Abstract
Background
Lung infection by various organisms is a characteristic feature of cystic fibrosis (CF). CFTR genotype effects acquisition of Pseudomonas aeruginosa (Pa), however the effect on acquisition of other infectious organisms that frequently precede Pa is relatively unknown. Understanding the role of CFTR in the acquisition of organisms first detected in patients may help guide symptomatic and molecular-based treatment for CF.
Methods
Lung infection, defined as a single positive respiratory tract culture, was assessed for 13 organisms in 1,381 individuals with CF. Subjects were divided by predicted CFTR function: 'Residual': carrying at least one partial function CFTR mutation (class IV or V) and 'Minimal' those who do not carry a partial function mutation. Kaplan-Meier estimates were created to assess CFTR effect on age of acquisition for each organism. Cox proportional hazard models were performed to control for possible cofactors. A separate Cox regression was used to determine whether defining infection with Pa, mucoid Pa or Aspergillus (Asp) using alternative criteria affected the results. The influence of severity of lung disease at the time of acquisition was evaluated using stratified Cox regression methods by lung disease categories.
Results
Subjects with 'Minimal' CFTR function had a higher hazard than patients with 'Residual' function for acquisition of 9 of 13 organisms studied (HR ranging from 1.7 to 3.78 based on the organism studied). Subjects with minimal CFTR function acquired infection at a younger age than those with residual function for 12 of 13 organisms (p-values ranging: < 0.001 to 0.017). Minimal CFTR function also associated with younger age of infection when 3 alternative definitions of infection with Pa, mucoid Pa or Asp were employed. Risk of infection is correlated with CFTR function for 8 of 9 organisms in patients with good lung function (>90%ile) but only 1 of 9 organisms in those with poorer lung function (<50%ile).
Conclusions
Residual CFTR function correlates with later onset of respiratory tract infection by a wide spectrum of organisms frequently cultured from CF patients. The protective effect conferred by residual CFTR function is diminished in CF patients with more advanced lung disease.
Background
Cystic fibrosis (CF) is the most common autosomal recessive life-shortening disorder in the Caucasian population and progressive obstructive lung disease is the primary cause of mortality[1,2]. Persistent respiratory infection by particular organisms, such as Pseudomonas aeruginosa (Pa) and Burkholderia cepacia complex (Bcc), are hallmarks of CF lung disease and lead to the progressive obstructive lung disease[2-5]. Infection with Pa is often preceded by infection with other organisms that are rarely cultured from healthy individuals such as Staphylococcus aureus. Additionally, a number of other pathogens such as methicillin resistant Staphylococcus aureus (MRSA)[6], Achromobacter xylosoxidans (Ax)[7] and Aspergillus fumigatus (Asp)[8] have been found to infect CF patients and affect the severity of lung disease in these patients. Mutations which permit residual CFTR function and confer a "mild" phenotypic effect have been associated with lower rates of Pa infection[9,10]. However, it is not clear whether residual CFTR function also influences acquisition of respiratory pathogens other than Pa [11].
Teasing out the role that CFTR genotype has on CF lung infection is important for several reasons. Recent success in the identification of small molecules augmenting CFTR function raises the possibility that delay of infection might be used as a measure for clinical efficacy of these new treatments, especially in children where few alternative options exist[12]. Secondly, patient populations have been accrued to find genetic modifiers of traits such as risk of infection and quantifying the contribution CFTR has to infection aids the search for genetic modifiers of this trait providing novel therapeutic targets for CF. Finally, neonatal CF pigs show acquisition of infection with a variety of organisms indicating that loss of CFTR function causes a pervasive defect in lung defense[13]. If this is the case in humans, then alteration in CFTR function should correlate with infection with a variety of respiratory pathogens. To address these issues, we evaluated the correlation between CFTR genotype and infection with a variety of pathogens using detailed infection data collected by the U.S. CF Twin and Sibling Study (CFTSS)[14].
Methods
Population
All subjects in the U.S. CFTSS were recruited on the basis of having a twin or sibling affected with CF. All enrolled patients met CF diagnostic criteria[15] and 99% attend CF care centers in the United States. Isolation of patient DNA, identification of CFTR mutations, and zygosity testing have been previously described[14,16]. Written, informed consent/assent was obtained from all subjects and/or their guardians, and this study was approved by the Johns Hopkins University Institutional Review Board.
Definitions of infection
Thirteen organisms were studied including Pseudomonas aeruginosa (Pa), mucoid Pa (MPa), Staphylococcus aureus (Sa), Methicillin resistant Sa (MRSa), Burkholderia cepacia complex (Bcc), Atypical mycobacteria (atyp), Aspergillus fumigatus (Asp), Stenotrophomonas maltophilia (Sm), Achromobacter xylosoxidans (Ax), Haemophilus influenzae (Hi), Streptococcus pneumoniae (Sp), Escherichia coli (Ec), and Klebsiella pneumoniae (Kp). Subjects were defined as having an "infection" with the organism in question if they had a positive oropharyngeal culture (throat, sputum, or bronchoalveolar lavage culture) for that particular organism (FirstInfx), regardless of whether the subject was positive for other organisms. An additional inclusion criterion was that a negative culture for the organism had to have been obtained a minimum of one week prior to the first positive culture to insure that subjects were in follow-up and routine cultures were being obtained. Patients treated at CF care centers in the U.S. have complete microbiological assessment of respiratory cultures at least annually and preferably quarterly. All subjects were passively followed-up after enrollment in the CFTSS until December 31, 2008 with culture data provided from their care centers and the CF Foundation Patient registry. Subjects were excluded if a respiratory culture was never performed prior to December 31, 2008.
A separate analysis of infection with Pa, MPa and Asp was performed using 3 additional criteria to define infection: 1) chronic infection (ChronInfx): 3 positive cultures within 6 months with each culture separated by at least 1 month (similar to that employed in Europe)[17] 2) multiple infection (MultiInfx): at least 3 positive cultures, but not meeting the definition for ChronInfx (as most patients in the U.S. do not attend CF clinic 3 times in 6 months); and 3) persistently infected (PersInfx): multiple cultures obtained in 3 consecutive years with positive cultures observed in at least 2 of the 3 years (recently used in a CF modifier study[18]).
CFTR genotypes and other variables
Subjects were grouped according to the functional class of CFTR mutations that he/she carried[10,19-22] (TABLE 1). Lung function variables were created using the maximum FEV1 within the year preceding the date of the first positive culture for a given organism referenced against the CF population (Kulich CF specific FEV1 percentile)[23] (FEV1BeforeInfx). The number of cultures per year for the subject was determined as the number of respiratory cultures obtained from the subject divided by the time (in years) from first documented culture to the last recorded culture (cultures per year). Age of acquisition was defined as the time from birth until recorded first positive culture for a given organism. Subjects who did not have a positive culture for the organism were assigned their age at the last obtained culture.
Table 1.
Classification of CFTR alleles
| Category | Mutation | Specific mutations |
|---|---|---|
| Class I | Defective Protein Synthesis (nonsense, frameshift, aberrant splicing) | 1078delT, 1154 insTC, 1525-2A > G, 1717-1G > A, 1898+1G > A, 2184delA, 2184 insA, 3007delG, 3120+1G > A, 3659delC, 3876delA, 3905insT, 394delTT, 4010del4, 4016insT, 4326delTC, 4374+1G > T, 441delA, 556delA, 621+1G > T, 621-1G > T, 711+1G > T, 875+1G > C, E1104X, E585X, E60X, E822X, G542X, G551D/R553X, Q493X, Q552X, Q814X, R1066C, R1162X, R553X, V520F, W1282X, Y1092X |
| Class II | Abnormal Processing and Trafficking | A559T, D979A, ΔF508, ΔI507, G480C, G85E, N1303K, S549I, S549N, S549R |
| Class III | Defective Channel Regulation/Gating | G1244E, G1349D, G551D, G551S, G85E, H199R, I1072T, I48T, L1077P, R560T, S1255P, S549(R75Q) |
| Class IV | Decreased Channel Conductance | A800G, D1152H, D1154G, D614G, delM1140, E822K, G314E, G576A, G622D, G85E, H620Q, I1139V, I1234V, L1335P, M1137V, P67L, R117C, R117P, R117H, R334W, R347H, R347P, R347P/R347H, R792G, S1251N, V232D |
| Class V | Reduced Synthesis and/or Trafficking | 2789+5G > A, 3120G > A, 3272-26A > G, 3849+10kbC > T, 5T variant, 621+3A > G, 711+3A > G, A445E, A455E, IVS8 poly T, P574H |
Statistical analysis and regression modeling
Time to acquisition of the first positive culture for an organism was assessed by the Kaplan Meier product limit estimator and the log-rank test. Cox proportional hazard regression models were used to account for covariates which could affect the relationship of CFTR genotype and age of acquisition for a particular organism. Covariates were identified a priori based on clinical experience and review of literature. Univariate regression was first performed with co-variates of gender (male vs. female), ethnicity (Caucasian vs. non-Caucasian), pancreatic status (pancreatic insufficient vs. pancreatic sufficient), FEV1BeforeInfx (linear co-variate), and cultures per year (linear co-variate) to identify significant co-variates. Co-variates with a p-value < 0.05 (pancreatic status, FEV1BeforeInfx, and cultures per year) were included in a multivariate Cox regression. In this regression, only FEV1BeforeInfx and cultures per year remained significant with p-values < 0.05. The final multivariate Cox regression model included FEV1BeforeInfx for the organism and cultures per year.
A second analysis was performed also using Cox regression to assess the relationship of CFTR with alternative ways of defining infection status for Pa, MPa and Asp. This regression was also adjusted for FEV1BeforeInfx for the organism and cultures per year. The time to "event" was defined as the age of the subject from birth until the first positive culture that would be included to meet the infection definition. In other words, for a subject meeting the definition of ChronInfx, the subject would need 3 positive cultures within 6 months but each 1 month from the other and the age of the subject at the first of the 3 positive cultures would be used. If subjects did not meet the definition of infection the age at the last obtained culture is used for censoring. All statistical calculations were performed using Intercooled Stata 10 (Stata Corp., College Station, TX).
Results
Clinical characteristics and infection rates in the study population
1,659 CF patients were enrolled in the U.S. CFTSS as of December 31, 2008. Thirty-five subjects were excluded for lack of infection data; 16 for lack of CFTR genotype information; 227 subjects for inability to classify CFTR mutations. The remaining 1,381 subjects were stratified according to predicted mutation effect upon CFTR function. 'Residual' function was defined as the presence of at least one class IV or V mutation (n = 163) while 'Minimal' function was defined as presence of only class I, II or III mutations (n = 1218). As expected from previous reports[10,11] subjects grouped according to predicted CFTR function differed for multiple characteristics (TABLE 2). Prevalence data for the organisms studied as well as comparability of the study sample to the general U.S. CF population is available in TABLE 3.
Table 2.
Overall prevalence of organisms in the U.S. CF Twin and Sibling Study, prevalence of infections in the U.S. CF Twin and Sibling Study in 2008 and the U.S. CF Foundation Registry[29] in 2008.
| U.S. CF Twin and Sibling Study Overall | U.S. CF Twin and Sibling Study 2008 | CF Foundation Registry 2008 | |
|---|---|---|---|
| Pa | 85.4% | 46.9% | 52.5% |
| MPa | 57.5% | Not estimated | Not estimated |
| Asp | 40.3% | Not estimated | Not estimated |
| Sa | 94.3% | 48.1% | 50.9% |
| MRSa | 39.2% | 23.6% | 22.6% |
| Bcc | 7.8% | 2.1% | 2.8% |
| Atyp | 7.3% | Not estimated | Not estimated |
| Sm | 41.6% | 11.6% | 12.5% |
| Ax | 19.6% | Not estimated | Not estimated |
| Hi | 74.2% | 15.0% | 16.3% |
| Sp | 35.8% | Not estimated | Not estimated |
| Ec | 19.4% | Not estimated | Not estimated |
| Kp | 19.8% | Not estimated | Not estimated |
Abbreviations: Pa: Pseudomonas aeruginosa MPa : mucoid Pseudomonas aeruginosa, Asp: Aspergillus fumigatus, Sa: Staphylococcus aureus, MRSa: Methicillin resistant Staphylococcus aureus, Bcc: Burkholderia cepacia complex, atyp: Atypical mycobacteria, Sm: Stenotrophomonas maltophilia, Ax: Achromobacter xylosoxidans, Hi: Haemophilus influenzae, Sp: Streptococcus pneumoniae, Ec: Escherichia coli and Kp: Klebsiella pneumoniae
Table 3.
Characteristics of study subjects
| 'Minimal' Function | 'Residual' Function | p value | |
|---|---|---|---|
| Individuals | 1218 | 163 | |
| Female | 579 (47.5%) | 81 (49.7%) | 0.605 a |
| Caucasian | 943 (95.5%) | 115 (83.9%) | <0.001b |
| Pancreatic Insufficient | 1180 (97.8%) | 46 (30.3%) | <0.001b |
| Age (yrs) last visit ± SD (range), (n) | 15.82 ± 8.60 (0.44-57.31) (1209) | 18.59 ± 12.87 (0.54-63.94) (159) | 0.133b |
| Maximal FEV1CF% since last clinic visit, (n) | 0.68 ± 0.26 (1111) | 0.75 ± 0.25 (145) | <0.001b |
| Average number of cultures per subject per year ± SD, (n) | 3.89 ± 1.92 (1201) | 3.93 ± 2.27 (159) | 0.530 b |
Definition of abbreviations: Maximal FEV1 CF%: CF specific percentile for the maximum forced expiratory volume in one second in the last year of data for the subject.
a: P-value reported from chi-squared, assumes equal variance in group means.
b: P-value reported from Kruskal-Wallis test due to unequal variances in group means.
Residual CFTR function decreases the risk of respiratory infection by multiple organisms
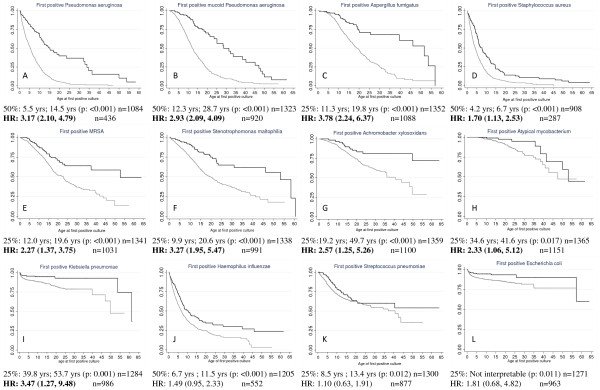
Kaplan Meier analysis was performed to assess the effect of CFTR genotype upon the age at which the infection was acquired for 13 organisms. The unadjusted median age of acquisition for Pa for subjects with 'Minimal' CFTR function was substantially lower (5.5 years) than those with 'Residual' CFTR function (14.5 years; log rank p-value: < 0.001). Overall prevalence of Asp in this study was less than 50%, we report the age at which 25% of the population would be affected. Patients with 'Minimal' function reached 25% prevalence at11.3 years while those with 'Residual' function reached this rate at 19.8 years (p-value < 0.001). Ten other organisms also had a lower age of acquisition in those subjects with 'Minimal' CFTR function when compared to 'Residual' function using log rank tests. (See FIGURE 1 for further details).
Figure 1.
Unadjusted Kaplan-Meier (K-M) plots for 12 of 13 infectious organisms studied. Gray lines represent 'Minimal' CFTR function subjects and black lines represent subjects with 'Residual' CFTR function. There are differing numbers of subjects used in each plot due to the inclusion criteria of having a previous negative culture prior to the first positive culture for the particular organism. Median age differences are presented for those infections not affected by censoring, otherwise ages corresponding to the point where 25% of the population is affected is presented. Corresponding t-tests for the unadjusted K-M and Cox proportional hazard ratios with 95% confidence interval adjusted for the FEV1BeforeInfx and the number of cultures per subject per year are presented. A. Pseudomonas aeruginosa. B. mucoid Pseudomonas aeruginosa. C. Aspergillus fumigatus. D. Staphylococcus aureus. E. Methicillin-resistant Staphylococcus aureus. F. Stenotrophomas maltophilia. G. Achromobacter xylosoxidans. H. Atypical mycobacterium. I. Klebsiella pneumoniae. J. Haemophilus influenzae. K. Streptococcus pneumoniae. L. Escherichia coli. Burkholderia cepacia complex is not presented as few subjects developed this infection and results were limited in interpretation.
As noted in TABLE 2, subjects with 'Residual' CFTR function and those with 'Minimal' CFTR function differ in a number of respects, therefore a Cox proportional hazards model was created that incorporated these difference as covariates. The final multivariate Cox regression included lung function (FEV1BeforeInfx) and cultures per year. Nine of the 13 organisms were found to have a significant hazard ratio (HR) indicating an increased risk of acquisition in subjects with 'Minimal' CFTR function (FIGURE 1). For Pa, the hazard ratio was increased 3 fold for those with 'Minimal' function when compared to those with 'Residual' function. Increased risk for the other 8 organisms ranged from 1.7 to 3.8. Bcc is not presented in the figure as it was found to not be associated with CFTR genotype by log rank or Cox regression.
Residual CFTR function decreases risk of infection with Pa, MPa and Asp regardless of infection definition
Various criteria have been employed in the literature to define infection status[17]. As this study is retrospective and possibly prone to misclassification, we determined whether the use of different criteria with more rigorous definitions for infection altered the results. Patients were classified for infection status for Pa, MPa and Asp using three other definitions (ChronInfx, MultiInfx and PersInfx). The number of subjects defined as positive using each definition is presented in TABLE 4. One-hundred nineteen subjects were positive for Pa by all 4 methods. Forty-one subjects were positive by 2 of 4 definitions, most commonly meeting the definition of FirstInfx and MultiInfx. Only 55 subjects were positive by the definition for FirstInfx. Similar concordance was found for MPa with 180 subjects positive using all 4 definitions and 78 only positive for FirstInfx. For Asp, fewer subjects were positive for all four definitions (n = 76) and 140 were only positive if FirstInfx was used to define infection.
Table 4.
Risk of infection is increased for 'Minimal' CFTR function using any definition of infection with Pseudomonas aeruginosa, mucoid Pseudomonas aeruginosa and Aspergillus fumigatus.
| Total Positive (n included in analysis) |
'Minimal' Function (% of class positive) |
'Residual' Function (% of class positive) |
Adjusted Hazards Ratioa | |
|---|---|---|---|---|
| FirstInfx Pa | 318 (436) | 278 (79.4%) | 40 (46.5%) | 3.17 (2.10, 4.78) |
| ChronInfx Pa | 127 (436) | 118 (33.7%) | 9 (10.5%) | 5.47 (2.20, 13.58) |
| MultiInfx Pa | 229 (436) | 206 (58.9%) | 23 (26.7%) | 3.81 (2.32, 6.28) |
| PersInfx Pa | 228 (436) | 203 (58.0%) | 25 (29.1%) | 3.32 (2.00, 5.50) |
| FirstInfx MPa | 492 (919) | 453 (56.3%) | 39 (33.9%) | 2.93 (2.09, 4.09) |
| ChronInfx MPa | 230 (919) | 217 (27.0%) | 13 (11.3%) | 3.85 (2.00, 7.39) |
| MultiInfx MPa | 381 (919) | 354 (44.0%) | 27 (23.5%) | 3.11 (2.09, 4.62) |
| PersInfx MPa | 371 (919) | 345 (42.9%) | 26 (22.6%) | 3.14 (2.06, 4.76) |
| FirstInfx Asp | 451 (1090) | 427 (44.0%) | 24 (20.0%) | 3.77 (2.24, 6.35) |
| ChronInfx Asp | 89 (1090) | 88 (9.1%) | 1 (0.8%) | 18.97 (3.73, 96.43) |
| MultiInfx Asp | 252 (1090) | 237 (24.4%) | 15 (12.5%) | 3.22 (1.73, 5.98) |
| PersInfx Asp | 254 (1090) | 243 (25.1%) | 11 (9.2%) | 4.83 (2.47, 9.46) |
Definition of abbreviations: Pa: Pseudomonas aeruginosa, MPa: mucoid Pseudomonas aeruginosa, Asp: Aspergillus fumigatus, FirstInfx: one positive culture for the organism; MultiInfx: three or more positive cultures for the organism; PersInfx: at least one positive culture for the organism in two of three consecutive years; ChronInfx: three positive cultures within 6 months each positive culture at least 1 month from the last positive culture.
a: Adjusted for FEV1BeforeInfx and number of cultures.
Multivariate Cox regression was used to assess the relationship of CFTR genotype with acquisition of infection using each of these different definitions as the primary outcome. Subjects with 'Minimal' CFTR function had a substantially higher risk of acquiring Pa, MPa and Asp than those with 'Residual' CFTR function for any definitions used (TABLE 4). Thus, use of an alternate definition of infection does not substantially alter the association between predicted CFTR function and acquisition of Pa, MPa or Asp.
CFTR associated risk of infection is influenced by lung function status at the time of acquisition
Lung function affected the relationship of CFTR and infection risk and was retained as a significant covariate in all Cox regression models. To better understand this relationship, we performed a formal test of interaction of lung function as a linear covariate and CFTR as a dichotomous variable. The interaction term of FEV1BeforeInfx and CFTR was not significant for each organism with the exception of Klebsiella which was found to be highly significant (p-value = 0.008). However, we did not feel that this adequately represented what we observed in our data and therefore wanted to confirm this observation using stratification. Four strata distributed subjects into roughly equal size groups for each infecting organism (FEV1BeforeInfx less than the 50th %ile, 50-74.9th %ile, 75-89.9th %ile and greater than the 90th %ile) and revealed that the effect of CFTR genotype upon risk of infection differed depending upon lung function and the organism involved (TABLE 5). CFTR genotype confers a significant risk to the acquisition of 8 of 9 organisms for patients in the highest strata of lung function (i.e. >90th %ile). However, at the lowest lung function strata (<50th %ile), CFTR genotype significantly alters infection risk with only 1 organism (Sa). These results suggest that protection from lung infection originally conferred by 'Residual' CFTR function is lost as lung function declines.
Table 5.
Lung strataa specific hazard ratios for the association of presence of an infection with class mutation status
| Multivariate regressionb | Lung Function <0.50c | Lung Function 0.50-0.75c | Lung Function 0.75-0.9c | Lung Function >0.9c | |
|---|---|---|---|---|---|
| Pa | 3.17 (2.10, 4.78) | 1.59 (0.78, 3.24) (n = 81) | 3.74 (1.70, 8.22) (n = 112) | 4.17 (1.45, 12.0) (n = 110) | 4.01 (1.92, 8.36) (n = 133) |
| MPa | 2.93 (2.09, 4.09) | 1.59 (0.78, 3.22) (n = 192) | 3.80 (2.07, 6.98) (n = 227) | 4.23 (2.01, 8.92) (n = 246) | 3.83 (2.06, 7.11) (n = 255) |
| Asp | 3.78 (2.24, 6.37) | 2.85 (0.77, 10.63) (n = 250) | 10.22 (3.25, 32.21) (n = 282) | 4.51 (1.46, 13.92) (n = 269) | 2.82 (1.34, 5.93) (n = 289) |
| Sa | 1.70 (1.13, 2.53) | 3.41 (1.43, 8.16) (n = 74) | 1.05 (0.54, 2.03) (n = 83) | 0.90 (0.40, 2.06) (n = 66) | 1.94 (1.02, 3.70) (n = 64) |
| MRSa | 2.27 (1.37, 3.75) | 2.04 (0.71, 5.87) (n = 237) | 2.06 (0.84, 5.05) (n = 262) | 1.80 (0.76, 4.30) (n = 260) | 3.40 (1.41, 8.21) (n = 273) |
| Atyp | 2.33 (1.06, 5.12) | 2.12 (0.27, 16.66) (n = 265) | 5.41 (0.42, 70.41) (n = 294) | 1.40 (0.52, 3.78) (n = 292) | 2.47 (1.06, 5.72) (n = 303) |
| Sm | 3.27 (1.95, 5.47) | 2.70 (0.88, 8.31) (n = 229) | 3.62 (1.52, 8.62) (n = 257) | 2.45 (1.12, 5.33) (n = 260) | 3.64 (1.31, 10.13) (n = 247) |
| Ax | 2.57 (1.25, 5.26) | 1.94 (0.42, 9.05) (n = 252) | 8.41 (1.83, 38.58) (n = 282) | 2.07 (0.59, 7.30) (n = 275) | 1.93 (0.64, 5.82) (n = 293) |
| Kp | 3.47 (1.27, 9.48) | 0.28 (0.07, 1.06) (n = 233) | 7.87 e15 (2.30 e15, 2.69 e16) (n = 250) | 7.55 e15 (2.60 e15, 2.19 e16) (n = 247) | 4.73 (1.15, 19.43) (n = 258) |
Definition of abbreviations: Pa: Pseudomonas aeruginosa MPa : mucoid Pseudomonas aeruginosa, Asp: Aspergillus fumigatus, Sa: Staphylococcus aureus, MRSa: Methicillin resistant Staphylococcus aureus, atyp: Atypical mycobacteria, Sm: Stenotrophomonas maltophilia, Ax: Achromobacter xylosoxidans, Kp: Klebsiella pneumoniae.
a: Individual subjects may be in different strata for each infection as the value for the lung function variable reflects the maximal value in the year preceding acquisition of each of the organisms.
b: Cox regression adjusted for FEV1BeforeInfx and number of cultures per year for the subject; comparing 'Residual' function to 'Minimal' function.
c: Stratified Cox regression adjusted for number of cultures per year for the subject; comparing 'Residual' function to 'Minimal' function in each stratum.
Discussion
Pulmonary disease in CF is complicated by patients acquiring numerous different organisms at different periods of time during his/her lifetime. These organisms tend to progress from Sa and Hi to Pa. While Pa has been extensively studied for its relationship with CFTR mutations, the underlying defect of CF, few studies have evaluated the effect that CFTR genotype has on infection risk for other organisms commonly cultured from CF respiratory tracts. Our study corroborates the finding that CFTR mutations which permit 'Residual" CFTR function (i.e. class IV or V mutations) are associated with reduced rates and delayed acquisition of Pa infection[10,11]. We substantially extend this observation to include numerous other organisms commonly cultured from CF patients (mucoid Pa, Staphylococcus aureus, MRSA, Aspergillus fumigatus, Stenotrophomonas maltophilia, Achromobacter xylosoxidans, atypical mycobacterium and Klebsiella pneumoniae) indicating that CFTR dysfunction appears to cause a global alteration in infection resistance in humans. This concept is consistent with recent studies which indicate loss of CFTR function causes a pervasive defect in resistance to respiratory infection in pig models of CF[13]. Additionally this study shows that the concept of "mild" CFTR mutations (often 'Residual' function mutations) versus "severe" CFTR mutations (often 'Minimal' function mutations) may extend beyond pancreatic status to other features such as lung infection. Furthermore, we show that the association between CFTR genotype and respiratory infection has a complex relationship with the severity of lung disease. CFTR mutations that permit 'Residual' function reduce the risk of infection with multiple organisms in patients with mild lung dysfunction evidenced by near normal forced expiratory volumes. However, in patients with compromised lung function, the protective effect of 'Residual' CFTR function is lost for most of the organisms studied.
To date, studies on the role of CFTR as a risk factor for acquisition of infectious organisms have concentrated on the most common mutation, F508del. A few studies have assessed the role of the CFTR genotype beyond the classification of F508del homozygotes versus all others. One study found that mis-sense and splice mutations predicted to confer "mild" effect were associated with lower rates and later acquisition of Pa infection[9]. However, mis-sense and splice-site mutations have different effects upon CFTR function; the former being associated with protein formation (and possible residual function) while the latter is usually associated with no protein being made (and no residual function)[24]. Two subsequent studies[10,11] demonstrated that stratifying CFTR mutations by putative function was associated with lower rates of Pa infection. One of these studies[11] attempted to associate CFTR mutation class with infection risk for other organisms but failed to show a reliable association. One reason this current study found an association of CFTR genotype with multiple respiratory pathogens may be due to the increased power afforded by a larger sample size when subjects with class IV or V mutations are combined (as opposed to assessing each mutation class separately[11]). Alternatively, non-genetic factors may play a role as the prior study was performed in Europe as opposed to the current U.S. study.
Furthermore, the role of CFTR in the acquisition of lung infection has been debated. One possibility is that CFTR on epithelial cell membranes binds organisms directly by recognizing the lipopolysaccharide coat of Pa [25] thus allowing endocytosis and eventual destruction of Pa. Loss of this bacterial clearance process due to mutations that do not permit residual CFTR function (class I, II or III) is postulated to underlie the chronic infectious state associated with Pa[26]. This however does not fully explain the relationship observed for CFTR and other organisms. A second hypothesis posits that defective clearance of infecting organisms due to altered properties of airway surface liquid and innate defense caused by CFTR dysfunction [27,28]. In this scenario, our results might suggest that mutations causing a moderate loss of CF function lead to less severe changes in epithelial barriers to infection and consequently lower rate of infection with a number of organisms commonly found in the CF lung. Regardless of mechanism, the findings of this study indicate that reduced CFTR function leads to a global disruption of infection control and acquisition.
We have to acknowledge that there are several weaknesses in this study. First, the study was retrospective and patients were enrolled on the basis of being diagnosed with CF and having a sibling diagnosed with CF. This inclusion criterion may have skewed our population as survival of both siblings until entry in our study was required. To assess for a possible bias toward milder CF patients who may have a lower rate of respiratory infection, we compared our data to the general U.S. CF population in the CFF registry and found rates of infection in this study to be similar to the unrelated U.S. CF population (TABLE 2). Another weakness is that we did not apply a correction for age of diagnosis of CF as this event may have been precipitated by a positive respiratory culture. To address this shortcoming, we ensured that subjects had at least one negative respiratory culture for the organism being studied prior to entry into the analysis thereby excluding individuals who were infected at the time of CF diagnosis.
Strengths of this study include sample size, the conservative use of alternative definitions of infection and assessment of lung function prior to the acquisition of the organism. Using a mean survival time of 5 years for the control group ('Minimal' function), 5 years for accrual of all subjects, 10 years of follow-up time and 80% power, we had an adequate sample size to detect a hazard ratio (HR) of 1.3. As shown in FIGURE 1, organisms that demonstrated a significant correlation with CFTR function had HRs that considerably exceeded this threshold. While our main outcome measure was the initial acquisition of an organism, we also stratified patients using 3 other definitions for infection. As highly similar results were obtained when using either of these definitions, we conclude that our results are independent of the criteria used to assign infection status. Inclusion of additional definitions of infection also helped overcome the issue of using a single throat culture (if that was the only culture information for a subject) to determine infection status since throat cultures are known to have poor negative predictive value. By including criteria which require assessment of multiple cultures, correct classification of subjects is more assured. Finally, by assessing lung function prior to the acquisition of the infectious organism analyzed, we were able to evaluate whether the association between predicted CFTR function and infection status was dependent on the severity of lung disease around the time of first infection.
Conclusions
In conclusion, genotypes predicting 'Minimal' CFTR function are associated with increased risk and earlier age of infection by multiple organisms. For those with decreased lung function, 'Residual' CFTR genotype no longer provides a protective effect on infection risk. These observations suggest that 1) adjustment for CFTR functional genotype may be necessary in genetic modifier studies of infection, 2) monitoring infection status in patients with mild lung disease, especially children, could be an outcome measure for therapies that augment CFTR function and 3) augmentation of CFTR function may not reduce the risk of infection in patients with severe lung disease.
List of Abbreviations
ASP: Aspergillus fumigatus; ATYP: Atypical mycobacteria; AX: Achromobacter xylosoxidans; BCC: Burkholderia cepacia complex; CF: cystic fibrosis; CFTR: cystic fibrosis transmembrane conductance regulator; CFTSS: U.S. CF Twin and Sibling Study; EC: Escherichia coli; HI: Haemophilus influenzae; KP: Klebsiella pneumoniae; MPa: mucoid Pseudomonas aeruginosa; MRSa: methicillin resistant Staphylococcus aureus; Pa: Pseudomonas aeruginosa; Sa: Staphylococcus aureus; Sm: Stenotrophomonas maltophilia; Sp: Streptococcus pneumoniae; FirstInfx: first positive culture; ChronInfx: chronic infection: 3 positive cultures within 6 months each 1 month from the previous; MultiInfx: multiple infection: 3 positive cultures in the subjects lifetime; PersInfx: persistently infected: 2 positive cultures each in a separate year within 3 consecutive years; PS: pancreatic sufficient; PI: pancreatic insufficient; FEV1: forced expiratory volume in one second; FEV1BeforeInfx: maximum CF-specific percentile forced expiratory volume in one second in the year before the first positive culture for an organism
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DMG had full access to all data, contributed to data collection, conception and design of the original idea, data analysis, and drafting of the manuscript. KEM was involved in conception and design of the original idea, and data analysis. SMB contributed to conception and design of the study, and data acquisition. PRS was involved in conception and design of the original study, and data analysis. LBH provided assistance on conception and design of the study, and data analysis. KMN provided input on conception and study design, data acquisition, and administrative, technical and material support. JMC was involved conception and design of the study, data analysis, data acquisition of data, and statistical analysis. GRC was involved in conception and design of the study, data analysis, drafting of the manuscript, and administrative, technical and material support as well as overall study supervision. All authors read, revised and approved the final manuscript.
Contributor Information
Deanna M Green, Email: dgreen50@jhmi.edu.
Kathryn E McDougal, Email: kmcdouga@jhsph.edu.
Scott M Blackman, Email: sblackman@jhmi.edu.
Patrick R Sosnay, Email: psosnay@jhmi.edu.
Lindsay B Henderson, Email: lab.henderson@jhmi.edu.
Kathleen M Naughton, Email: knaught1@jhmi.edu.
J Michael Collaco, Email: mcollac1@jhmi.edu.
Garry R Cutting, Email: gcutting@jhmi.edu.
Acknowledgements
The authors would like to acknowledge the numerous CF patients, families, research coordinators and clinicians who participate in the CF Twin and Sibling Study. The authors would also like to thank Neeraj Sharma, DVM, PhD, and Vishal Doshi, MBBS, MPH for their thoughtful discussions; Patricia Cornwall for her administrative assistance; Sara Ritter, BA and Darci Ferrer, BS for technical expertise; Terri Beaty, PhD and Jennifer Mulle, PhD for their expert advice on epidemiology and genetic epidemiology. This work was supported by the National Institutes of Health Grants (NHLBI T32 HL072748 (D.M.G.); NIDDK DK99003 (G.R.C.); NHLBI HL068927 (G.R.C.); NIDDK DK076446 (S.M.B.)). This work was partially presented in abstract form in Pediatric Pulmonology 2008 Supplement 31: Abstract 232 and Pediatric Pulmonology 2009 Supplement 32: Abstract 170.
References
- Halliburton CS, Mannino DM, Olney RS. Cystic fibrosis deaths in the United States from 1979 through 1991. An analysis using multiple-cause mortality data. Arch Pediatr Adolesc Med. 1996;150:1181–1185. doi: 10.1001/archpedi.1996.02170360071012. [DOI] [PubMed] [Google Scholar]
- Emerson J, Rosenfeld M, McNamara S, Ramsey B, Gibson RL. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatr Pulmonol. 2002;34:91–100. doi: 10.1002/ppul.10127. [DOI] [PubMed] [Google Scholar]
- Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol. 2001;32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- Nixon GM, Armstrong DS, Carzino R, Carlin JB, Olinsky A, Robertson CF, Grimwood K. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J Pediatr. 2001;138:699–704. doi: 10.1067/mpd.2001.112897. [DOI] [PubMed] [Google Scholar]
- Courtney JM, Dunbar KE, McDowell A, Moore JE, Warke TJ, Stevenson M, Elborn JS. Clinical outcome of Burkholderia cepacia complex infection in cystic fibrosis adults. J Cyst Fibros. 2004;3:93–98. doi: 10.1016/j.jcf.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Dasenbrook EC, Merlo CA, Diener-West M, Lechtzin N, Boyle MP. Persistent methicillin-resistant Staphylococcus aureus and rate of FEV1 decline in cystic fibrosis. Am J Respir Crit Care Med. 2008;178:814–821. doi: 10.1164/rccm.200802-327OC. [DOI] [PubMed] [Google Scholar]
- De Baets F, Schelstraete P, Van Daele S, Haerynck F, Vaneechoutte M. Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J Cyst Fibros. 2007;6:75–78. doi: 10.1016/j.jcf.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Kanthan SK, Bush A, Kemp M, Buchdahl R. Factors effecting impact of Aspergillus fumigatus sensitization in cystic fibrosis. Pediatr Pulmonol. 2007;42:785–793. doi: 10.1002/ppul.20656. [DOI] [PubMed] [Google Scholar]
- Kubesch P, Dörk T, Wulbrand U, Kälin N, Neumann T, Wulf B, Geerlings H, Weibbrodt H, von der Hardt H, Tümmler B. Genetic determinants of airways' colonization with pseudomonas aeruginosa in cystic fibrosis. Lancet. 1993;341:189–193. doi: 10.1016/0140-6736(93)90062-L. [DOI] [PubMed] [Google Scholar]
- McKone EF, Emerson SS, Edwards KL, Aitken ML. Effect of genotype on phenotype and mortality in cystic fibrosis: a retrospective cohort study. Lancet. 2003;361:1671–1676. doi: 10.1016/S0140-6736(03)13368-5. [DOI] [PubMed] [Google Scholar]
- Koch C, Cuppens H, Rainisio M, Madessani U, Harms H, Hodson M, Mastella G, Navarro J, Strandvik B, McKenzie S. European Epidemiologic Registry of Cystic Fibrosis (ERCF): comparison of major disease manifestations between patients with different classes of mutations. Pediatr Pulmonol. 2001;31:1–12. doi: 10.1002/1099-0496(200101)31:1<1::AID-PPUL1000>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, Turnbull A, Singh A, Joubran J, Hazlewood A. et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, Hanfland RA, Wohlford-Lenane C, Dohrn CL, Bartlett JA. et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci Transl Med. 2010;2:29ra31. doi: 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, Algire M, McWilliams R, Beck S, Hoover-Fong J. et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med. 2007;175:1036–1043. doi: 10.1164/rccm.200608-1164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PM, Rosenstein BJ, White TB, Accurso FJ, Castellani C, Cutting GR, Durie PR, LeGrys VA, Massie J, Parad RB. et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. J Pediatr. 2008;153:S4–S14. doi: 10.1016/j.jpeds.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman SM, Hsu S, Vanscoy LL, Collaco JM, Ritter SE, Naughton K, Cutting GR. Genetic modifiers play a substantial role in diabetes complicating cystic fibrosis. J Clin Endocrinol Metab. 2009;94:1302–1309. doi: 10.1210/jc.2008-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring G, Conway SP, Heijerman HG, Hodson ME, Hoiby N, Smyth A, Touw DJ. Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur Respir J. 2000;16:749–767. doi: 10.1034/j.1399-3003.2000.16d30.x. [DOI] [PubMed] [Google Scholar]
- Taylor-Cousar JL, Zariwala MA, Burch LH, Pace RG, Drumm ML, Calloway H, Fan H, Weston BW, Wright FA, Knowles MR. Histo-blood group gene polymorphisms as potential genetic modifiers of infection and cystic fibrosis lung disease severity. PLoS ONE. 2009;4:e4270. doi: 10.1371/journal.pone.0004270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenski J, Tsui LC. Cystic fibrosis: Genotypic and phenotypic variations. Annu Rev Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]
- Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-R. [DOI] [PubMed] [Google Scholar]
- Ahmed N, Corey M, Forstner G, Zielenski J, Tsui LC, Ellis L, Tullis E, Durie P. Molecular consequences of cystic fibrosis transmembrane regulator (CFTR) gene mutations in the exocrine pancreas. Gut. 2003;52:1159–1164. doi: 10.1136/gut.52.8.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowntree RK, Harris A. The phenotypic consequences of CFTR mutations. Ann Hum Genet. 2003;67:471–485. doi: 10.1046/j.1469-1809.2003.00028.x. [DOI] [PubMed] [Google Scholar]
- Kulich M, Rosenfeld M, Campbell J, Kronmal R, Gibson RL, Goss CH, Ramsey B. Disease-specific Reference Equations for Lung Function in Patients with Cystic Fibrosis. Am J Respir Crit Care Med. 2005;172:885–891. doi: 10.1164/rccm.200410-1335OC. [DOI] [PubMed] [Google Scholar]
- Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR. In: The Metabolic and Molecular Bases of Inherited Disease. Eighth. Scriver CR, Beaudet AL, Valle D, Sly WS, editor. III. New York: McGraw-Hill, Inc; 2001. Cystic Fibrosis; pp. 5121–5188. [Google Scholar]
- Schroeder TH, Lee MM, Yacono PW, Cannon CL, Gerceker AA, Golan DE, Pier GB. CFTR is a pattern recognition molecule that extracts Pseudomonas aeruginosa LPS from the outer membrane into epithelial cells and activates NF-kappa B translocation. Proc Natl Acad Sci USA. 2002;99:6907–6912. doi: 10.1073/pnas.092160899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajmoczi M, Gadjeva M, Alper SL, Pier GB, Golan DE. Cystic fibrosis transmembrane conductance regulator and caveolin-1 regulate epithelial cell internalization of Pseudomonas aeruginosa. Am J Physiol Cell Physiol. 2009;297:C263–C277. doi: 10.1152/ajpcell.00527.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes TJ, Plumb J, Martin R, Vachon E, Cherepanov V, Koh A, Loeve C, Jongstra-Bilen J, Zurawska JH, Kus JV. et al. Abnormalities in the pulmonary innate immune system in cystic fibrosis. Am J Respir Cell Mol Biol. 2006;34:364–374. doi: 10.1165/rcmb.2005-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor RM, Superfine R, Rubinstein M, Iglewski BH. et al. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci USA. 2006;103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation: Patient Registry 2008 Annual Report. Bethesda, MD; 2008. [Google Scholar]