996; Enomoto, 1996). Calpain is required for microtubule-mediated disassembly of adhesive complex sites after nocodazole washout (Bhatt et al., 2002). Calpain is a calcium-dependent protease that catalyzes the cleavage of several cytoskeleton and FA proteins, including filamin, talin, FAK, paxillin, and Src. This proteolytic activity is required for the promotion of FA turnover (Potter et al., 1998) and induction of cell motility (Yan et al., 2001; Carragher et al., 2003; Robles et al., 2003).
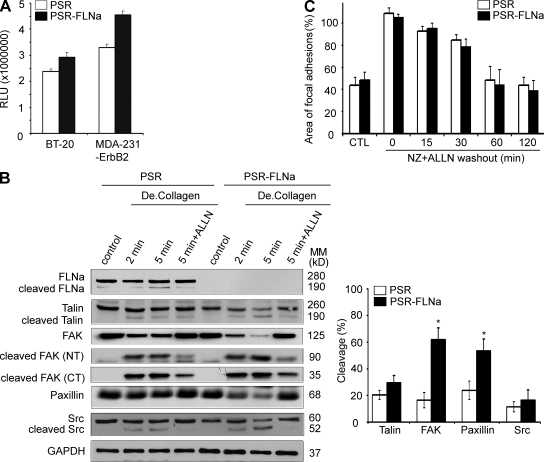
Using the luminescence Calpain-Glo assay, we observed that normalized calpain activity was increased in both FLNa-silenced MDA-231-ErbB2 and BT-20 cells compared with their respective control cells. This effect was significant for MDA-231-ErbB2, in which a 25% increase in calpain activity was observed in FLNa-silenced cells compared with control cells (P < 0.01; Fig. 6 A). Next, we used degraded type I collagen to stimulate calpain-mediated cleavage of FA proteins (Carragher et al., 1999). We confirmed that calpain induced a rapid cleavage of FLNa, talin, FAK, Src, and paxillin after incubation with degraded collagen (Fig. 6 B). Calpain induced cleavage products of ∼190 kD for FLNa, ∼190 kD for talin, ∼90 and ∼35 kD for FAK, and ∼52 and 47 kD for Src. Because the initial cleavage products of FA proteins can be further processed by calpain (e.g., cleavage of the 90-kD product of FAK can generate 50- and 40-kD products; Fig. S4; Carragher et al., 1999), we compared levels of calpain-induced cleavage of full-length proteins. In comparison with control cells, FLNa-silenced cells (PSR-FLNa) expressed less native calpain substrates, particularly FAK and paxillin. Although we could not detect the cleavage products of paxillin (55 kD) because of the limited antibody’s specificity, reduced intensity of the 68-kD paxillin was seen upon collagen stimulation of FLNa-silenced cells compared with their matched control cells. Moreover, pretreatment of MDA-231-ErbB2 cells with 50 µM of the calpain inhibitor ALLN for 22 h followed by a subsequent 5-min stimulation with degraded collagen revealed a significant inhibition of FA protein cleavages (Fig. 6 B, right). These results confirmed an increase of FA protein cleavage upon FLNa silencing (e.g., percentage cleaved FAK was 16.68 ± 5.77% and 61.83 ± 8.87% for control and FLNa-silenced cells, respectively, whereas the percentage of cleaved paxillin was 23.88 ± 6.79% and 53.57 ± 7.48% for control and FLNa-silenced cells, respectively; P < 0.001). Similar results were observed when using BT-20 cell line (Fig. S5).
Figure 6.
Regulation of FA turnover by FLNa is calpain dependent. (A) Control (PSR) or FLNa-silenced (PSR-FLNa) BT-20 and MDA-231-ErbB2 cells were grown to confluence, and total lysates were used for the luminescent Calpain-Glo protease assay. The relative luminescence units (RLU) were averaged and normalized to the amount of proteins in cell lysates. The experiments were repeated four times, and the results are expressed as mean ± SD. (B) Control (PSR) or FLNa-silenced (PSR-FLNa) MDA-231-ErbB2 cells were cultured on 10-cm dishes and stimulated or not with degraded type I collagen (De.Collagen) for the indicated times. Where indicated, cells were treated with 50 µM of calpain inhibitor ALLN (22 h). Lysates were prepared at the indicated time points and analyzed by Western blotting using the corresponding antibodies. Control indicates unstimulated. Bar graph shows quantification and comparison of FA protein cleavage (mean ± SD) at the 5-min time point after degraded collagen stimulation from Western blot results generated from five independent experiments (*, P < 0.001). CT, C terminus; MM, molecular mass; NT, N terminus. (C) Control (PSR) and FLNa-silenced (PSR-FLNa) BT-20 cells were incubated with 10 µM nocodazole (NZ) plus 100 µM ALLN for 4 h followed by a washout. Bar graph shows the area of FAs per DAPI-stained nucleus (mean ± SD) obtained from five fields per condition and from three independent nocodazole washout assays. CTL, untreated.
We then investigated the impact of calpain on FLNa-mediated FA turnover in BT-20 cells using the calpain inhibitor ALLN. Cell exposure to 100 µM ALLN and nocodazole for 4 h followed by a washout reduced FA turnover in control cells (Fig. 6 C and Fig. S6), which is in agreement with a previous finding by Bhatt et al. (2002). Interestingly, no difference in FA turnover was seen between FLNa-silenced and control cells preexposed to ALLN. Under a similar condition, Western blot analysis confirmed a restoration of phospho-protein levels of FAK-pY397 and paxillin-pY118 upon exposure to ALLN, which abolished the difference in FA turnover seen between FLNa-silenced and control cells (unpublished data). Together, these results indicate that calpain is required for microtubule-mediated FA complex disassembly and that inhibition of calpain activity abolished the regulatory effect of FLNa on FA turnover.
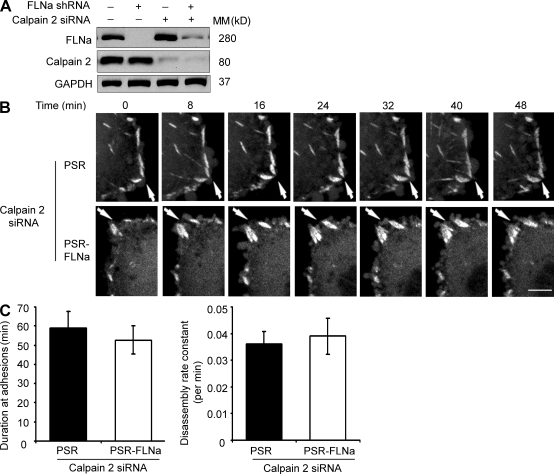
ALLN is a general calpain inhibitor that targets both the µ-calpain isoform (calpain 1) and m-calpain (calpain 2). Previous studies by Franco et al. (2004a,b) demonstrated that calpain 2 but not calpain 1 is critical for the cleavage of FA and cytoskeletal proteins, including FAK, paxillin, talin, and spectrin, as well for the regulation of membrane protrusion dynamics. We silenced calpain 2 in control and FLNa-silenced cells using a specific small interfering RNA (siRNA) sequence (Fig. 7 A and Fig. S2 B) and then followed FA formation and disassembly in live cells. Live fluorescence imaging of EYFP-paxillin revealed a persistence of FAs for extended durations as well as decreased disassembly rates of FA after calpain 2 silencing in both control and FLNa-silenced cells (Fig. 7, B and C; and Videos 3 and 4). These results indicate that inhibition of calpain 2 by siRNA or ALLN (Videos 5 and 6) can prevent the difference seen in FA disassembly between FLNa-silenced and control cells, which further supports a calpain-dependent mechanism for FLNa function in the regulation of FA turnover.
Figure 7.
Time-lapse quantification of FA dynamics after calpain 2 inhibition. (A) Western blot showing calpain 2 siRNA (sequence 1) efficiency in control and FLNa-silenced BT-20 cells. GAPDH was used as an internal control. MM, molecular mass. (B) Control (PSR) or FLNa-silenced (PSR-FLNa) BT-20 cells stably expressing EYFP-paxillin were transfected with calpain 2 siRNA, stimulated with EGF, and then analyzed by time-lapse spinning disk confocal microscopy. White arrows show the positions of paxillin-containing FAs. Four independent experiments were performed, and representative frames are shown. Bar, 5 µm. (C, left) Quantification of the persistence of EYFP-paxillin in control and FLNa knockdown cells. (right) Quantification of EYFP-paxillin disassembly. Quantifications show the mean ± SD from three independent experiments.
FLNa promotes calpain activation via mitogen-activated protein kinase (MAPK) signaling
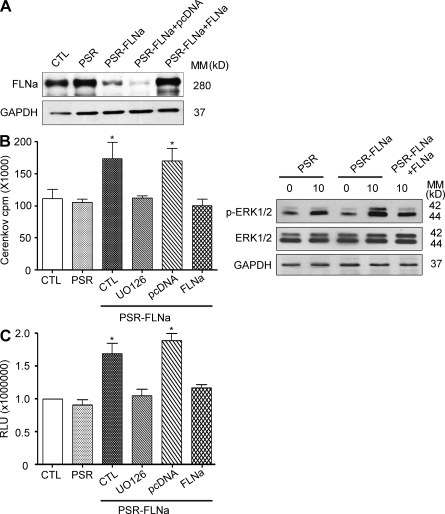
The MAPK cascade is central to both filamin and calpain signaling. We investigated the impact of MAPKs on FLNa-induced calpain 2 activation using parental BT-20 cells (CTL), cells expressing empty viral particles (PSR), FLNa shRNA–expressing cells (PSR-FLNa), and their matched cells rescued with shRNA-resistant FLNa (PSR-FLNa+FLNa) and control FLNa-shRNA cells expressing empty plasmid used for FLNa rescue (PSR-FLNa+pcDNA; Fig. 8 A). Cells were serum starved and then stimulated with 5 ng/ml epidermal growth factor (EGF) for 10 min (predetermined to be the maximal induction of MAPK activity in this model; unpublished data). Interestingly, MAPK–extracellular signal-regulated kinase (ERK) activity and ERK phosphorylation (Fig. 8 B) were found to be significantly elevated in FLNa-silenced cells compared with control cells. The induction of MAPK activity was prevented by preexposure of cells to the MEK (MAPK–ERK kinase) inhibitor UO126 or after reexpression of shRNA-resistant WT FLNa (P < 0.005; Fig. 8 B). Under similar conditions, FLNa silencing induced calpain activation that was prevented after cell pretreatment with UO126 or after reexpression of shRNA-resistant WT FLNa (P < 0.005; Fig. 8 C). Because FLNa has been shown to be required for calpain 2 activation via MAPK-dependent stimulation of calcium fluxes and calcium-sensing receptor (Zhang and Breitwieser, 2005), we investigated whether a possible change in intracellular calcium after FLNa knockdown could account for calpain activation. However, no differences in intracellular calcium were detected between control cells and their matched FLNa-silenced cells in three independent experiments using the Fluo-4 NW calcium assay from Invitrogen (unpublished data). Together, these data argue for MAPK being a central regulator for coupling FLNa and calpain signaling to FA protein cleavage.
Figure 8.
FLNa down-regulation stimulates calpain activity via MAPK activation. (A) Lysates of parental BT-20 cells (CTL), cells expressing empty viral particles used for FLNa-shRNA expression (PSR), FLNa shRNA–expressing cells (PSR-FLNa), and their matched cells rescued with shRNA-resistant FLNa (PSR-FLNa+FLNa) or empty plasmid used for FLNa rescue (PSR-FLNa+pcDNA) were immunoblotted with FLNa-specific antibody. GAPDH was used as an internal control. (B) Control BT-20 cells (CTL), BT-20 cells expressing empty viral particles (PSR), BT-20 cells expressing FLNa-shRNA alone (PSR-FLNa, CTL), FLNa-shRNA cells pretreated for 2 h with 10 µM UO126 (PSR-FLNa, UO126), FLNa-rescued FLNa shRNA cells (PSR-FLNa, FLNa), and PSR-FLNa cells expressing empty plasmid (PSR-FLNa, pcDNA) were serum starved and then stimulated or not with 10 ng/ml EGF for 10 min. Total cell extracts were assayed for kinase activity (left) by immunocomplex kinase assay using anti-ERK2 for immunoprecipitation and MBP as kinase substrate in presence of γ-[32P]ATP as described in Materials and methods or by Western blotting using anti–phospho-ERK1/2 (right). The radioactivity incorporated into MBP was quantified from the excised MBP bands by Cerenkov counting. (C) Samples as in B were used to determine calpain activity using the luminescent Calpain-Glo protease assay as described in Materials and methods. (B and C) Results are expressed as the mean ± SD of three independent experiments (*, P < 0.005). MM, molecular mass.
FLNa and talin may compete for calpain-mediated cleavage, and this impacts FA disassembly
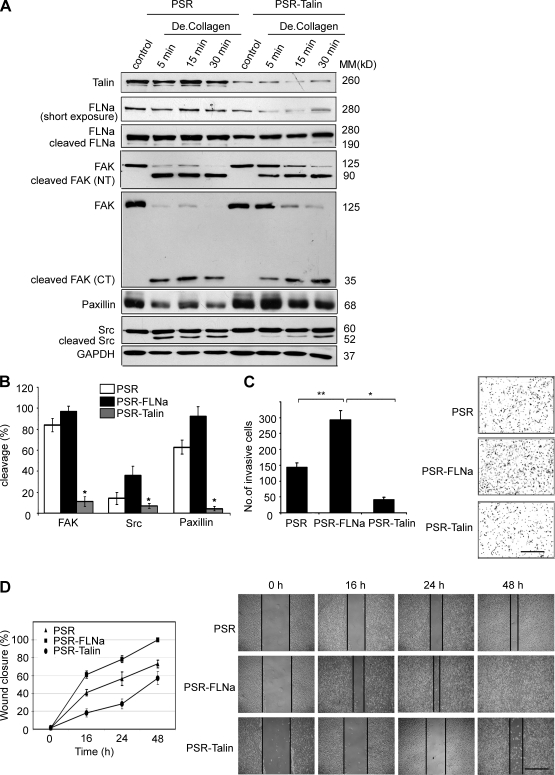
It has been demonstrated that calpain regulates cell spreading (Potter et al., 1998) and motility (Carragher et al., 2003) via modulation of cleavage of FA and cytoskeletal protein substrates (Glading et al., 2002). For instance, Franco et al. (2004b) reported that proteolysis of talin by calpain is the rate-limiting step for FA turnover. FLNa and talin have been shown to regulate integrin activation via a competitive binding to integrin (Kiema et al., 2006). Therefore, we investigated whether FLNa competes with other FA proteins for calpain cleavage.
We examined calpain-mediated cleavage activity on control and talin-silenced BT-20 cells stimulated with degraded collagen. The levels of full-length FAK, Src, and paxillin were found to be elevated in talin-silenced cells compared with control cells (Fig. 9 A). These results indicate that inhibition of talin can prevent the cleavage of FAK, Src, and possibly paxillin. Noticeable, FLNa level was reduced upon knockdown of talin, suggesting an induction of FLNa cleavage. Quantification of the cleavage products of FAK, Src, and paxillin after stimulation with degraded collagen for 5 min confirmed an increase in protein cleavage upon knockdown of FLNa and a decrease in protein cleavage upon knockdown of talin (P < 0.005; Fig. 9 B). Moreover, FLNa silencing promoted both cell invasion and migration, whereas talin silencing inhibited these phenotypes (Fig. 9, C and D; also reported in Fig. 3, B–D).
Figure 9.
FLNa and talin are competitive substrates for cleavage by calpain and differentially regulate breast cancer cell invasion. (A) Control (PSR) and talin-silenced (PSR-Talin) BT-20 cells were cultured on 10-cm dishes and stimulated with degraded type I collagen (De.Collagen) to promote calpain-mediated cleavage of FA proteins. Control indicates unstimulated. Cell lysates were immunoblotted with the indicated antibodies. CT, C terminus; MM, molecular mass; NT, N terminus. (B) Quantification and comparison of FA protein cleavage (mean ± SD) at 5-min time point after degraded collagen stimulation from Western blot results of three independent experiments as in A and Fig. S5 B (*, P < 0.005). (C) Invasion of control (PSR), FLNa-silenced (PSR-FLNa), and talin-silenced (PSR-Talin) BT-20 cells using the Boyden chamber assay as described in Materials and methods. The bar graph shows the quantification of cell invasion results as the mean ± SD of four independent experiments and five fields per condition (*, P < 0.005; **, P < 0.01). Representative images of invasive cells on the membrane are shown. (D) Bar graph shows the quantification of cell migration analyzed by the wound healing assay. Control (PSR), FLNa-silenced (PSR-FLNa), and talin-silenced (PSR-Talin) BT-20 cells were wounded and monitored over 48 h to determine the rate of migration into the scratched area. The experiment was repeated four times, and results are expressed as mean ± SD. Representative images of the wound captured at different time point are shown. The vertical black lines delimit the wounded regions at different time points. Bars: (C) 150 µm; (D) 200 µm.
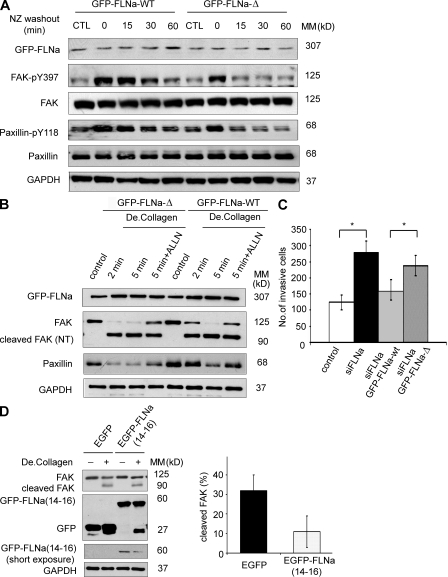
To further investigate whether FLNa may exert an indirect effect on the dynamics of FA through competition for calpain-mediated proteolysis, we used a noncleavable form of GFP-FLNa in which the calpain-targeting sequence (TYA: 1762–1764 aa) was deleted (GFP–FLNa-Δ). Although this mutation did not completely prevent FLNa cleavage, the levels of smaller molecular weight degradation products were found to be significantly reduced (Gorlin et al., 1990; Mammoto et al., 2007). We compared FA disassembly in FLNa-silenced BT-20 cells transfected with either GFP-FLNa WT (GFP–FLNa-WT) or GFP–FLNa-Δ mutant. Interestingly, BT-20 cells expressing siFLNa GFP–FLNa-Δ mutant showed a greater reduction of phosphorylation levels of FAK-pY397 and paxillin-pY118 compared with cells expressing siFLNa GFP–FLNa-WT (more evident at 15 and 30 min after nocodazole washout; Fig. 10 A). We then compared the cleavage of FA proteins after rescue of FLNa-silenced cells with either GFP–FLNa-WT or GFP–FLNa-Δ mutant. BT-20 siFLNa-silenced cells rescued with GFP–FLNa-Δ mutant showed a pronounced reduction of native FAK compared with BT-20 siFLNa rescued with GFP–FLNa-WT. Moreover, transfection of GFP–FLNa-WT in FLNa knockdown BT-20 cells successfully rescued cell invasion to control levels, whereas GFP–FLNa-Δ had no effect on rescue of this phenotype (Fig. 10 C). These data further indicate that cleavage of FLNa by calpain impacts the cleavage of other FA proteins such as FAK and can affect cell invasion. Moreover, transient expression of FLNa 14–16 repeats (enhanced GFP [EGFP]–FLNa 14–16), which includes hinge 1 (calpain-targeting sequence), partially rescued the phenotype in FLNa-silenced BT-20 cells with a slight but reproducible inhibition of calpain-mediated cleavage of FA proteins (Fig. 10 D) and FA turnover (Fig. S7) compared with FLNa-silenced BT-20 cells expressing EGFP alone.
Figure 10.
Cleavage of FLNa regulates FA turnover and cell invasion. (A) BT-20 siFLNa cells were transfected with construct either expressing GFP-tagged FLNa WT (GFP–FLNa-WT) or GFP-tagged FLNa mutant resistant to calpain cleavage (GFP–FLNa-Δ) and assayed for nocodazole (NZ)-based FA disassembly. Cells were untreated (control) or incubated with 10 µM nocodazole for 4 h. Cell lysates were collected at the indicated times and immunoblotted with anti–FAK-pY397 or anti–paxillin-pY118. (B) BT-20 siFLNa cells transfected with either GFP–FLNa-WT or GFP–FLNa-Δ were cultured on 10-cm dishes and stimulated or not with degraded type I collagen (De.Collagen) for the indicated times. Where indicated, cells were treated with 50 µM of calpain inhibitor ALLN (22 h). Lysates were prepared at the indicated time points and analyzed by Western blotting using the corresponding antibodies. Control indicates unstimulated. NT, N terminus. (C) Control BT-20 cells, BT-20 siFLNa cells, and BT-20 siFLNa cells expressing GFP–FLNa-WT or GFP–FLNa-Δ were placed in the top layer of Boyden migration chamber, and after 48 h, cells migrated through the chamber to the bottom membrane were measured. The graph represents the mean ± SD from four independent experiments (*, P < 0.005). (D) FLNa-silenced BT-20 cells were transfected with construct either expressing EGFP-tagged FLNa 14–16 repeats (EGFP-FLNa (14–16)) or expressing EGFP alone, stimulated or not stimulated with degraded type I collagen. Cell lysates were immunoblotted with the indicated antibodies. Bar graph shows quantification of cleavage of FAK (mean ± SD) from three independent experiments. MM, molecular mass.
DISCUSSION
The function of filamin in cancer cell migration and invasion is not fully understood. In this study, we report a correlation between low FLNa levels and breast cancer progression using a cancer TMA from patients with progressive disease, including lymph node–negative and –positive breast cancer. The inverse correlation between FLNa expression and cancer progression was further supported by results of the migration and invasion assays in alternative breast cancer cell lines, as well as in orthotopic breast cancer xenograft models in which down-regulation of FLNa is found to promote cancer cell invasion and metastasis formation. These results were unexpected in light of previous studies showing a positive association between FLNa expression levels and cell migration (Cunningham et al., 1992; Alper et al., 2009; Baldassarre et al., 2009). However, they are in agreement with other studies showing that FLNa overexpression in FLNa-deficient M2 cells (Cunningham et al., 1992; Nagano et al., 2004) or in mouse cortical neurons (Sarkisian et al., 2006) inhibited cell migration, and an association of FLNa with carcinoembryonic antigen-related cell adhesion molecule has been shown to strengthen cell–cell contact and thus acts negatively on tumor cell invasion and metastasis formation (Klaile et al., 2005). In humans, FLNa gene duplications rather than loss of function mutations were found in a patient with a severe form of X-linked periventricular heterotopia (Fink et al., 1997). Moreover, Lin et al. (2007) reported decreased intracellular levels of an endogenous fragment of FLNa (repeat 7–15) in patients with advanced prostate cancer, whereas another study found low levels of endogenous FLNa in advanced and aggressive prostate cancer compared with benign tissues (Varambally et al., 2005).
Our results on breast cancer tissues support a potential utility of filamins as signature biomarkers for invasive cancers because FLNa levels were significantly decreased in invasive breast cancer. However, they bring up challenging questions on how the FLNa signaling nexus regulates breast cancer cell invasion. Using alternate cancer models, we observed that down-regulation of FLNa can increase cancer cell migration via regulation of FA turnover, at least in part because of activation of calpain activity and FA protein cleavage. A previous study demonstrated that cell spreading associated with FLNa knockdown is partially compensated by competing proteins that bind to β-integrin, including talin, which can activate integrin (Kiema et al., 2006). Suppression of FLNa could increase talin binding to integrin β tail and thus enhance integrin activation and cell motility. In addition to the competition between FLNa and talin for integrin β tail, our data suggest a second level of competition between FLNa, talin, and other FA proteins such as FAK and paxillin via calpain cleavage; this could impact the regulation of FA turnover (Fig. S8).
Molecular mechanisms of FA disassembly, unlike FA assembly (Raftopoulou and Hall, 2004), are not fully defined. Our biochemical and real-time imaging results revealed that FLNa-silenced cells have elevated calpain activity and accelerated FA disassembly compared with their matched control cells. This observation is significant because FA formation at the leading edge and disassembly at the trailing edge of a motile cell are established to be critical events for the initiation and maintenance of cell locomotion. Previous studies have reported that binding of FLNa to integrin can inhibit cell migration without affecting FA formation or fibronectin matrix assembly (Pfaff et al., 1998; Calderwood et al., 2001). Moreover, a study by Bhatt et al. (2002) reported that calpain inhibition had no effect on microtubule targeting of FA sites but rather stabilized peripheral adhesive complexes and inhibited microtubule-mediated turnover of FA after nocodazole washout. Therefore, although our data support a functional connection between FLNa levels and FA disassembly kinetics, it remains to be determined whether such a connection occurs in vivo. In particular, a recent study using live-cell microscopy reported that FAs in cells embedded in a three-dimensional matrix are not readily detected and that FA proteins such as vinculin, paxillin, talin, FAK, p130Cas, α-actinin, and zyxin do not seem to form aggregates but are diffusely distributed throughout the cytoplasm (Fraley et al., 2010). Despite the absence of detectable FAs, FA proteins were still able to modulate cell motility by affecting protrusion activity and extracellular matrix remodeling in a manner that is distinct from cells grown on two-dimensional substrates.
Our data also support a role of calpain in FLNa-induced regulation of FA cleavage because FLNa silencing led to increased proteolysis of talin and other FA proteins such as FAK and paxillin. Calpains regulate the dynamics of both integrin-mediated adhesions and actin-based membrane protrusions (Perrin and Huttenlocher, 2002), but the molecular mechanisms underlying this regulation remain partially understood. Cleavage of specific FA proteins by calpain can alter the composition of adhesion complexes, thus allowing dynamic recruitment and anchoring properties of the cell cytoskeleton. Of relevance to our finding, Franco et al. (2004b) reported a role for calpain in the turnover of integrin-mediated cell adhesion via talin proteolysis. In particular, this study demonstrated that calpain-mediated proteolysis of talin is a rate-limiting step during FA turnover. In line with these findings, our results support that FLNa and talin also compete for cleavage by the calpain protease because FLNa silencing led to increased proteolysis of talin, as well as of other FA proteins such as FAK and paxillin. It is intriguing to note that calpain action is primarily caused by its ability to modify rather than degrade its substrates. Previous studies indicated that calpain cleavage products such as the talin head domain can regulate integrin activation and FA turnover (Franco et al., 2004b; Goksoy et al., 2008; Huang et al., 2009). More recently, a study reported that cleavage of FAK by calpain can also regulate adhesion dynamics (Chan et al., 2010). In the case of FLNa, calpain induced a rapid cleavage of the protein at hinge 1 (between aa 1762 and 1764) generating fragments of 190 and 100 kD (Kiema et al., 2006). A 10-kD small fragment can also be generated from the end of the 100 kD (cleavage of FLNa subfragment at hinge 2) after longer calpain digestion. These cleavage kinetics indicated that hinge 1 is a predominant first cleavage site (1 min after incubation with calpain, whereas hinge 2 cleavage was seen only 5 min after incubation with calpain; Gorlin et al., 1990). In our study, transfection of FLNa knockdown cells with the noncleavable form of GFP–FLNa-Δ (which has a deletion in the calpain-targeting sequence located in hinge 1) failed to rescue FA turnover or cell invasion as we observed with the WT FLNa. Therefore, mutation of this cleavage site is critical to regulate the competition between FLNa and other calpain substrates. In support of this hypothesis, expression of FLNa 14–16 repeats (which includes hinge 1) in FLNa-silenced cells rescued, although only partially, the reduced cleavage of FA proteins and FA turnover observed after FLNa silencing. This partial rescue could possibly be explained by low efficiency of transient transfection or a requirement of full FLNa protein for optimal interaction with its partners. Moreover, it is not excluded that the partial rescue may be contributed by the short half-life of the exogenous EGFP–FLNa 14–16 fusion protein, which we found to be very unstable in BT-20 cells compared with control cells expressing EGFP alone (unpublished data).
Moreover, recent studies postulated that FLNa cleavage fragments can also function as a signaling integrator of proteolytic peptides and their generation could be regulated by phosphorylation/dephosphorylation (Jay et al., 2004; García et al., 2006). FLNa phosphorylation on Ser2152 by Pak1 (p21-activated kinase 1; Vadlamudi et al., 2002), protein kinase A (Jay et al., 2004), and ribosomal S6 kinase (Woo et al., 2004) has been implicated in inducing FLNa resistance to calpain cleavage and in mediating stimulation of membrane ruffle formation and cell migration, respectively. Furthermore, Mammoto et al. (2007) demonstrated that FLNa regulates p190RhoGAP translocation to lipid rafts and Rho activation, actions which were mediated by calpain cleavage of FLNa. Therefore, FLNa and talin can allow the reorganization of integrin links to actin and/or dynamic recruitment of selective substrates for calpain cleavage; the resulting fragments can modulate the process of actin reorganization, cell spreading, and cell migration.
The regulation of calpain 2 by FLNa opens up a challenging question on the mechanism by which this is accomplished. FLNa and several FA proteins can act as scaffolding proteins via multiple protein–protein interactions. One candidate we have investigated is the MAPK–ERK pathway, which regulates both FLNa (Awata et al., 2001; Hjälm et al., 2001; Zhang and Breitwieser, 2005; Onoprishvili et al., 2008) and calpain 2 (Glading et al., 2001, 2004) activities, as well as calpain 2 colocalization with FA proteins in FA sites (Beckerle et al., 1987). FLNa regulates MAPK activity via interaction with several mitogen-activated proteins, including MAPK kinase 4 (MKK-4/SEK-1; Marti et al., 1997), or modulation of the activity of guanine nucleotide exchange factors of the Ras–MAPK pathway (Gorlin et al., 1990; Zhu et al., 2007). We observed a high MAPK activity and ERK phosphorylation in FLNa-silenced cells compared with control cells. This induction of MAPK–ERK activity was sufficient to stimulate calpain activity and was prevented by cell exposure to the MEK inhibitor UO126 or reexpression of FLNa in FLNa-silenced cells. In contrast, immunostaining of phospho-ERK did not reveal any obvious changes in subcellular localization, suggesting a global activation of ERK pathway upon FLNa silencing.
MAPK–ERK activation has a broad implication on cellular processes and can impact multiple signaling pathways in addition to calpains. For example, FLNa down-regulation has been reported to promote matrix metalloproteinase secretion (Zhu et al., 2007), an additional mechanism that can contribute to explain the association of FLNa down-regulation and cancer progression. In conclusion, our data further support FLNa participation as a scaffolding signaling molecule to link the cell cytoskeleton remodeling to FA dynamics.
MATERIALS AND METHODS
Antibodies and reagents.
Antibody to paxillin was purchased from Thermo Fisher Scientific. Monoclonal antibody to FLNa N-terminal portion (MCA464ST) was obtained from AbD Serotec. A polyclonal antibody to FLNb was obtained from Abcam. Antibodies to phospho-FLNa-Ser2152 and phospho-ERK1/2 were purchased from Cell Signaling Technology. Antibodies to FAK (A-17) and FAK (C-20) were purchased from Santa Cruz Biotechnology, Inc. Antibodies to FAK (clone 4.47), FAK-pY397, and paxillin-pY118 were purchased from Invitrogen. Antibody to Src (clone GD11) was purchased from Millipore. Antibodies to vinculin (clone hVLN-1) and talin (clone 8D4) were obtained from Sigma-Aldrich. Antibody to GAPDH was obtained from Cedarlane, and antibody to α-tubulin was purchased from Abcam. Fibronectin, collagenase type I, 10× RPMI 1640, and nocodazole were purchased from Sigma-Aldrich. Secondary antibodies for immunofluorescence were obtained from Jackson ImmunoResearch Laboratories, Inc. Collagen was obtained from BD. Calpain inhibitor experiments were performed using calpain inhibitor 1 (EMD).
Cell culture.
Breast adenocarcinoma cell lines MCF-7, MDA-231, and BT-20 were obtained from American Type Culture Collection and maintained in RPMI 1640 (Mediatech, Inc.) supplemented with 10% FBS and supplemented with penicillin and streptomycin antibiotics. ErbB-2 receptor was overexpressed using a bicistronic retrovector that coexpressed ErbB2 receptor with EGFP (Yen et al., 2002).
Collagen matrix preparation.
Polymerized collagen gels (1.4 mg/ml final concentration) were prepared by neutralization of an 8/10 vol of collagen solution with 1/10 vol of 10× RPMI 1640 concentrate, 1/10 vol of 0.1 M NaOH, and 1/40 vol of 7.5% NaHCO3, and then 1 vol of 1× DME was added and incubated at 37°C for 24 h. Degraded type I collagen was prepared by incubating polymerized collagen gels with 2.5 mg/ml collagenase type I for 30 min at 37°C. After digestion, collagenase activity was inhibited by the addition of an equal volume of 1× DME containing 10% FBS (Carragher et al., 1999).
Generation of stable FLNa and talin knockdown cells.
Stable FLNa and talin knockdown cells were generated in a polyclonal population as described previously (Benlimame et al., 2005). FLNa (available from GenBank/EMBL/DDBJ under accession no. NM_001456.3) sequence 1, 5′-GGGCTGACAACAGTGTGGTGC-3′ (targeting nt 3323–3343); and sequence 2, 5′-GGTGCTGCCTACTCATGATGC-3′ (targeting nt 4663–4683; Huang et al., 2006); and talin (available from GenBank under accession no. NM_006289.3) sequence 1, 5′-GACTGTCACTGACATGCTCAT-3′ (targeting nt 672–692, not reported before); and sequence 2, 5′-TCGTGAGGGTACTGAAACT-3′ (targeting nt 6169–6187; Manevich et al., 2007) are the target sequences used for stable knockdown of FLNa and talin. Each sequence was cloned as inverted repeats into PSR puromycin vector according to the manufacturer’s instructions (Oligoengine).
Construction of plasmid.
The DNA segment encoding human FLNa residues 1534–1875 (repeats 14–16) was amplified from pEGFP-C1-FLNa WT. The 5′ primer 5′-GCAAGCTTGCCCCTTCAAGGTCAAGG-3′ and 3′ primer 5′-GCTCTAGAGAGGCCAGGCCCATA-3′ were used to amplify a 1-kbp PCR product that was inserted into the HindIII and XbaI sites of the pEGFP-C1 plasmid.
For WT FLNa rescue, we designed an siRNA-resistant FLNa by introducing a silent mutation at position 3330 (gac→gat) using standard site-specific mutagenesis assay. The cDNA clone was confirmed by sequencing using a dye primer cycle sequencing kit (PerkinElmer) and DNA sequencer (ABI 377; PerkinElmer).
Transfection of siRNA.
Calpain 2 (available from GenBank under accession no. NM_001748.4) sequence 1, 5′-GACTTCACCGGAGGCATTG-3′ (targeting nt 840–858, not reported before); and sequence 2, 5′-GTACCTCAACCAGGACTAC-3′ (targeting nt 302–320; Franco et al., 2004b; Benetti et al., 2005) are the target sequences used for transient knockdown of calpain 2. 100 nM of siRNA oligonucleotides (Thermo Fisher Scientific) was incubated with DharmaFECT1 (Thermo Fisher Scientific) in Opti-MEM I reduced serum medium (Invitrogen) according to the manufacturer’s instructions.
Western blot analysis.
Western blot analyses were performed on total cell extracts from exponentially growing cells collected by scrapping into modified radioimmunoprecipitation assay lysis buffer supplemented with 20 mg/ml pepstatin A, 1 mM PMSF, and protease inhibitor cocktail (Roche) as we described previously (Benlimame et al., 2005). Blots were detected using the appropriate antibodies, and signal was detected with horseradish peroxidase–conjugated secondary antibodies and enhanced chemiluminescence detection system. When indicated, membranes were subsequently stripped for reprobing. Immunoblots were quantified by using ImageJ (National Institutes of Health) software.
FA disassembly assay.
The assay for FA disassembly and reformation after nocodazole treatment has been described previously (Ezratty et al., 2005; Yeo et al., 2006). Serum-starved cells were grown on glass coverslips and treated with 10 µM nocodazole for 4 h to completely depolymerize microtubules. The drug was washed out with serum-free media, and cells were fixed and processed for immunofluorescence at different intervals. Cells were stained with vinculin, FAK-pY397, paxillin-pY118, and/or α-tubulin.
Indirect immunofluorescence analysis.
The cells were fixed with paraformaldehyde, permeabilized, and labeled for immunofluorescence essentially as we described previously (Yen et al., 2002; Benlimame et al., 2005). Slides were viewed in a fluorescent microscope (Axiophot; Carl Zeiss, Inc.) equipped with a 63× Plan-Apochromat objective and selective filters. Images were acquired from a cooled charge-coupled device camera (Retiga 1300; QImaging) and displayed on a high resolution monitor. Images were analyzed by a Northern Eclipse Image analysis system (Carl Zeiss, Inc.).
Invasion assay and migration assay.
Cell invasion assays were performed using 8-µm porous chambers coated with Matrigel (BD) according to the manufacturer’s recommendations. Cell migration was assayed using the qualitative wound healing assay (Benlimame et al., 2005). Each experiment was performed at least three times, and results are expressed as mean ± SD. Statistical significance was analyzed using the Student’s t test.
Phagokinetic tracks assay.
To analyze cell motility, the phagokinetic track assay was conducted as described previously (Albrecht-Buehler, 1977). Gold colloid particles were prepared, and glass coverslips were coated with the particles. BT-20 cells were plated on the gold particle–coated glass coverslips, cultured for 24 h, and then fixed with 3.5% formaldehyde. The phagokinetic range was observed and photographed by brightfield microscopy. The area of migration was calculated by ImageJ software.
Calpain activity.
Cells were grown to confluence and lysed with radioimmunoprecipitation assay lysis buffer. The lysate was cleared of debris and used for the Calpain-Glo protease assay (Promega) according to the manufacturer’s protocol. The relative luminescence was averaged over 10 s, background-subtracted, and normalized to the amount of protein in the lysate (Boehmerle et al., 2007). Statistical significance was analyzed using the Student’s t test.
MAPK kinase assay.
Serum-starved cells at 70% confluence were stimulated with 5 ng/ml EGF for 10 min (this time point was chosen based on a preliminary time-response study indicating a maximum MAPK activity 10 min after EGF stimulation followed by a rapid decrease in activity). Cells were then washed with cold PBS and lysed with a lysis buffer (containing 50 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, 0.1% Triton X-100, 10% glycerol, 0.5 mM DTT, 1 mM Na3VO4, 1 mM PMSF, 5 mg/ml aprotinin, and 5 mg/ml leupeptin) and immunoprecipitated using a polyclonal anti-ERK2 antibody (Santa Cruz Biotechnology, Inc.). The immunoprecipitates were washed in Triton X-100–only lysis buffer, followed by kinase buffer (10% glycerol, 20 mM Hepes, 10 mM MgCl2, 10 mM MnCl2, and 100 mM NaCl). Immunoprecipitates were then incubated for 15 min at 32°C in 30 µl of kinase buffer containing 4 µg of myelin basic protein (MBP), 20 µM ATP, and 5 µCi/nmol γ-[32P]ATP (3,000 Ci/mmol). Reactions were stopped with 2× SDS-PAGE sample buffer, resolved by SDS-PAGE, stained with Coomassie blue, and visualized by autoradiography, and the radioactivity incorporated into MBP was determined by Cerenkov counting of the excised MBP bands. The data shown represent the mean ± SD of three independent experiments.
Live fluorescence microscopy.
For live fluorescent imaging, the BT-20 cells were stably transfected with EYFP-vinculin or EYFP-paxillin and plated in RPMI 1640 on a multiwell chambered coverglass (LabTek) and stimulated with 20 ng/ml EGF. Cells were placed directly on a heated stage and supplemented with 5% CO2. Fluorescent images were captured every 4 min for 2 h using a heated 63× NA 1.40 objective at the optimized Nipkow spinning disk confocal microscope (WaveFx spinning disk; Quorum Technologies Inc.). Cooled charge-coupled device camera control and image acquisition were performed using Volocity imaging software (PerkinElmer). At the extremely high speed, we investigated different cells in ∼15 s intervals. The experimental results show representative cells from a minimum of five different experiments (Xu et al., 2009).
Quantification of FA protein dynamics.
Fluorescence intensities of individual adhesions from background-subtracted images were measured over time using Volocity imaging software and quantified according to the previously described protocol (Franco et al., 2004b; Webb et al., 2004). Measurements were made on at least 25 individual adhesions in five separate cells for both FLNa silenced and control cells. Duration measurements were made for these same adhesions by counting the amount of time lapsed between the first and last frames in which an individual adhesion was observed. Data are presented as mean ± SD. Statistical significance was analyzed using the Student’s t test.
In vivo xenograft model of breast cancer metastasis.
In vivo experiments were approved by the McGill University Animal Care Committee (protocol number 4101). Control MDA-231-ErbB2 cells expressing scrambled shRNA and their matched polyclonal population of cells stably expressing FLNa shRNA (106 cells/each mouse) were transplanted into the mammary fat pad of SCID mice. Tumor size was measured using a caliper, and tumor volume was calculated as π/6 (length × width2). Primary tumors were excised surgically once they had reached a mean size of 0.8 cm3. The wound was then closed with a single layer of surgical clips. Mice were sacrificed 120 d after the cell injection. The lungs were fixed in 10% Bouin’s fixative, and lung surface metastases were counted using a stereomicroscope (Optimax; Leica). Statistical analysis was performed using the multivariance analysis unpaired Student’s t test.
TMA construction and immunohistochemistry.
The human tissue study and access to clinical and pathological data were approved by the institutional review boards of the Jewish General Hospital (Montreal, Quebec, Canada) and Institut fur klinische Pathologie of Universitäts Spital (Zurich, Switzerland). Three TMAs from 222 samples from 75 patients with progressive breast cancer were assembled using the manual tissue array (Beecher Instruments, Inc.). In addition, an independent TMA was assembled from a total of 258 lymph node–positive and –negative breast cancer patients’ samples. Tissue cores from circled areas were targeted for transfer to the recipient array blocks. Three to five tissue cores were sampled from each patient sample. The 1.0-mm diameter TMA cores were each spaced at 1.5 mm from core center to core center. All patients’ blocks were assembled without prior knowledge of associated clinical or pathology staging information. After construction, 4-µm sections were cut and stained with hematoxylin and eosin on the initial slides to verify the histological diagnosis. To analyze the expression of FLNa in breast cancer TMAs, 4-µm sections were cut and placed on silane-coated slides (Sigma-Aldrich) and dried overnight at 37°C. TMA sections were deparaffinized in graded alcohols, rehydrated, and boiled (microwave) in 10 mM citrate buffer, pH 6.0, for antigen retrieval. The TMA slides were subsequently incubated for 40 min at 37°C with primary monoclonal mouse anti-FLNa (1:4,000; MCA464ST) or anti–phospho-FLNa (Ser2152) antibody, using the NexES automated immunostainer (Ventana Medical Systems, Inc.). The automated Ventana Medical Systems, Inc. system uses an indirect biotin–avidin system with a universal biotinylated immunoglobulin secondary antibody. Diaminobenzidine was used as a chromogen, and slides were counterstained with hematoxylin before mounting. All staining procedures were performed according to the manufacturer’s recommendations. A multitissue control TMA slide containing samples of prostate, colon, and breast was screened using the primary antibody or normal mouse 1:200 prediluted serum (Ventana Medical Systems, Inc.) as positive and negative controls, respectively.
Manual scoring of FLNa and data analysis.
All TMA cores were assigned a diagnosis (i.e., benign, in situ carcinoma, invasive breast cancer, and lymph node metastatic breast cancer) by the study pathologist. All manual scoring was performed on a microscope (×100; BX41; Olympus). The protein expression was evaluated using a categorical scoring method ranging from 0 to 3 (0, negative; 1, weak; 2, moderate; and 3, strong). Moreover, the extent of expression was quantified into four subgroups of <25, 25–50, 50–75, and 75–100%. A final score was obtained by multiplying the intensity and extent of immunostaining within each sample. All data analyses were performed by means of the computer-based statistical package of SPSS version 5.1 (SPSS, Inc.) for Windows. Results were reported as mean ± SEM. Differences among the three groups (benign, in situ carcinoma, and invasive carcinoma) were compared with one-way analysis of variance and the post-hoc Tukey honestly significant difference test for multiple comparisons. A p-value of ≤0.05 was considered statistically significant.
Online supplemental material.
Fig. S1 shows the expression of FLNb and phospho-FLNa in breast cancer patients with progressive disease. Fig. S2 depicts Western blot analysis showing efficient knockdown of FLNa, talin, and calpain 2 by targeting different sequences. Fig. S3 shows that FLNa regulates FA disassembly. Fig. S4 shows cleavage of FAK in response to treatment with degraded type I collagen. Fig. S5 shows the effect of FLNa inhibition on calpain activity. Fig. S6 shows that calpain inhibitor ALLN abolishes the effect of FLNa in regulation of FA disassembly. Fig. S7 shows that expression of FLNa 14–16 repeats partially rescues the effect of FLNa knockdown in regulation of FA disassembly. Fig. S8 shows a schematic representation of FLNa regulation of cell migration. Videos 1–3 show FA dynamics of BT-20 control, FLNa-silenced, and calpain 2–silenced cells, respectively. Video 4 shows FA dynamics of BT-20 cells with FLNa and calpain double knockdown. Videos 5 and 6 show FA dynamics of ALLN-treated BT-20 control and FLNa-silenced cells, respectively. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100433/DC1.
Acknowledgments
We thank Krikor Bijian for the help with the design of FLNa resistant to siRNA, Christian Young and Naciba Benlimame for assistance with the spinning disc confocal microscopy, and Dr. Koren Mann for reading the manuscript.
This work was supported by the Canadian Institutes of Health Research (CIHR) and in part by the Canadian Cancer Society. M.A. Alaoui-Jamali is a Fonds de la recherche en santé du Québec Scholar and a recipient of the Dundi and Lyon Sachs Scientist Award. Y. Xu is a recipient of a CIHR training award and Maysie MacSporran Studentship from the Faculty of Medicine, McGill University. D.E. Ingber is a recipient of a Department of Defense Breast Cancer Research Program Innovator Award (#BC074986).
The authors declare that they have no conflicting financial interests.
Footnotes
Abbreviations used:
- EGF
- epidermal growth factor
- EGFP
- enhanced GFP
- ERK
- extracellular signal-regulated kinase
- EYFP
- enhanced YFP
- FA
- focal adhesion
- FAK
- FA kinase
- MAPK
- mitogen-activated protein kinase
- MBP
- myelin basic protein
- PSR
- pSuper retro
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- TMA
- tissue microarray
References
- Albrecht-Buehler G. 1977. The phagokinetic tracks of 3T3 cells. Cell. 11:395–404 10.1016/0092-8674(77)90057-5 [DOI] [PubMed] [Google Scholar]
- Alper O., Stetler-Stevenson W.G., Harris L.N., Leitner W.W., Ozdemirli M., Hartmann D., Raffeld M., Abu-Asab M., Byers S., Zhuang Z., et al. 2009. Novel anti-filamin-A antibody detects a secreted variant of filamin-A in plasma from patients with breast carcinoma and high-grade astrocytoma. Cancer Sci. 100:1748–1756 10.1111/j.1349-7006.2009.01244.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata H., Huang C., Handlogten M.E., Miller R.T. 2001. Interaction of the calcium-sensing receptor and filamin, a potential scaffolding protein. J. Biol. Chem. 276:34871–34879 10.1074/jbc.M100775200 [DOI] [PubMed] [Google Scholar]
- Baldassarre M., Razinia Z., Burande C.F., Lamsoul I., Lutz P.G., Calderwood D.A. 2009. Filamins regulate cell spreading and initiation of cell migration. PLoS One. 4:e7830 10.1371/journal.pone.0007830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle M.C., Burridge K., DeMartino G.N., Croall D.E. 1987. Colocalization of calcium-dependent protease II and one of its substrates at sites of cell adhesion. Cell. 51:569–577 10.1016/0092-8674(87)90126-7 [DOI] [PubMed] [Google Scholar]
- Benetti R., Copetti T., Dell’Orso S., Melloni E., Brancolini C., Monte M., Schneider C. 2005. The calpain system is involved in the constitutive regulation of beta-catenin signaling functions. J. Biol. Chem. 280:22070–22080 10.1074/jbc.M501810200 [DOI] [PubMed] [Google Scholar]
- Benlimame N., He Q., Jie S., Xiao D., Xu Y.J., Loignon M., Schlaepfer D.D., Alaoui-Jamali M.A. 2005. FAK signaling is critical for ErbB-2/ErbB-3 receptor cooperation for oncogenic transformation and invasion. J. Cell Biol. 171:505–516 10.1083/jcb.200504124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershadsky A., Chausovsky A., Becker E., Lyubimova A., Geiger B. 1996. Involvement of microtubules in the control of adhesion-dependent signal transduction. Curr. Biol. 6:1279–1289 10.1016/S0960-9822(02)70714-8 [DOI] [PubMed] [Google Scholar]
- Bhatt A., Kaverina I., Otey C., Huttenlocher A. 2002. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J. Cell Sci. 115:3415–3425 [DOI] [PubMed] [Google Scholar]
- Boehmerle W., Zhang K., Sivula M., Heidrich F.M., Lee Y., Jordt S.E., Ehrlich B.E. 2007. Chronic exposure to paclitaxel diminishes phosphoinositide signaling by calpain-mediated neuronal calcium sensor-1 degradation. Proc. Natl. Acad. Sci. USA. 104:11103–11108 10.1073/pnas.0701546104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D.A., Huttenlocher A., Kiosses W.B., Rose D.M., Woodside D.G., Schwartz M.A., Ginsberg M.H. 2001. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3:1060–1068 10.1038/ncb1201-1060 [DOI] [PubMed] [Google Scholar]
- Carragher N.O., Levkau B., Ross R., Raines E.W. 1999. Degraded collagen fragments promote rapid disassembly of smooth muscle focal adhesions that correlates with cleavage of pp125FAK, paxillin, and talin. J. Cell Biol. 147:619–630 10.1083/jcb.147.3.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher N.O., Westhoff M.A., Fincham V.J., Schaller M.D., Frame M.C. 2003. A novel role for FAK as a protease-targeting adaptor protein: regulation by p42 ERK and Src. Curr. Biol. 13:1442–1450 10.1016/S0960-9822(03)00544-X [DOI] [PubMed] [Google Scholar]
- Chan K.T., Bennin D.A., Huttenlocher A. 2010. Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J. Biol. Chem. 285:11418–11426 10.1074/jbc.M109.090746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C.C., Gorlin J.B., Kwiatkowski D.J., Hartwig J.H., Janmey P.A., Byers H.R., Stossel T.P. 1992. Actin-binding protein requirement for cortical stability and efficient locomotion. Science. 255:325–327 10.1126/science.1549777 [DOI] [PubMed] [Google Scholar]
- Danowski B.A. 1989. Fibroblast contractility and actin organization are stimulated by microtubule inhibitors. J. Cell Sci. 93:255–266 [DOI] [PubMed] [Google Scholar]
- Eksioglu Y.Z., Scheffer I.E., Cardenas P., Knoll J., DiMario F., Ramsby G., Berg M., Kamuro K., Berkovic S.F., Duyk G.M., et al. 1996. Periventricular heterotopia: an X-linked dominant epilepsy locus causing aberrant cerebral cortical development. Neuron. 16:77–87 10.1016/S0896-6273(00)80025-2 [DOI] [PubMed] [Google Scholar]
- Enomoto T. 1996. Microtubule disruption induces the formation of actin stress fibers and focal adhesions in cultured cells: possible involvement of the rho signal cascade. Cell Struct. Funct. 21:317–326 10.1247/csf.21.317 [DOI] [PubMed] [Google Scholar]
- Ezratty E.J., Partridge M.A., Gundersen G.G. 2005. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7:581–590 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Feng Y., Walsh C.A. 2004. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6:1034–1038 10.1038/ncb1104-1034 [DOI] [PubMed] [Google Scholar]
- Feng Y., Chen M.H., Moskowitz I.P., Mendonza A.M., Vidali L., Nakamura F., Kwiatkowski D.J., Walsh C.A. 2006. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. USA. 103:19836–19841 10.1073/pnas.0609628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J.M., Dobyns W.B., Guerrini R., Hirsch B.A. 1997. Identification of a duplication of Xq28 associated with bilateral periventricular nodular heterotopia. Am. J. Hum. Genet. 61:379–387 10.1086/514863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.W., Lamperti E.D., Eksioglu Y.Z., Hong S.E., Feng Y.Y., Graham D.A., Scheffer I.E., Dobyns W.B., Hirsch B.A., Radtke R.A., et al. 1998. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 21:1315–1325 10.1016/S0896-6273(00)80651-0 [DOI] [PubMed] [Google Scholar]
- Fraley S.I., Feng Y., Krishnamurthy R., Kim D.H., Celedon A., Longmore G.D., Wirtz D. 2010. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 12:598–604 10.1038/ncb2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S., Perrin B., Huttenlocher A. 2004a. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp. Cell Res. 299:179–187 10.1016/j.yexcr.2004.05.021 [DOI] [PubMed] [Google Scholar]
- Franco S.J., Rodgers M.A., Perrin B.J., Han J., Bennin D.A., Critchley D.R., Huttenlocher A. 2004b. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6:977–983 10.1038/ncb1175 [DOI] [PubMed] [Google Scholar]
- García E., Stracher A., Jay D. 2006. Calcineurin dephosphorylates the C-terminal region of filamin in an important regulatory site: a possible mechanism for filamin mobilization and cell signaling. Arch. Biochem. Biophys. 446:140–150 10.1016/j.abb.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Glading A., Uberall F., Keyse S.M., Lauffenburger D.A., Wells A. 2001. Membrane proximal ERK signaling is required for M-calpain activation downstream of epidermal growth factor receptor signaling. J. Biol. Chem. 276:23341–23348 10.1074/jbc.M008847200 [DOI] [PubMed] [Google Scholar]
- Glading A., Lauffenburger D.A., Wells A. 2002. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol. 12:46–54 10.1016/S0962-8924(01)02179-1 [DOI] [PubMed] [Google Scholar]
- Glading A., Bodnar R.J., Reynolds I.J., Shiraha H., Satish L., Potter D.A., Blair H.C., Wells A. 2004. Epidermal growth factor activates m-calpain (calpain II), at least in part, by extracellular signal-regulated kinase-mediated phosphorylation. Mol. Cell. Biol. 24:2499–2512 10.1128/MCB.24.6.2499-2512.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goksoy E., Ma Y.Q., Wang X., Kong X., Perera D., Plow E.F., Qin J. 2008. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell. 31:124–133 10.1016/j.molcel.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin J.B., Yamin R., Egan S., Stewart M., Stossel T.P., Kwiatkowski D.J., Hartwig J.H. 1990. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J. Cell Biol. 111:1089–1105 10.1083/jcb.111.3.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514 10.1126/science.279.5350.509 [DOI] [PubMed] [Google Scholar]
- Hart A.W., Morgan J.E., Schneider J., West K., McKie L., Bhattacharya S., Jackson I.J., Cross S.H. 2006. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum. Mol. Genet. 15:2457–2467 10.1093/hmg/ddl168 [DOI] [PubMed] [Google Scholar]
- Hjälm G., MacLeod R.J., Kifor O., Chattopadhyay N., Brown E.M. 2001. Filamin-A binds to the carboxyl-terminal tail of the calcium-sensing receptor, an interaction that participates in CaR-mediated activation of mitogen-activated protein kinase. J. Biol. Chem. 276:34880–34887 10.1074/jbc.M100784200 [DOI] [PubMed] [Google Scholar]
- Huang C., Wu Z., Hujer K.M., Miller R.T. 2006. Silencing of filamin A gene expression inhibits Ca2+ -sensing receptor signaling. FEBS Lett. 580:1795–1800 10.1016/j.febslet.2006.02.035 [DOI] [PubMed] [Google Scholar]
- Huang C., Rajfur Z., Yousefi N., Chen Z., Jacobson K., Ginsberg M.H. 2009. Talin phosphorylation by Cdk5 regulates Smurf1-mediated talin head ubiquitylation and cell migration. Nat. Cell Biol. 11:624–630 10.1038/ncb1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay D., García E.J., de la Luz Ibarra M. 2004. In situ determination of a PKA phosphorylation site in the C-terminal region of filamin. Mol. Cell. Biochem. 260:49–53 10.1023/B:MCBI.0000026052.76418.55 [DOI] [PubMed] [Google Scholar]
- Kiema T., Lad Y., Jiang P., Oxley C.L., Baldassarre M., Wegener K.L., Campbell I.D., Ylänne J., Calderwood D.A. 2006. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 21:337–347 10.1016/j.molcel.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Klaile E., Müller M.M., Kannicht C., Singer B.B., Lucka L. 2005. CEACAM1 functionally interacts with filamin A and exerts a dual role in the regulation of cell migration. J. Cell Sci. 118:5513–5524 10.1242/jcs.02660 [DOI] [PubMed] [Google Scholar]
- Lin J.F., Xu J., Tian H.Y., Gao X., Chen Q.X., Gu Q., Xu G.J., Song J.D., Zhao F.K. 2007. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int. J. Cancer. 121:2596–2605 10.1002/ijc.23016 [DOI] [PubMed] [Google Scholar]
- Mammoto A., Huang S., Ingber D.E. 2007. Filamin links cell shape and cytoskeletal structure to Rho regulation by controlling accumulation of p190RhoGAP in lipid rafts. J. Cell Sci. 120:456–467 10.1242/jcs.03353 [DOI] [PubMed] [Google Scholar]
- Manevich E., Grabovsky V., Feigelson S.W., Alon R. 2007. Talin 1 and paxillin facilitate distinct steps in rapid VLA-4-mediated adhesion strengthening to vascular cell adhesion molecule 1. J. Biol. Chem. 282:25338–25348 10.1074/jbc.M700089200 [DOI] [PubMed] [Google Scholar]
- Marti A., Luo Z., Cunningham C., Ohta Y., Hartwig J., Stossel T.P., Kyriakis J.M., Avruch J. 1997. Actin-binding protein-280 binds the stress-activated protein kinase (SAPK) activator SEK-1 and is required for tumor necrosis factor-alpha activation of SAPK in melanoma cells. J. Biol. Chem. 272:2620–2628 10.1074/jbc.272.5.2620 [DOI] [PubMed] [Google Scholar]
- Nagano T., Morikubo S., Sato M. 2004. Filamin A and FILIP (Filamin A-Interacting Protein) regulate cell polarity and motility in neocortical subventricular and intermediate zones during radial migration. J. Neurosci. 24:9648–9657 10.1523/JNEUROSCI.2363-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura F., Osborn E., Janmey P.A., Stossel T.P. 2002. Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J. Biol. Chem. 277:9148–9154 10.1074/jbc.M111297200 [DOI] [PubMed] [Google Scholar]
- Onoprishvili I., Ali S., Andria M.L., Shpigel A., Simon E.J. 2008. Filamin A mutant lacking actin-binding domain restores mu opioid receptor regulation in melanoma cells. Neurochem. Res. 33:2054–2061 10.1007/s11064-008-9684-y [DOI] [PubMed] [Google Scholar]
- Perrin B.J., Huttenlocher A. 2002. Calpain. Int. J. Biochem. Cell Biol. 34:722–725 10.1016/S1357-2725(02)00009-2 [DOI] [PubMed] [Google Scholar]
- Pfaff M., Liu S., Erle D.J., Ginsberg M.H. 1998. Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J. Biol. Chem. 273:6104–6109 10.1074/jbc.273.11.6104 [DOI] [PubMed] [Google Scholar]
- Potter D.A., Tirnauer J.S., Janssen R., Croall D.E., Hughes C.N., Fiacco K.A., Mier J.W., Maki M., Herman I.M. 1998. Calpain regulates actin remodeling during cell spreading. J. Cell Biol. 141:647–662 10.1083/jcb.141.3.647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., Hall A. 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265:23–32 10.1016/j.ydbio.2003.06.003 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Hall A. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 70:389–399 10.1016/0092-8674(92)90163-7 [DOI] [PubMed] [Google Scholar]
- Robles E., Huttenlocher A., Gomez T.M. 2003. Filopodial calcium transients regulate growth cone motility and guidance through local activation of calpain. Neuron. 38:597–609 10.1016/S0896-6273(03)00260-5 [DOI] [PubMed] [Google Scholar]
- Sarkisian M.R., Bartley C.M., Chi H., Nakamura F., Hashimoto-Torii K., Torii M., Flavell R.A., Rakic P. 2006. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 52:789–801 10.1016/j.neuron.2006.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen V.L., Feng Y., Graham D., Takafuta T., Shapiro S.S., Walsh C.A. 2002. Filamin A and Filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum. Mol. Genet. 11:2845–2854 10.1093/hmg/11.23.2845 [DOI] [PubMed] [Google Scholar]
- Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., Shapiro S.S. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138–145 10.1038/35052082 [DOI] [PubMed] [Google Scholar]
- Vadlamudi R.K., Li F., Adam L., Nguyen D., Ohta Y., Stossel T.P., Kumar R. 2002. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 4:681–690 10.1038/ncb838 [DOI] [PubMed] [Google Scholar]
- Varambally S., Yu J.J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A., Shah R.B., Chandran U., Monzon F.A., Becich M.J., et al. 2005. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 8:393–406 10.1016/j.ccr.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Parsons J.T., Horwitz A.F. 2002. Adhesion assembly, disassembly and turnover in migrating cells — over and over and over again. Nat. Cell Biol. 4:E97–E100 10.1038/ncb0402-e97 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., Horwitz A.F. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Woo M.S., Ohta Y., Rabinovitz I., Stossel T.P., Blenis J. 2004. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol. Cell. Biol. 24:3025–3035 10.1128/MCB.24.7.3025-3035.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Benlimame N., Su J., He Q., Alaoui-Jamali M.A. 2009. Regulation of focal adhesion turnover by ErbB signalling in invasive breast cancer cells. Br. J. Cancer. 100:633–643 10.1038/sj.bjc.6604901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Calderwood D.A., Yaspan B., Ginsberg M.H. 2001. Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 276:28164–28170 10.1074/jbc.M104161200 [DOI] [PubMed] [Google Scholar]
- Yen L., Benlimame N., Nie Z.R., Xiao D., Wang T., Al Moustafa A.E., Esumi H., Milanini J., Hynes N.E., Pages G., Alaoui-Jamali M.A. 2002. Differential regulation of tumor angiogenesis by distinct ErbB homo- and heterodimers. Mol. Biol. Cell. 13:4029–4044 10.1091/mbc.E02-02-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M.G., Partridge M.A., Ezratty E.J., Shen Q., Gundersen G.G., Marcantonio E.E. 2006. Src SH2 arginine 175 is required for cell motility: specific focal adhesion kinase targeting and focal adhesion assembly function. Mol. Cell. Biol. 26:4399–4409 10.1128/MCB.01147-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Breitwieser G.E. 2005. High affinity interaction with filamin A protects against calcium-sensing receptor degradation. J. Biol. Chem. 280:11140–11146 10.1074/jbc.M412242200 [DOI] [PubMed] [Google Scholar]
- Zhu T.N., He H.J., Kole S., D’Souza T., Agarwal R., Morin P.J., Bernier M. 2007. Filamin A-mediated down-regulation of the exchange factor Ras-GRF1 correlates with decreased matrix metalloproteinase-9 expression in human melanoma cells. J. Biol. Chem. 282:14816–14826 10.1074/jbc.M611430200 [DOI] [PubMed] [Google Scholar]